Open Access Article
Open Access ArticleNanoparticle shape is the game-changer for blood–brain barrier crossing and delivery through tunneling nanotubes among glioblastoma cells†
Giulia
Sierri‡
 *a,
Ines
Saenz-de-Santa-Maria‡
*a,
Ines
Saenz-de-Santa-Maria‡
 b,
Antonio
Renda
a,
Marcus
Koch
c,
Patrizia
Sommi
b,
Antonio
Renda
a,
Marcus
Koch
c,
Patrizia
Sommi
 d,
Umberto
Anselmi-Tamburini
d,
Umberto
Anselmi-Tamburini
 e,
Mario
Mauri
e,
Mario
Mauri
 a,
Alessia
D'Aloia
a,
Alessia
D'Aloia
 f,
Michela
Ceriani
f,
Michela
Ceriani
 f,
Domenico
Salerno
a,
Francesco
Mantegazza
f,
Domenico
Salerno
a,
Francesco
Mantegazza
 a,
Chiara
Zurzolo‡
a,
Chiara
Zurzolo‡
 bg and
Francesca
Re‡
bg and
Francesca
Re‡
 a
a
aSchool of Medicine and Surgery, University of Milano-Bicocca, Monza, Italy. E-mail: g.sierri@campus.unimib.it
bUnité de Trafic Membranaire et Pathogenèse, Institut Pasteur, Université Paris Cité, CNRS UMR 3691, Paris, France
cINM-Leibniz Institute for New Materials, Saarbrücken, Germany
dDepartment of Molecular Medicine, University of Pavia, Pavia, Italy
eDepartment of Chemistry, University of Pavia, Pavia, Italy
fDepartment of Biotechnology and Biosciences, University of Milano-Bicocca, Milano, Italy
gDepartment of Molecular Medicine and Medical Biotechnology, University of Naples Federico II, Naples, Italy
First published on 9th November 2024
Abstract
Tunneling nanotubes (TNTs) are thin, dynamic, long membrane protrusions that allow intercellular exchanges of signaling clues, molecules and organelles. The presence of TNTs and their involvement as drug delivery channels have been observed in several types of cancer, including glioblastoma. Recently, increased attention has been directed toward nanoparticles (NPs) that can be transported in TNTs. However, few data are available on the role of physical parameters of nanoparticles, such as size, shape, charge and flexibility, in determining their transfer efficiency between cells by TNTs. Here, we focused our attention on NP shape, manufacturing spherical, discoidal and deformable negatively charged lipid-based NPs with sizes <120 nm and similar stiffness. The TNT-mediated transfer of NPs was investigated in 2D and 3D culture models of human glioblastoma cells. The permeability and biocompatibility of the blood–brain barrier (BBB) were also assessed. Results showed that discoidal NPs displayed the highest TNT-mediated transfer efficiency between cancer cells, with a maximum velocity of 69 nm s−1, and a higher endothelial permeability (1.29 × 10−5 cm min−1) across the BBB in an in vitro model. This depends on the NP shape because discoidal NPs have a larger surface area exposed to the flow along the TNT channel. Overall, the results suggest that the shape of NPs is the game-changer for more efficient TNT-mediated transfer between cancer cells, thus introducing a sustainable solution to improve the diffusion rate at which the NPs spread in the tumour microenvironment, opening the possibility of ameliorating drug distribution to difficult-to-reach cancer cell populations.
1. Introduction
Glioblastoma multiforme (GBM) is classified as a grade IV astrocytoma by the World Health Organization (WHO), and it represents one of the most aggressive and prevalent forms of malignant primary tumour in the central nervous system (CNS). GBM constitutes approximately half of all malignant CNS tumours, the median survival time being approximately half a year and the 5-year survival rate being less than 7%. The occurrence of GBM increases with age, with a higher prevalence observed in men compared to women. The conventional therapy according to the Stupp protocol is neurosurgery followed by radiotherapy and chemotherapy with temozolomide (TMZ).1Despite these aggressive treatment strategies, patients’ prognosis remains poor. One of the major causes of therapeutic failure is the blood–brain barrier (BBB), which is a natural protective membrane that limits the influx of biomolecules, drugs and toxins from the bloodstream into neural tissues. Additionally, the heterogeneity of the tumour and its associated microenvironment, including the presence of GBM stem cells, make GBM highly refractory to therapies.2 The BBB is usually disrupted in the necrotic core regions but remains intact at the invasive edges, so it is evident that designing drug delivery systems to overcome the BBB could be a promising strategy in GBM therapy.3
In this context, nanomedicine holds great promise for the treatment of brain tumours.4 In particular, the possibility of modifying the surface of NPs with ligands makes them suitable tools for delivering drugs across the BBB and, simultaneously, selectively targeting GBM cells. Moreover, their chemical versatility represents a key feature to design individualized nanomedicines to address GBM inter-patient heterogeneity.5 A precision-medicine approach for GBM treatment has been developed that involves the use of brain-penetrant RNA interference (RNAi)-based spherical nucleic acids, where gold NP cores are covalently conjugated with siRNA oligonucleotides. A phase 0 first-in-human trial has been conducted to determine the safety, pharmacokinetics, intra-tumoural accumulation and gene suppressive activity of a specific GBM oncogene. Results indicated that NPs reached the tumours and their uptake into glioma cells correlated with significant reduction in oncogene expression, demonstrating the utility of these nanocarriers in a brain-penetrant precision-medicine approach for the systemic treatment of GBM.6
However, another issue to be considered is the distribution of the drug when the NPs reach the brain,7 so intercellular connections between cells could be a promising tool. The communication between cells is fundamental to brain function because it permits dialogue between all of them, and this is necessary for the proper development and function of the brain. Often, under diseased conditions, the crosstalk between cells is dysregulated, thus contributing to tumour progression.8 Within the tumour microenvironment, tunneling nanotubes (TNTs) are particular routes for communication among cancer cells,9 normal cells,10 and immune cells.11 They are open membranous channels that are able to transfer calcium, molecules, and organelles such as mitochondria and lysosomes. It has been shown that GBM cells form TNTs upon being subjected to oxidative stress and TMZ treatment.12 These intercellular exchanges play a crucial role in promoting the survival, metastasis, and chemo-resistance of cancer cells.13 In particular, recent studies suggest that TNTs play a crucial role in the growth and invasion of GBM by establishing a cooperative network among tumour cells and their surrounding cells. Furthermore, TNTs contribute to tumour relapse and the adaptation of GBM cells to TMZ and ionizing radiation therapy. Studies have revealed that the interaction between astrocytes and glioma cells can modulate GBM cell functions, such as enhancing their proliferation and inducing a drug-resistant state through structures resembling TNTs.14 Glioblastoma-like stem cells have been described as exhibiting different properties in terms of the formation of TNTs and their ability to transfer mitochondria in 2D and 3D cultures in response to irradiation, depending on their metabolic activity.15
A possible strategy that can be pursued is the inhibition of TNT formation. Since specific markers for TNTs have not yet been identified, inhibitors of actin polymerization or of DNA synthesis were used. In 2012, Lou et al. and in 2016, Desir et al. evaluated metformin and everolimus, two FDA-approved drugs for other indications, as TNT formation inhibitors in human mesothelioma cells and in ovarian cancer cells, respectively. Results showed that these compounds were able to interfere with TNT formation; however, in vivo preclinical data on the applicability of these drugs are not yet available.16,17 On the other hand, a promising alternative could be the exploitation of intercellular communication via TNTs. Indeed, TNTs have been demonstrated to facilitate the transfer of NPs, drugs, and viruses between cells.18 The exploitation of TNTs might improve the localized cellular distribution of nanomedicines due to their ability to connect even distant cells, suggesting that metastatic cells in different tumour niches could also be targeted.18,19
Until now, different types of NPs have been identified within TNTs, from inorganic to lipid-based NPs.20,21 Very little is known about the role of fundamental physical parameters of NPs, such as size, shape, charge and flexibility, in determining their penetration across the BBB and their transfer between cells by TNTs. Positively charged NPs trigger the formation of TNTs due to a higher toxicity compared to those that are negatively charged.20 It has been shown that 100–200 nm particles with a rod-like shape exhibited higher BBB transport compared to larger and spherical ones.22 Considering that the thickness of TNTs is in the range of 0.2–1 μm, we can speculate that size should not be a critical parameter, while the shape of NPs could be important. As far as we know, no data are available on the effect of the shape of NPs on transfer efficiency between TNTs.
Therefore, we designed spherical, discoidal and deformable lipid-based NPs with a size of <120 nm and similar stiffness. The BBB permeability, biocompatibility, and TNT-mediated transfer of NPs were investigated in 2D and 3D in vitro models composed of human GBM cells. Results showed that discoidal NPs displayed a higher BBB permeability and transfer efficiency between GBM TNTs. This could be attributable to the shape of the NPs because discoidal NPs have a larger surface area exposed to the flow along the TNT channel.
2. Experimental
2.1 Cell lines
Gli36ΔEGFR-2 cells (here referred to as GBM cells), provided by Prof. Rosa Maria Moresco (University of Milano-Bicocca, Italy), were used as a GBM in vitro model. Gli36ΔEGFR cells,23 carrying the EGFRvIII mutation present in ∼50% of all EGFR-amplified GBM cases, were made resistant to TMZ after 1 month of in vitro exposure to 50 μM TMZ.24 These cells were selected because their TMZ sensitivity was repeatedly tested in vitro and in vivo in orthotopic GBM models.25 Gli36ΔEGFR-2 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) high glucose w/o sodium pyruvate (ECM0101L, Euroclone, Milan, Italy) supplemented with 10% fetal bovine serum (FBS, ECS0180L, Euroclone, Milan, Italy), 4 mM L-glutamine (ECB3000D, Euroclone, Milan, Italy), and 100 U ml−1 penicillin/streptomycin (P/S) (ECB3001B, Euroclone, Milan, Italy). They were maintained at 37 °C under 5% CO2 and saturated humidity.Immortalized human cerebral microvascular endothelial cells (hCMEC/D3) were provided by Prof. S. Bourdoulous (Institut Cochin, Inserm, Paris, France) and were used as a model of the brain capillary endothelium.26 Cells between passages 27 and 35 were seeded on tissue culture flasks, pre-treated with rat tail collagen type I (0.05 mg mL−1). Cells were grown in complete culture medium (EBM-2 supplemented with 10% FBS, 1% chemically defined lipid concentrate (CDLC), 1% penicillin/streptomycin (P/S), 10 mM HEPES, 5 μg mL−1 ascorbic acid, 1 ng mL−1 bFGF, and 1.4 μM hydrocortisone) and maintained at 37 °C under 5% CO2. The culture medium was changed every 2 days.
2.2 Characterization of Gli36ΔEGFR-2 TNTs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Sigma), SPY555-tubulin (1
1000, Sigma), SPY555-tubulin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Spirochrome or Tubulin Tracker Green 1
1000, Spirochrome or Tubulin Tracker Green 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, ThermoFisher) and Hoechst (1
1000, ThermoFisher) and Hoechst (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, ThermoFisher) to label actin, α-tubulin, and nuclei, respectively, as previously described.27 Pictures were acquired with the Spinning Disk W1 Nikon Eclipse Ti2 confocal system (SD Ti2) (Nikon Instruments, Melville, NY, USA) using a 60× oil immersion objective. Alternatively, cells were analyzed in live imaging over time, up to 24 h, using the Operetta CLS High Content Analysis Systemconfocal microscope (40×) at different time points. The entire volume of the cells was acquired with a Z-stack interval of 0.5 μm. About 200 cells per experiment were analyzed. Experiments were performed in triplicate. Images were analyzed using ImageJ or ICY software.
000, ThermoFisher) to label actin, α-tubulin, and nuclei, respectively, as previously described.27 Pictures were acquired with the Spinning Disk W1 Nikon Eclipse Ti2 confocal system (SD Ti2) (Nikon Instruments, Melville, NY, USA) using a 60× oil immersion objective. Alternatively, cells were analyzed in live imaging over time, up to 24 h, using the Operetta CLS High Content Analysis Systemconfocal microscope (40×) at different time points. The entire volume of the cells was acquired with a Z-stack interval of 0.5 μm. About 200 cells per experiment were analyzed. Experiments were performed in triplicate. Images were analyzed using ImageJ or ICY software.
2.3 Synthesis and characterization of NPs
Spherical NPs (sNPs) composed of cholesterol and sphingomyelin in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio were prepared using the extrusion procedure as previously described.28,29 Briefly, cholesterol and sphingomyelin were dissolved in chloroform/methanol (2
1 molar ratio were prepared using the extrusion procedure as previously described.28,29 Briefly, cholesterol and sphingomyelin were dissolved in chloroform/methanol (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), mixed together and the solvents removed to form a thin film. The lipid film was hydrated in phosphate-buffered saline (PBS) and then extruded through 100 nm polycarbonate membrane filters at 60 ± 4 °C under 20 bar nitrogen pressure. Discoidal NPs (diNPs) composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, cholesterol, and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000] (Sigma Aldrich, Milano, Italy) mixed in a molar ratio of 35
1, v/v), mixed together and the solvents removed to form a thin film. The lipid film was hydrated in phosphate-buffered saline (PBS) and then extruded through 100 nm polycarbonate membrane filters at 60 ± 4 °C under 20 bar nitrogen pressure. Discoidal NPs (diNPs) composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, cholesterol, and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000] (Sigma Aldrich, Milano, Italy) mixed in a molar ratio of 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 (POPC
25 (POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol
Chol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEG-DSPE) were prepared and solvents removed using a vacuum pump in order to form a thin lipidic film. They were subsequently prepared by sonication of the hydrated solution for 40 min in an ice-bath using a Sonics Vibra-Cell (Pasquali Ettore S.R.L., Milano, Italy) as previously described.30 Deformable NPs (deNPs) were composed of POPC, cholesterol, and lysophosphatidylcholine in the molar ratio of 85
PEG-DSPE) were prepared and solvents removed using a vacuum pump in order to form a thin lipidic film. They were subsequently prepared by sonication of the hydrated solution for 40 min in an ice-bath using a Sonics Vibra-Cell (Pasquali Ettore S.R.L., Milano, Italy) as previously described.30 Deformable NPs (deNPs) were composed of POPC, cholesterol, and lysophosphatidylcholine in the molar ratio of 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5. Lipids were dissolved in chloroform/methanol (2
5. Lipids were dissolved in chloroform/methanol (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), dried under vacuum, and the lipid film was then rehydrated in PBS at pH 7.4. They were prepared using the extrusion method, as described above.31 NPs were fluorescently labelled by adding 1 mol% Bodipy SM (Sigma-Aldrich, Milano, Italy) to the lipid mixture. They were characterized in terms of lipid recovery using Stewart's assay;32 their size, stability, ζ-potential, and polydispersity index were obtained using the dynamic light scattering (DLS) technique (Brookhaven Instruments Corporation, Holtsville, NY, USA, equipped with a ZetaPALS device33); and their morphology was observed through cryoEM imaging.34
1, v/v), dried under vacuum, and the lipid film was then rehydrated in PBS at pH 7.4. They were prepared using the extrusion method, as described above.31 NPs were fluorescently labelled by adding 1 mol% Bodipy SM (Sigma-Aldrich, Milano, Italy) to the lipid mixture. They were characterized in terms of lipid recovery using Stewart's assay;32 their size, stability, ζ-potential, and polydispersity index were obtained using the dynamic light scattering (DLS) technique (Brookhaven Instruments Corporation, Holtsville, NY, USA, equipped with a ZetaPALS device33); and their morphology was observed through cryoEM imaging.34
Briefly, a 3 μL droplet of the sample was vitrified in liquid ethane at −165 °C using a Gatan CP3 Plunge Freezer and transferred to a Gatan model 914 cryo-EM holder under liquid nitrogen. Bright field TEM images were obtained using a JEOL JEM-2100 LaB6 microscope equipped with a Gatan Orius SC1000 CCD camera at 200 kV accelerating voltage under low-dose conditions.
To verify the deformability of deNPs under external forces, we assessed their penetration ability into polycarbonate films with pore sizes of 100 and then 80 nm, under an applied extrusion force. After the extrusions, the number of NPs per mL was evaluated through nanoparticle tracking analysis (NanoSight NS300, Malvern Panalytical).35
sNPs 2 and sNPs 3 were prepared as described in ref. 36 and 37, respectively.
2.4 BBB permeability to NPs
The in vitro BBB model was prepared and characterized, as previously described,38 using hCMEC/D3 cells. Briefly, cells were seeded at a density of 5.6 × 104 cells per well onto collagen-coated (150 μg mL−1 rat tail collagen type 1; Gibco, ThermoFisher) Transwell® filters (polyester 12-well, pore size 0.4 μm, translucent membrane inserts 1.12 cm2; Costar) to establish a polarized monolayer. Cells were grown for 3 days in complete EBM-2 medium, and after 3 days, the medium was replaced with EBM-2 supplemented with 5% FBS, 1% CDLC, 1% P/S, 10 mM HEPES, 5 μg mL−1 ascorbic acid, 1.4 μM hydrocortisone, and 10 mM LiCl. The formation of a valid monolayer was evaluated by measuring transendothelial electrical resistance (TEER), monitored using an STX2 electrode epithelial volt-ohm meter (World Precision Instruments, Sarasota, FL, USA), the permeability to TRITC-dextran 4400 Da (λecc = 557 nm, λem = 572 nm) and FITC-dextran 40 000 Da (λecc = 495 nm; λem = 525 nm) (Sigma Aldrich, Milano, Italy), and by visualization of the tight junction between cells, in particular ZO-1. Cells were fixed with 4% PFA, then permeabilized with 3% BSA and 0.1% Triton X-100 in PBS 1× and then incubated with the primary antibody anti-ZO-1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, ThermoFisher) for 3 hours. After 3 h, the secondary anti-mouse antibody conjugated with Alexa Fluor 488 was added for 1 h. In the end, DAPI (1
50, ThermoFisher) for 3 hours. After 3 h, the secondary anti-mouse antibody conjugated with Alexa Fluor 488 was added for 1 h. In the end, DAPI (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 Sigma-Aldrich) was added and images were acquired using an inverted point scanner confocal microscope (Airyscan LSM170, Zeiss) with a 63× oil immersion objective and additional digital zooming when required.
5000 Sigma-Aldrich) was added and images were acquired using an inverted point scanner confocal microscope (Airyscan LSM170, Zeiss) with a 63× oil immersion objective and additional digital zooming when required.
On the tenth day of the hCMEC/D3 culture, at the highest TEER values and lowest endothelial permeability (EP) of TRITC-dextran, fluorescent NPs were added to the apical compartment (400 nmol mL−1) and then incubated at 37 °C for up to 3 h. At different time points, the SM-Bodipy fluorescence in the basolateral compartment was measured at λecc = 500 nm and λem = 515 nm using a spectrofluorometer (Spectrofluorometer FP-8500, Jasco),39 and the EP was calculated as described.40 NP integrity after BBB crossing was evaluated using Nanoparticle Tracking Analysis (NanoSight NS300, Malvern Panalytical).41
2.5 Effect of NPs on Gli36ΔEGFR-2 cells
Gli36ΔEGFR-2 cells were plated at a density of 4 × 104 cells per cm2. After 24 h, they were incubated for 1 h with the 3 different NPs and then fixed in a solution of 2% PFA, 0.05% glutaraldehyde and 0.2 M HEPES in PBS for 15 min, followed by 15 min in 4% PFA and 0.2 M HEPES in PBS, both at 37 °C in order to preserve TNT integrity. The plasma membrane was labelled with fluorescent wheat germ agglutinin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 in PBS, Life Technologies) for 20 min at RT. Nuclei were stained with DAPI (1
500 in PBS, Life Technologies) for 20 min at RT. Nuclei were stained with DAPI (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 Sigma-Aldrich). Confocal images were acquired using a Zeiss LSM 700 controlled by ZEN software. All the images were processed using ICY software in order to semi-automatically count the number of TNT-connected cells.
5000 Sigma-Aldrich). Confocal images were acquired using a Zeiss LSM 700 controlled by ZEN software. All the images were processed using ICY software in order to semi-automatically count the number of TNT-connected cells.
2.6 Tracking of NPs inside the TNTs and co-localization with lysosomes
Gli36RΔEGFR-2 cells were seeded on a 24-well Ibidi plate (IbiTreat: #1.5 polymer coverslip, 82426) at a density of 4 × 104 cells per well and incubated with fluorescently labelled NPs (400 nmol ml−1) for 30 minutes at 37 °C. Then they were washed with PBS and time-lapse microscopy images were acquired at time intervals between 30 s and 1 min per Z-stack using the Spinning Disk W1 Nikon Eclipse Ti2 confocal system (SD Ti2) with a 60× oil immersion objective (Plan Apo 60×OIL NA = 1.42) or a 40× water immersion objective (Apo LWD 40× NA = 1.15). The acquisition of Z-stacks is crucial to visualize TNT structures, which hover above the substrate.27Brightfield and 488 channels were acquired to visualize the membrane of the cell and NPs, respectively, with the minimum possible power of laser illumination to avoid phototoxicity.
The maximum intensity projection of the time-lapse movies of NPs was processed using ImageJ/Fiji software. Analysis of the velocity of NP movement inside the TNTs was performed using the TrackMate plugin in Fiji software.
To determine if the NPs co-localize with acidic intracellular organelles inside the TNT, Gli36RΔEGFR-2 cells were incubated with fluorescently labelled NPs for 30 minutes at 37 °C (400 nmol mL−1) and then with lysotracker red for 30 minutes at 37 °C (Sigma), following the manufacturer's instructions. Images were acquired using Brightfield, 546 and 488 channels with a Spinning Disk 60× oil immersion objective and then analyzed with ImageJ using the JacoP plug-in to determine the co-localization of acidic organelles and NPs. The degree of colocalization was calculated using the thresholded Manders’ correlation coefficient of global statistical analysis, considering pixel intensity distributions. At least 10 Z-stacks were analysed per sample, and the percentage of the fraction of each type of NPs in the acidic organelles was expressed as the mean ± standard error of the mean.42
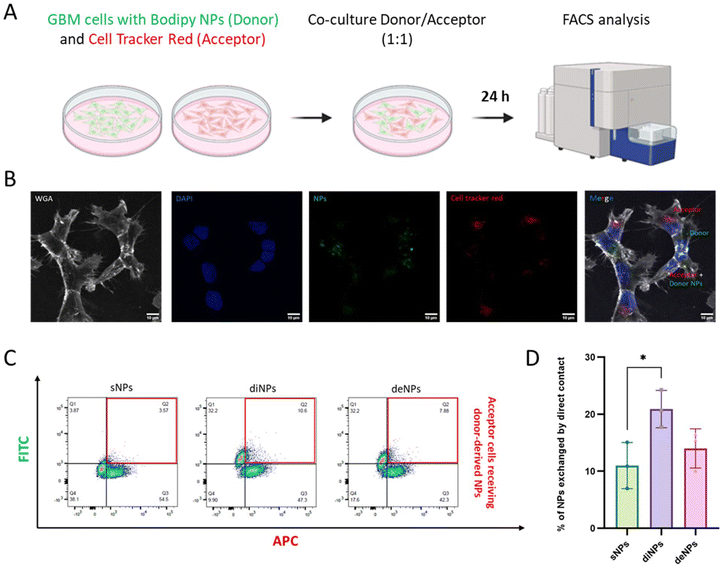
2.7 Quantification of NP transfer by flow cytometry (FACS)
Transfer assays were performed using a population of Gli36RΔEGFR-2 cells incubated with fluorescent NPs for 30 minutes at 37 °C as donor cells and GBM cells labelled with Cell Mask RED (Sigma-Aldrich) as acceptor cells. The cells were mixed in a 1 ÷ 1 ratio at a density of 8 × 104 in a 24-well plate, as described in ref. 27. To monitor transfer by secretion, donor and acceptor cells were co-cultured, separated by a 1 μm filter, which allows passage of the NPs in case they are secreted but prevents direct contact between the two populations. After 24 h of co-culture, cells were detached, fixed in 2% PFA and then passed through a cell strainer to separate cell aggregates for FACS analysis. Flow cytometry data were acquired using a BD LSR Fortessa flow cytometer. Fluorescence intensities were measured at 488 nm and 651 nm excitation wavelengths, respectively. Ten thousand events were acquired for each condition and data were analyzed using FlowJo analysis software.2.8 Three-dimensional (3D) GBM cell culture model
Tumour organoids were prepared according to the protocol published in ref. 43 and cultured in DMEM Glutamax (Euroclone) supplemented with 1% P/S (100× Gibco), 10% FBS (Capricorn) and GelTrex (ThermoFisher) at 37 °C in 5% CO2 humidified incubators for up to 10 days. Part of the cultured medium was removed and replaced with fresh medium every 2–3 days. Tumour organoids were imaged with an inverted point scanner microscope (CellDiscoverer7, Zeiss) using a 2.5 or 50×, 1.2 NA water immersion objective performing a Z-stack reconstruction of all organoids or focusing on specific regions of interest.2.9 Statistical analysis
Statistical analysis was performed with GraphPad Prism 8 using the following tests: two-way ANOVA, one-way ANOVA, unpaired t test, Dunnett's multiple comparisons test, and Tukey's multiple comparisons test. Statistical significance was considered at p < 0.05.3. Results and discussion
NPs are promising tools for the treatment of brain tumours because of their performance in delivering drugs across the BBB, selectively targeting cancer cells. However, the distribution of NPs in the tumour parenchyma still remains a challenge that needs to be addressed in order to improve their efficacy. In this context, the intercellular connections between cells, such as TNTs, represent a promising approach to enhance NP distribution among connected cells.To deepen our understanding of the role of physical parameters of NPs in determining their penetration across the BBB and their transfer by TNTs, spherical, discoidal and deformable lipid-based NPs have been synthetized and tested in vitro.
3.1 Structural and morphological characterization of TNTs formed by Gli36ΔEGFR-2 cells
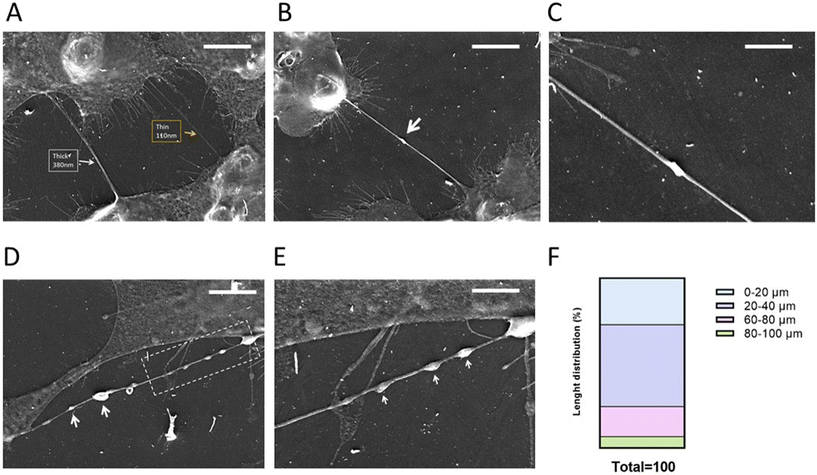
Gli36ΔEGFR-2 cells (here referred to as GBM cells) have been chosen as a GBM in vitro model because EGFR mutations occur in ∼50% of all EGFR-amplified GBM cases (Fig. S1†) and correlate with bad prognoses.44In order to verify if this cellular model was able to form TNTs and thus be suitable for this investigation, GBM cells were seeded at different densities and observed at different time points by optical microscopy (Fig. S2†). The chosen conditions to better visualize TNTs were 4 × 104 cells per cm2 density and 24 h of seeding. After this, the cells were fixed and samples were visualized by SEM to characterize the morphology and structure of TNTs.
Analyzing the SEM images, we can observe that GBM cells are able to form mainly thin TNTs (Fig. 1A) with a thickness of 0.2–0.7 μm,45 while thick TNTs are usually considered the ones with a diameter in the range of 0.7–1 μm. These dimensions make TNTs formed by GBM cells suitable for the transfer of NPs, in accordance with data in the literature where drug-loaded liposomes were transferred between U87MG GBM cells via TNTs.46
The biogenesis of TNTs is a difficult issue to address because of cell heterogeneity and the different mechanisms involved.45,46 Even if it is not possible to assess the TNT formation mechanism in a fixed sample, we can suppose that GBM cells can form TNTs through a “hand-shake” mechanism,45 as shown Fig. 1B and C.
Moreover, TNTs formed by GBM cells displayed a “pearling” morphology (Fig. 1D and E), attributable to the presence of intracellular vesicles, suggesting that they are capable of intercellular transfer of cargoes45,46 and that thin TNTs can exhibit dilatation of the membrane, enabling the passage of cargoes with a larger diameter.
Classifying the TNTs formed by GBM cells on the basis of length (Fig. 1F), we can assert that these cells are able to form short TNTs (∼25%, 0–20 μm), medium-length TNTs (∼65%, 20–80 μm) and long TNTs (∼10%, 80–100 μm). This variability could be a key factor in improving the distribution of NPs between near and far-away cells in brain tumour niches, microanatomical regions displaying morphological and functional differences that are considered communication centers where cell populations interact.47
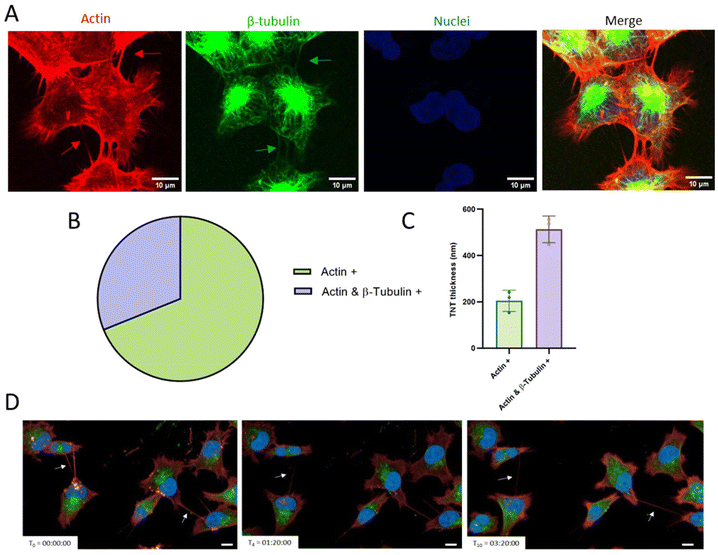
Considering that the presence of F-actin cytoskeletal filaments and/or tubulin filaments is important to allow the transfer of cargoes and to ensure the TNT stability, we analysed the internal composition of TNTs formed by GBM cells. Cells were plated and labelled with fluorescent probes in order to visualize actin, β-tubulin and nuclei. Pictures (Fig. 2A) were acquired through spinning disk confocal microscopy and analyzed through the ImageJ software. Results (Fig. 2B and C) showed that 69% of the total TNTs were only actin-based, while 31% were actin and β-tubulin positive. Regarding the correlation between the thickness of TNTs and their composition,20 we found that actin-based TNTs had a thickness of around 0.2 μm, while actin- and β-tubulin-based TNTs were thicker, at around 0.5 μm. Moreover, it is notable that a single cell is able to form multiple TNTs, which can connect it with one or more different cells.
To evaluate TNT dynamics, cells were plated, fluorescently labelled and then observed live over time for up to 24 h using the Operetta CLS High Content Analysis System confocal microscope. Results (Fig. 2D; time-lapse in Fig. S3†) showed that they are transient structures that possess different half-life, ranging from minutes to hours, and in the same area different TNTs can be formed.
3.2 Characterization, BBB crossing and biocompatibility of spherical, discoidal and deformable lipid-based NPs
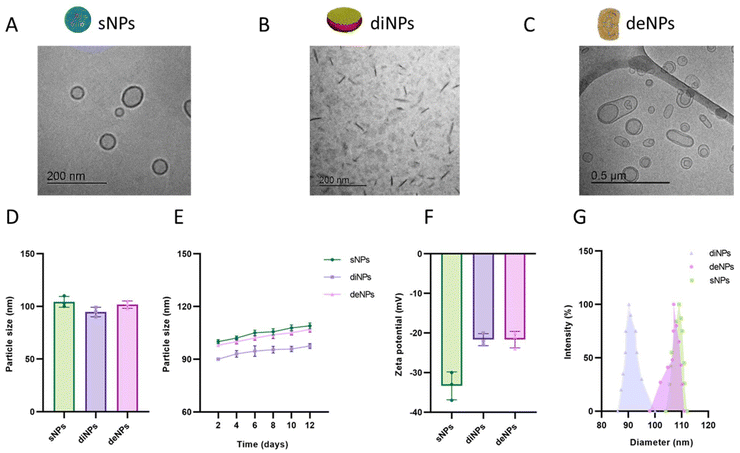
Since the purpose of this study was to investigate the contribution of physical parameters in determining the BBB permeability and the TNT-mediated transfer between cells, spherical, discoidal and deformable lipid-based NPs have been synthesized and characterized. All the NPs tested were composed of a matrix of cholesterol and POPC, with the exception of sNPs where POPC was substituted with SM, a lipid with the same polar head but different melting temperature (−4 °C and +42 °C, respectively). This modification is critical to obtaining NPs with a stable spherical shape and very low deformability at 37 °C,48,49 a temperature at which all the in vitro experiments have been performed.NPs were characterized morphologically by cryoEM imaging: sNPs displayed a spherical-shaped structure homogeneously distributed in vitreous ice, with diameters ranging from 50 to 150 nm (Fig. 3A); diNPs showed a flat, circular disk-shaped structure (Fig. 3B). The ellipsoid shape in the background represents the face-on or front side of the discoidal NPs, while the darkest elongated shapes represent their flank side, depending on their orientation.30 deNPs exhibited variably dimpled hemispheres, spinning tops, bowls, discs, and multilamellar bodies (Fig. 3C).
The diameter, stability, surface charge and polydispersity of NPs were determined using the dynamic light scattering (DLS) technique and Z-pals device. NPs displayed similar size, with a diameter of ∼100 nm (Fig. 3D). All the NPs demonstrated high stability over time, as indicated by the negligible increase in their diameter (<10% in 12 days) (Fig. 3E). NPs are negatively charged, as demonstrated by ζ-potential values <−20 mV; this suggests that the dispersions are stable and not prone to aggregation (Fig. 3F). The size distribution analysis (Fig. 3G) showed that diNP and sNP samples have a narrow size distribution around the mean diameter, while deNPs have a broader size distribution due to their deformable features. Their deformability was confirmed by measuring the number of NPs after serial filtration through membranes with different pore sizes. Results (Fig. S4†) showed that the number of deNPs did not change significantly after filtrations through 100 nm and 80 nm pore-sized membranes, in contrast to conventional liposomes. All the preparations were fairly monodispersed, with a PDI < 0.2.
Considering that the brain is protected by the BBB, it is important to evaluate the permeability of NPs designed as drug delivery systems across the BBB. Several engineered NPs have been developed to target and cross the BBB with promising results,50 but the effect of shape on crossing biological barriers is still under investigation. In this context, sNPs, diNPs and deNPs were tested in vitro using a Transwell® system made with hCMEC/D3 cells as a BBB model.
The structural, electrical and functional properties of hCMEC/D3 monolayers were determined by immunofluorescence, TEER and EP to fluorescent probes, respectively. Results showed that hCMEC/D3 monolayers were able to form tight junctions, as indicated by the detection of the ZO-1 signal (Fig. 4A). TEER values progressively increased over time, reaching maximum values of ≥50 Ω × cm2 by the 9th day after seeding (Fig. 4B), in agreement with data reported in the literature.50 The EP values for the fluorescent probes FITC-dextran (40 kDa) and TRITC-dextran (4.4 kDa) were 6.31 × 10−6 cm min−1 and 1.97 × 10−4 cm min−1 (Fig. 4C), respectively, demonstrating the tightness feature of the hCMEC/D3 monolayers.
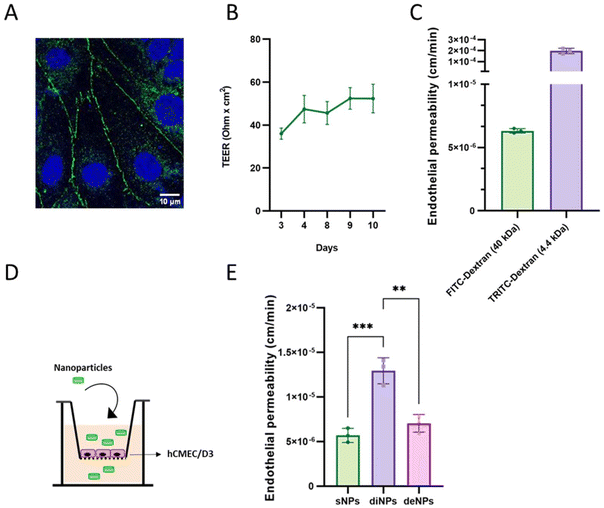 | ||
| Fig. 4 Characterization of the BBB in vitro model and evaluation of the passage of NPs. (A) hCMEC/D3 cells were plated and after 72 h cells were fixed and then labelled with DAPI and the primary antibody anti-ZO-1. Tight junctional proteins were visualized in the cell monolayers through confocal microscopy. The scale bar is embedded in the figure. (B) TEER values (Ω cm2) from day 3 to day 10 of growth of cells on the Transwell® system. (C) FITC- and TRITC-dextran permeability across hCMEC/D3 monolayers. (D) Representative drawing of the experimental model to measure the BBB permeability to sNPs, diNPs and deNPs. (E) Concentrations of 400 nmol mL−1 of total lipids for each type of fluorescently labelled NPs were added in the apical compartment; the fluorescence in the basolateral compartment was measured over time for up to 3 h. EP was calculated as described.40 Data are presented as the mean of three independent experiments ± SD. Each graph represents the result of three independent experiments. ***, p < 0.001 calculated by Tukey's multiple comparisons test. | ||
Fluorescently labelled NPs were added to the apical compartment of the Transwell® system, and the fluorescence in the basolateral compartment was measured over time for up to 3 h (Fig. 4D).
Results showed that diNPs exhibited a 2-fold higher EP value (1.29 × 10−5 cm min−1) compared to those of sNPs and deNPs (5.69 × 10−6 cm min−1 and 7.04 × 10−6 cm min−1, respectively) (Fig. 4E), suggesting that the discoidal shape allows a BBB crossing of higher efficiency by the NPs. This is comparable to the enhanced BBB permeability of discoidal high-density lipoproteins (dHDL) in comparison with the spherical ones, as previously reported.51 Moreover, Fu et al.52 found that the BBB permeability of NPs was closely related to particle shape, showing that rod-shaped polystyrene NPs were better performing than the spherical NPs on cerebral microvasculature cells in vitro. The superior performance of discoidal NPs compared to spherical ones can be attributed to their lower aspect ratio and orientation, which affect the rate of cellular internalization, a prerequisite for crossing the BBB, as suggested by Sabourian et al., who highlighted the importance of engineered physical and mechanical characteristics of NPs in facilitating their internalization into the targeted cells.53 This is analogous to the findings of Deng et al., who demonstrated that the particle entry mode into cells is highly dependent on particle shape, with ellipsoidal NPs exhibiting different entry modes based on their aspect ratio.54
It is also important to point out that, to the best of our knowledge, there is no evidence of TNT formation between endothelial cells composing the BBB. However, there is limited evidence regarding the involvement of TNT in the communication between endothelial cells and the other cells composing the BBB (such as pericytes and astrocytes).55 Whether TNT plays a functional role in controlling the BBB crossing of NPs is still unknown.
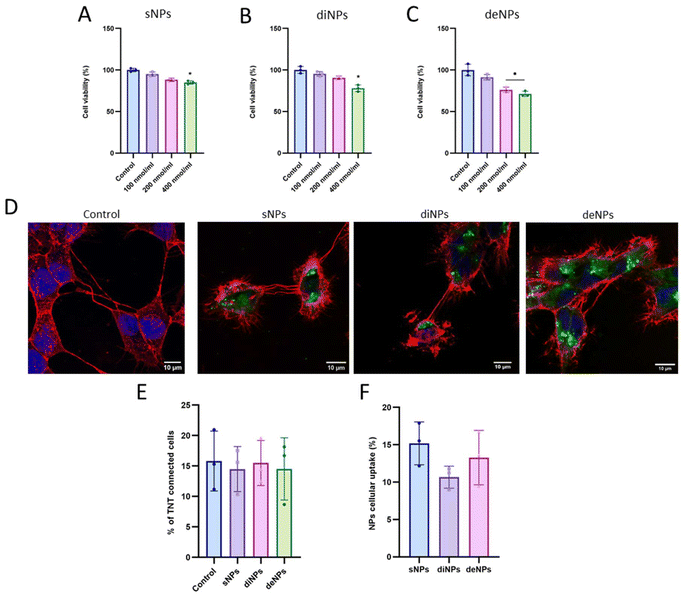
To assess the biocompatibility of NPs, cell viability and the % of TNT-connected cells were evaluated using the MTT assay and confocal microscopy, respectively.
Results (Fig. 5A–C) showed that the treatment of GBM cells with 100 nmol mL−1 (total lipids) of NPs did not affect cell viability. A slight decrease of cell viability was detected at the highest NP concentration tested; however, the cell viability was always ≥70%, suggesting that all NPs used are well tolerated. This was confirmed by measuring the % of TNT-connected cells after incubation with the highest dose of NPs (400 nmol mL−1). Results (Fig. 5D and E) showed that the % of TNT-connected cells (∼15%) was comparable to that of untreated cells, demonstrating that NP treatment did not interfere with TNT formation.
The assumption useful to make NPs transferable between cells via TNTs is the cellular uptake. Accordingly, the cellular uptake of sNPs, diNPs and deNPs was evaluated by fluorescence spectroscopy. Results (Fig. 5F) showed no significant difference in cellular uptake between the three NPs tested, but it can be noticed that spherical NPs tended to have higher uptake in comparison with discoidal and deformable NPs. This agrees with data present in the literature, showing that the uptake of rod-like NPs is lower in comparison with spherical NPs. This is probably because cells require more time to encapsulate rod-like NPs56 and/or because of the binding of surfactant molecules on the longitudinal axis of nanorods, thus affecting their cellular uptake.57
3.3 NP tracking inside TNTs
Gli36ΔEGFR-2 cells were treated with fluorescently labelled sNPs, diNPs or deNPs to study their TNT-mediated transfer between cells.Results showed that NPs co-localized with TNTs and were tracked along the TNTs (Fig. S5†).
To assess the direction and the velocity of the NPs inside the TNTs, GBM cells were incubated with fluorescently labelled NPs and videos were acquired over time through spinning disk confocal microscopy. Results demonstrated that NPs can be transferred along TNTs in a bidirectional way (Fig. 6A and movie S1†), probably due to the presence of β-tubulin in the TNTs,58 as suggested for the transfer of organelles via TNTs. The average velocity was similar for all the NPs tested (∼14 nm s−1), while the highest maximum velocity was reached by diNPs (69.2 nm s−1) (Fig. 6B). To the best of our knowledge, there are no data in the literature concerning the NP velocity inside TNTs.
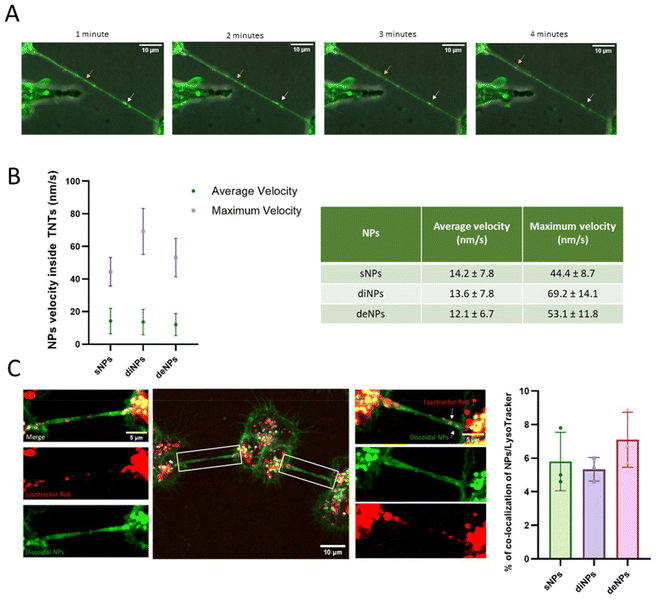 | ||
| Fig. 6 Tracking of NPs inside TNTs. In order to assess the direction and the velocity of the NPs inside the TNTs, GBM cells were incubated with fluorescently labelled NPs (400 nmol mL−1) for 30 minutes. Then, they were washed with PBS and videos (movie S1†) were acquired over time through the spinning disk 60× oil immersion objective to determine their direction. (A) Representative images extrapolated from GBM cells loaded with diNPs. (B) Maximum velocity and average velocity of NPs inside the TNTs, calculated by analysing 6 TNTs per NP, from three independent experiments. (C) To determine if the NPs co-localize with lysosomes inside the TNT, GBM cells were incubated with NPs (400 nmol mL−1) and then with lysotracker red. The graph represents the results of colocalization analysis using the JacoP plugin from ImageJ using Manders’ correlation coefficient. The Mander's M values as threshold values corresponding to the fraction of NPs in the acidic organelles following 1 h exposure are shown. Results are presented as the mean ± SD. The white squares indicate the magnified area for a better visualisation of the TNT connections. Scale bars are embedded in the figures. | ||
To determine if NPs were transferred alone or in association with organelles, GBM cells were incubated with fluorescently labelled NPs, incubated with lysotracker red to track acidic organelles in live cells and images were acquired using spinning disk confocal microscopy. Images were analyzed using ImageJ with the JacoP plugin to quantify the co-localization of green and red signals. Results (Fig. 6C) showed that the rate of co-localization of the NPs tested with acidic organelles was ∼6%, suggesting that the taken up NPs did not follow degradative pathways, raising the possibility of TNT-mediated transfer between cells. Considering that several strategies are currently under investigation to increase the diffusion of NPs in the tumour parenchyma,59 the exploitation of TNTs as NP delivery systems, as suggested herein, represents a promising approach to improve NP spread in the tumour microenvironment. Moreover, considering the already reported structural differences between the TNTs formed by GBM cells (thicker and longer) vs. those formed by normal human astrocytes (thinner and shorter),46 it is also possible to exploit these channels to increase the treatment specificity. Accordingly, more efficient NP transport has been observed between GBM cells compared to non-tumour brain cells.46
To confirm whether TNTs allowed the transfer of the NPs between connected cells, a co-culture assay between a donor population, loaded with fluorescently labelled NPs, and an acceptor cell population marked with Cell Tracker Red was performed. After 24 h of co-culture, cells were analyzed by confocal microscopy and FACS (Fig. 7A and B). Results showed that the % of acceptor cells receiving NPs from donor cells was 11% for sNPs, 21% for diNPs and 14% for deNPs (Fig. 7C and D), suggesting that the TNT-mediated transfer efficacy was highest for diNPs. This can be attributed to the shape of NPs because the disk-like shape has a larger surface area exposed to the flow along the TNT channel in comparison with spherical NPs, similar to the improved delivery observed for rod-like NPs in a mimetic blood vessel.60 Considering that the NPs tested herein possess a different lipid composition, we performed similar experiments comparing diNPs with sNPs 236 and sNPs 337 with the same lipid composition. Even in this case, diNPs showed the highest transfer efficiency (Fig. S6†).
The NP exchange between cells was exclusively due to a contact-dependent mechanism since negligible transfer (<3%) was observed when the two cell populations were separated by a filter but shared the same medium (Fig. S7†). Moreover, to study the transport dynamics of NPs, similar experiments were conducted from 16 hours to 24 hours at 2-hour intervals. The results showed that the transport efficiency (%) of the NPs did not change significantly over time. Interestingly, the differences in transport efficiency among the three NPs tested remained consistent, with diNPs showing the highest values. Unfortunately, it was not possible to perform the experiment at earlier time points due to the necessary waiting period for cell adhesion.
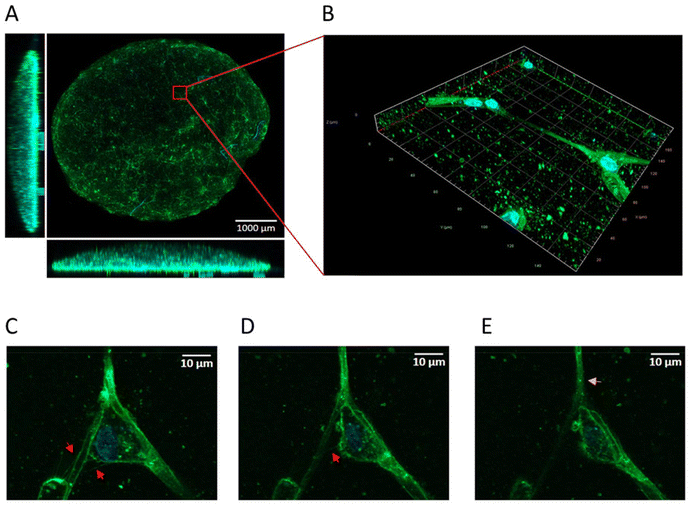
To verify whether GBM cells are able to form TNTs embedding NPs in a context more representative of the tumour, a 3D tumoroid culture was set up according to the protocol published by Hubert et al.43 The 3D-cell model is a culture method, which allows long-term growth, preserving the cell morphology. Five days after culturing, tumoroids were treated with fluorescently labelled NPs and cell nuclei were labelled. The tumoroids were visualized by confocal microscopy. Even if it is extremely challenging to identify these transient and delicate structures in 3D, it is possible to visualize TNT-like cell protrusions in confocal images (Fig. 8A and B) already within the first days of culture. From the acquired images, it is possible to visualize NPs inside the cell protrusions, suggesting that even in a 3D model, GBM cells are able to form functional TNTs for NP transfer (Fig. 8C–E), consistent with the results obtained in 2D.
4. Conclusion
In conclusion, this study introduces a sustainable solution to improve the diffusion rate at which the NPs can spread in the brain tumour region, based on the presence of intercellular connections (TNTs). Several strategies are currently under investigation to increase the diffusion of NPs, such as modifications to particle size and surface charge, but limited data are available on NP shape and TNTs. Here, we have shown that discoidal NPs can be transferred by TNTs formed between tumour cells with high efficiency, suggesting that the shape, especially the aspect ratio, is also a key parameter in determining NP diffusion among cells. These results open the possibility of exploiting TNTs as drug-delivery channels, thus improving cancer therapy by promoting drug distribution to difficult-to-reach cell populations. Moreover, we have shown that the shape of NPs is a crucial factor affecting their permeability across the BBB, and the discoidal shape allows for a greater efficiency in crossing. Overall, these results highlight the significant role of the shape of NPs in crossing the BBB and enhancing local cellular distribution.Author contributions
G. S. prepared the NPs, performed all the in vitro experiments, analyzed the data and prepared the manuscript; I. S. performed FACS analysis and supervised the experimental activities of G. S.; A. R. performed the characterization of NPs; M. K. performed cryoEM analysis of NPs; P. S. and U. A. performed the SEM experiments; M. M. performed confocal microscopy analysis of 3D tumoroids; D. S. and F. M. performed the characterization of NPs; A. D. performed FACS analysis; F. R conceived the project; F. R. and C. Z. supervised the research and revised the manuscript with contribution from all co-authors.Data availability
The data supporting this article have been included as part of the ESI and are available in the interactive notebook [Experimental data_Sierri G, Google Drive Folder] at https://drive.google.com/drive/folders/1Q2qqHVA97X6EvNVKLe54kxNH2xgx4gjz?usp=sharing.†Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was partially financially supported by PRIN Research Italy, grant number ID 2022YJSMBX, CUP H53D23006320006, awarded to F. R.; the Institut National du Cancer (INCa, Canceropole IDF 2018-1-PL-03-IP-1); the Equipe Fondation Recherche Médicale (FRM EQU202103012692); the Agence Nationale de la Recherche (ANR-20-CE13-0032); the Programme Explore Tumeurs Cérébrales de l’Institut Pasteur; and the late Marguerite Michel, whose bequest to the Institut Pasteur made this project possible, awarded to CZ. I.SSM. was supported by fellowships from the Fondation de France, in memory of the late G. Michellet Neejacquet (00100228), and the Pasteur-Roux-Cantarini fellowship from the Institut Pasteur. A. D. was supported by the Fondazione Umberto Veronesi.References
- A. Czarnywojtek, M. Borowska, K. Dyrka, S. Van Gool, N. Sawicka-Gutaj and J. Moskal, et al., Glioblastoma Multiforme: The Latest Diagnostics and Treatment Techniques, Pharmacology, 2023, 108(5), 423–431 CrossRef CAS PubMed.
- R. Deshmukh, M. F. Allega and S. Tardito, A map of the altered glioma metabolism, Trends Mol. Med., 2021, 27(11), 1045–1059 CrossRef CAS PubMed.
- I. Khan, M. H. Baig, S. Mahfooz, M. A. Imran, M. I. Khan and J. J. Dong, et al., Nanomedicine for glioblastoma: Progress and future prospects, Semin. Cancer Biol., 2022, 86, 172–186 CrossRef CAS.
- D. J. Lundy, H. Nguyễn and P. C. H. Hsieh, Emerging Nano-Carrier Strategies for Brain Tumor Drug Delivery and Considerations for Clinical Translation, Pharmaceutics, 2021, 13(8), 1193 CrossRef CAS.
- L. Tang, Y. Feng, S. Gao, Q. Mu and C. Liu, Nanotherapeutics Overcoming the Blood-Brain Barrier for Glioblastoma Treatment, Front. Pharmacol., 2021, 12, 786700 CrossRef CAS.
- P. Kumthekar, C. H. Ko, T. Paunesku, K. Dixit, A. M. Sonabend and O. Bloch, et al., A first-in-human phase 0 clinical study of RNA interference–based spherical nucleic acids in patients with recurrent glioblastoma, Sci. Transl. Med., 2021, 13, 584 Search PubMed.
- R. Moreira, C. Nóbrega, L. P. de Almeida and L. Mendonça, Brain-targeted drug delivery - nanovesicles directed to specific brain cells by brain-targeting ligands, J. Nanobiotechnol., 2024, 22(1), 260 CrossRef CAS PubMed . Available from: https://www.ncbi.nlm.nih.gov/pubmed/38760847.
- K. E. Khattar, J. Safi, A. M. Rodriguez and M. L. Vignais, Intercellular Communication in the Brain through Tunneling Nanotubes, Cancers, 2022, 14(5), 1207 CrossRef CAS.
- G. Pinto, C. Brou and C. Zurzolo, Tunneling Nanotubes: The Fuel of Tumor Progression?, Trends Cancer, 2020, 6(10), 874–888 CrossRef CAS PubMed.
- R. Z. Lin, Mitochondrial transfer mediates endothelial cell engraftment through mitophagy, Nature, 2024, 629, 660–668 CrossRef CAS PubMed.
- J. G. Baldwin, Intercellular nanotube-mediated mitochondrial transfer enhances T cell metabolic fitness and antitumor efficacy, Cell, 2024, 187, 6614–6630.e21 CrossRef CAS PubMed.
- S. Valdebenito, A. Audia, K. P. L. Bhat, G. Okafo and E. A. Eugenin, Tunneling Nanotubes Mediate Adaptation of Glioblastoma Cells to Temozolomide and Ionizing Radiation Treatment, iScience, 2020, 23(9), 101450 CrossRef CAS PubMed.
- G. Pinto, C. Brou and C. Zurzolo, Tunneling Nanotubes: The Fuel of Tumor Progression?, Trends Cancer, 2020, 6, 874–878 CrossRef CAS.
- A. K. Sisakht, M. Malekan, F. Ghobadinezhad, S. N. M. Firouzabadi, A. Jafari and S. M. A. Mirazimi, et al., Cellular Conversations in Glioblastoma Progression, Diagnosis and Treatment, Cell. Mol. Neurobiol., 2023, 43(2), 585–603 CrossRef PubMed.
- G. Pinto, I. Sáenz-de-Santa-María, P. Chastagner, E. Perthame, C. Delmas and C. Toulas, et al., Patient-derived glioblastoma stem cells transfer mitochondria through tunneling nanotubes in tumor organoids, Biochem. J., 2021, 478(1), 21–39 CrossRef CAS.
- S. Desir, E. L. Dickson, R. I. Vogel, V. Thayanithy, P. Wong and D. Teoh, et al., Tunneling nanotube formation is stimulated by hypoxia in ovarian cancer cells, Oncotarget, 2016, 7(28), 43150–43161 CrossRef PubMed.
- E. Lou, S. Fujisawa, A. Morozov, A. Barlas, Y. Romin and Y. Dogan, et al., Tunneling Nanotubes Provide a Unique Conduit for Intercellular Transfer of Cellular Contents in Human Malignant Pleural Mesothelioma, PLoS One, 2012, 7(3), e33093 CrossRef CAS PubMed.
- P. K. Melwani and B. N. Pandey, Tunneling nanotubes: The intercellular conduits contributing to cancer pathogenesis and its therapy, Biochim. Biophys. Acta, Rev. Cancer, 2023, 1878(6), 189028 CrossRef CAS PubMed.
- L. Taiarol, B. Formicola, S. Fagioli, G. Sierri, A. D’Aloia and M. Kravicz, et al., The 3.0 Cell Communication: New Insights in the Usefulness of Tunneling Nanotubes for Glioblastoma Treatment, Cancers, 2021, 13(16), 4001 CrossRef CAS PubMed.
- I. Ottonelli, R. Caraffi, G. Tosi, M. A. Vandelli, J. T. Duskey and B. Ruozi, Tunneling Nanotubes: A New Target for Nanomedicine?, Int. J. Mol. Sci., 2022, 23(4), 2237 CrossRef CAS PubMed.
- I. Sáenz-de-Santa-María, C. Bernardo-Castiñeira, E. Enciso, I. García-Moreno, J. L. Chiara and C. Suarez, et al., Control of long-distance cell-to-cell communication and autophagosome transfer in squamous cell carcinoma via tunneling nanotubes, Oncotarget, 2017, 8(13), 20939–20960 CrossRef.
- M. Nowak, T. D. Brown, A. Graham, M. E. Helgeson and S. Mitragotri, Size, shape, and flexibility influence nanoparticle transport across brain endothelium under flow, Bioeng. Transl. Med., 2020, 5(2), e10153 CrossRef CAS PubMed.
- T. Abe, H. Wakimoto, R. Bookstein, D. C. Maneval, E. A. Chiocca and J. P. Basilion, Intra-arterial delivery of p53-containing adenoviral vector into experimental brain tumors, Cancer Gene Ther., 2002, 9(3), 228–235 CrossRef CAS.
- T. Viel, S. Schelhaas, S. Wagner, L. Wachsmuth, K. Schwegmann and M. Kuhlmann, et al., Early Assessment of the Efficacy of Temozolomide Chemotherapy in Experimental Glioblastoma Using [18F]FLT-PET Imaging, PLoS One, 2013, 8(7), e67911 CrossRef CAS PubMed.
- S. Valtorta, A. Lo Dico, I. Raccagni, C. Martelli, V. Pieri and P. Rainone, et al., Imaging Metformin Efficacy as Add-On Therapy in Cells and Mouse Models of Human EGFR Glioblastoma, Front Oncol, 2021, 3, 11 Search PubMed.
- B. Poller, H. Gutmann, S. Krähenbühl, B. Weksler, I. Romero and P. Couraud, et al., The human brain endothelial cell line hCMEC/D3 as a human blood–brain barrier model for drug transport studies, J. Neurochem., 2008, 107(5), 1358–1368 CrossRef CAS.
- I. Sáenz-de-Santa-María, J. M. Henderson, A. Pepe and C. Zurzolo, Identification and Characterization of Tunneling Nanotubes for Intercellular Trafficking, Curr. Protoc., 2023, 3(11), e939 CrossRef PubMed.
- E. Conti, M. Gregori, I. Radice, F. Da Re, D. Grana and F. Re, et al., Multifunctional liposomes interact with Abeta in human biological fluids: Therapeutic implications for Alzheimer's disease, Neurochem. Int., 2017, 108, 60–65 CrossRef CAS PubMed.
- S. Sesana, F. Re, A. Bulbarelli, D. Salerno, E. Cazzaniga and M. Masserini, Membrane Features and Activity of GPI-Anchored Enzymes: Alkaline Phosphatase Reconstituted in Model Membranes, Biochemistry, 2008, 47(19), 5433–5440 CrossRef CAS PubMed.
- G. Sierri, R. Dal Magro, B. Vergani, B. E. Leone, B. Formicola and L. Taiarol, et al., Reduced Levels of ABCA1 Transporter Are Responsible for the Cholesterol Efflux Impairment in β-Amyloid-Induced Reactive Astrocytes: Potential Rescue from Biomimetic HDLs, Int. J. Mol. Sci., 2021, 23(1), 102 CrossRef.
- S. Jebari-Benslaiman, K. B. Uribe, A. Benito-Vicente, U. Galicia-Garcia, A. Larrea-Sebal and I. Alloza, et al., Cholesterol Efflux Efficiency of Reconstituted HDL Is Affected by Nanoparticle Lipid Composition, Biomedicines, 2020, 8(10), 373 CrossRef CAS PubMed.
- J. C. M. Stewart, Colorimetric determination of phospholipids with ammonium ferrothiocyanate, Anal. Biochem., 1980, 104(1), 10–14 CrossRef CAS.
- M. Gobbi, F. Re, M. Canovi, M. Beeg, M. Gregori and S. Sesana, et al., Lipid-based nanoparticles with high binding affinity for amyloid-β1–42 peptide, Biomaterials, 2010, 31(25), 6519–6529 CrossRef CAS.
- L. Taiarol, C. Bigogno, S. Sesana, M. Kravicz, F. Viale and E. Pozzi, et al., Givinostat-Liposomes: Anti-Tumor Effect on 2D and 3D Glioblastoma Models and Pharmacokinetics, Cancers, 2022, 14(12), 2978 CrossRef CAS PubMed.
- Y. Zhu, Y. Song, Z. Cao, L. Dong, Y. Lu and X. Yang, et al., Magnetically Actuated Active Deep Tumor Penetration of Deformable Large Nanocarriers for Enhanced Cancer Therapy, Adv. Funct. Mater., 2021, 31(35), 2103655 CrossRef CAS.
- I. V. Zhigaltsev, N. Belliveau, I. Hafez, A. K. K. Leung, J. Huft and C. Hansen, et al., Bottom-Up Design and Synthesis of Limit Size Lipid Nanoparticle Systems with Aqueous and Triglyceride Cores Using Millisecond Microfluidic Mixing, Langmuir, 2012, 28(7), 3633–3640 CrossRef CAS PubMed . Available from: https://pubs.acs.org/doi/10.1021/la204833h.
- A. Lundquist, P. Wessman, A. R. Rennie and K. Edwards, Melittin–Lipid interaction: A comparative study using liposomes, micelles and bilayerdisks, Biochim. Biophys. Acta, Biomembr., 2008, 1778(10), 2210–2216 CrossRef CAS PubMed . Available from: https://linkinghub.elsevier.com/retrieve/pii/S0005273608001685.
- M. A. Rizzuto, R. Dal Magro, L. Barbieri, L. Pandolfi, A. Sguazzini-Viscontini and M. Truffi, et al., H-Ferritin nanoparticle-mediated delivery of antibodies across a BBB in vitro model for treatment of brain malignancies, Biomater. Sci., 2021, 9(6), 2032–2042 RSC.
- B. Formicola, R. Dal Magro, C. V. Montefusco-Pereira, C. M. Lehr, M. Koch and L. Russo, et al., The synergistic effect of chlorotoxin-mApoE in boosting drug-loaded liposomes across the BBB, J. Nanobiotechnol., 2019, 17(1), 115 CrossRef.
- R. Cecchelli, B. Dehouck, L. Descamps, L. Fenart, V. Buée-Scherrer and C. Duhem, et al., In vitro model for evaluating drug transport across the blood-brain barrier, Adv. Drug Delivery Rev., 1999, 36(2–3), 165–178 CrossRef CAS PubMed.
- M. Pizzocri, F. Re, E. Stanzani, B. Formicola, M. Tamborini and E. Lauranzano, et al., Radiation and adjuvant drug-loaded liposomes target glioblastoma stem cells and trigger in situ immune response, Neuro-oncol. Adv., 2021, 3(1), vdab076 CrossRef PubMed.
- B. B. Manshian, T. F. Martens, K. Kantner, K. Braeckmans, S. C. De Smedt and J. Demeester, et al., The role of intracellular trafficking of CdSe/ZnS QDs on their consequent toxicity profile, J. Nanobiotechnol., 2017, 15(1), 1–14 CrossRef.
- C. G. Hubert, M. Rivera, L. C. Spangler, Q. Wu, S. C. Mack and B. C. Prager, et al., A Three-Dimensional Organoid Culture System Derived from Human Glioblastomas Recapitulates the Hypoxic Gradients and Cancer Stem Cell Heterogeneity of Tumors Found In Vivo, Cancer Res., 2016, 76(8), 2465–2477 CrossRef CAS PubMed.
- Y. Gao, W. R. Vallentgoed and P. J. French, Finding the Right Way to Target EGFR in Glioblastomas; Lessons from Lung Adenocarcinomas, Cancers, 2018, 10(12), 489 CrossRef CAS PubMed.
- D. Cordero Cervantes and C. Zurzolo, Peering into tunneling nanotubes-The path forward, EMBO J., 2021, 40(8), e105789 CrossRef CAS PubMed.
- B. Formicola, A. D'Aloia, R. Dal Magro, S. Stucchi, R. Rigolio and M. Ceriani, et al., Differential Exchange of Multifunctional Liposomes Between Glioblastoma Cells and Healthy Astrocytes via Tunneling Nanotubes, Front. Bioeng. Biotechnol., 2019, 7, 403 CrossRef PubMed.
- D. Hambardzumyan and G. Bergers, Glioblastoma: Defining Tumor Niches, Trends Cancer, 2015, 1(4), 252–265 CrossRef . Available from: https://www.ncbi.nlm.nih.gov/pubmed/27088132.
- M. Saeedimasine, A. Montanino, S. Kleiven and A. Villa, Role of lipid composition on the structural and mechanical features of axonal membranes: a molecular simulation study, Sci. Rep., 2019, 9(1), 8000 CrossRef . Available from: https://www.ncbi.nlm.nih.gov/pubmed/31142762.
- D. Drabik, G. Chodaczek, S. Kraszewski and M. Langner, Mechanical Properties Determination of DMPC, DPPC, DSPC, and HSPC Solid-Ordered Bilayers, Langmuir, 2020, 36(14), 3826–3835 CrossRef CAS . Available from: https://www.ncbi.nlm.nih.gov/pubmed/32176506.
- S. Zha, H. Liu, H. Li, H. Li, K. L. Wong and A. H. All, Functionalized Nanomaterials Capable of Crossing the Blood-Brain Barrier, ACS Nano, 2024, 18(3), 1820–1845 CrossRef CAS PubMed.
- R. Dal Magro, S. Simonelli, A. Cox, B. Formicola, R. Corti and V. Cassina, et al., The Extent of Human Apolipoprotein A-I Lipidation Strongly Affects the β-Amyloid Efflux Across the Blood-Brain Barrier in vitro, Front. Neurosci., 2019, 13, 419 CrossRef.
- L. Fu, B. Shi, S. Wen, M. Morsch, G. Wang and Z. Zhou, et al., Aspect Ratio of PEGylated Upconversion Nanocrystals Affects the Cellular Uptake In Vitro and In Vivo, Acta Biomater., 2022, 147, 403–413 CrossRef CAS.
- P. Sabourian, G. Yazdani, S. S. Ashraf, M. Frounchi, S. Mashayekhan and S. Kiani, et al., Effect of Physico-Chemical Properties of Nanoparticles on Their Intracellular Uptake, Int. J. Mol. Sci., 2020, 21(21), 8019 CrossRef CAS , Available from: https://www.ncbi.nlm.nih.gov/pubmed/33126533.
- H. Deng, P. Dutta and J. Liu, Entry modes of ellipsoidal nanoparticles on a membrane during clathrin-mediated endocytosis, Soft Matter, 2019, 15(25), 5128–5137 RSC . Available from: https://www.ncbi.nlm.nih.gov/pubmed/31190048.
- F. Pisani, V. Castagnola, L. Simone, F. Loiacono, M. Svelto and F. Benfenati, Role of pericytes in blood-brain barrier preservation during ischemia through tunneling nanotubes, Cell Death Dis., 2022, 13(7), 582 CrossRef CAS PubMed . Available from: https://www.ncbi.nlm.nih.gov/pubmed/35790716.
- N. Ma, C. Ma, C. Li, T. Wang, Y. Tang and H. Wang, et al., Influence of nanoparticle shape, size, and surface functionalization on cellular uptake, J. Nanosci. Nanotechnol., 2013, 13(10), 6485–6498 CrossRef CAS PubMed.
- B. D. Chithrani and W. C. W. Chan, Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes, Nano Lett., 2007, 7(6), 1542–1550 CrossRef CAS.
- A. L. Jolly and V. I. Gelfand, Bidirectional intracellular transport: utility and mechanism, Biochem. Soc. Trans., 2011, 39(5), 1126–1130 CrossRef CAS.
- T. Yuan, L. Gao, W. Zhan and D. Dini, Effect of Particle Size and Surface Charge on Nanoparticles Diffusion in the Brain White Matter, Pharm. Res., 2022, 39(4), 767–781 CrossRef CAS PubMed.
- C. G. Uhl, Y. Gao, S. Zhou and Y. Liu, The Shape Effect on Polymer Nanoparticle Transport in a Blood Vessel, RSC Adv., 2018, 8(15), 8089–8100 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr03174a |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2025 |