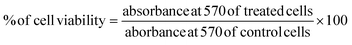Open Access Article
Open Access ArticlePeptide-derived gold nanoparticles as a promising delivery system for Src targeting siRNA in breast cancer cells†
Uday
Suryakanta
a,
Bijayananda
Panigrahi
 ab,
Sumana
Pal
a,
Swatilekha
Das
a,
Soumyadeep
Biswas
a and
Dindyal
Mandal
ab,
Sumana
Pal
a,
Swatilekha
Das
a,
Soumyadeep
Biswas
a and
Dindyal
Mandal
 *a
*a
aSchool of Biotechnology, KIIT deemed to be University, Campus 11, Patia, Bhubaneswar, Odisha 751024, India. E-mail: dmandal@kiitbiotech.ac.in; Fax: +91-674-2725732; Tel: +91-674-2725466
bInternational Centre for Genetic Engineering and Biotechnology (ICGEB), Aruna Asaf Ali Marg, Vasant Kunj, 110067, New Delhi, India
First published on 29th January 2025
Abstract
Src, a non-receptor tyrosine kinase, is involved in various cellular processes including cell division, motility, adhesion, angiogenesis, and survival. RNAi therapy, particularly siRNA, aims to silence genes essential for tumor growth, metastasis, and therapy resistance. In this study, previously developed linear peptides containing tryptophan and histidine/arginine were screened for synthesizing gold nanoparticles. The efficiency of the derived nanoparticles was investigated for nucleic acid delivery. The synthesized AuNPs were characterized using UV-visible spectroscopy, TEM, and DLS. Gel retardation assays demonstrated strong siRNA binding (90%) by gold nanoparticles compared to peptides alone (40%), specifically for peptide W4R4. FACS results revealed a 10-fold enhancement in the cellular uptake of fluorescence-tagged siRNA when delivered via nanoparticles compared to that of naked siRNA. Confocal microscopy confirmed siRNA localization primarily in the cytosol and partially in the nucleus. Western blot analysis indicated 78% downregulation of the Src protein in MCF-7 cells using AuNP/siRNA complexes. These results collectively indicate that the synthesized AuNPs are promising delivery systems for siRNA and might be a potential candidate for RNAi therapeutics.
Introduction
Recent WHO (2022) reports indicate that 2.3 million individuals have been diagnosed with breast cancer globally, resulting in over half a million deaths. This life-threatening disease, primarily affecting women, has limited risk factors beyond sex and age.1 Certain genes, such as Src from the non-receptor tyrosine kinase family, are known to enhance the risk of breast cancer progression. Src, a proto-oncogene in mammalian cells, is known to play a crucial role in the development of cancer by promoting various malignant behaviors like cell proliferation, survival, invasion, angiogenesis, tumorigenesis, and metastasis.2 Targeting Src for therapeutic purposes could offer a promising approach for inhibiting tumor growth and diminishing metastatic potential.3,4 Various cancer treatment approaches exist in this field such as radiotherapy, traditional surgery and chemotherapy; however, they have their own drawbacks such as long-term financial, physical and mental burden. Thus, targeted therapy is favoured over traditional therapy to overcome their drawbacks.5Recently, the RNA interference (RNAi) technique has emerged as a promising targeted therapeutic approach, leveraging therapeutic nucleic acids such as microRNA (miRNA),6,7 small interfering RNA (siRNA),8–11 messenger RNA (mRNA),12 and antisense oligonucleotides (ASOs).9 These molecules specifically target and degrade disease-related genes, thereby inhibiting post-translational protein synthesis and facilitating effective disease management. Among the nucleic acid molecules, siRNA exhibits favourable attributes such as specificity, lack of immunogenicity, biocompatibility, and high efficacy, positioning it as a promising therapeutic agent. Cancer, a disease often triggered by gene over-expression, can be targeted by non-coding siRNA molecules, which are usually 20–25 nucleotide double-stranded RNA sequences designed to complementarily bind to disease-causing genes. These siRNAs consist of two strands – guide and passenger. Following internalization into the cytosol, the passenger strand dissociates while the guide strand associates with the RNA-induced silencing complex (RISC). The RISC complex facilitates the binding of the guide strand to the target mRNA, resulting in mRNA cleavage and degradation, ultimately leading to protein silencing.13 The clinical application of siRNA technology has always been in question due to several challenges, including poor cellular uptake, immunogenicity and enzymatic degradation under in vivo conditions. Additionally, the presence of phosphate groups renders the molecule negatively charged, and its hydrophilic nature hinders its ability to cross the lipid bilayer of the cancer cell membrane to reach the cytosol.14,15
The challenges of siRNA delivery can be addressed by developing suitable carriers that preserve the structural integrity of siRNA, ensure controlled and targeted release, and minimize cytotoxicity. Among the vectors, non-viral vectors are a preferred choice for researchers because they do not exhibit the same drawbacks as viral vectors, such as immunogenicity, low transgene expression, and high cost. Currently several non-viral siRNA vectors have been developed, which include lipid nanoparticles,16 exosomes,17 dendrimers,18 protein nanoparticles,19 linear and cyclic peptides,20,21 polymer-based nanomaterials22,23 and peptide derived-metal nanoparticles.24,25 Among non-viral vectors, polymers like PEI (Polyethylenimine) and lipids are often avoided due to their higher cationic properties, which lead to severe toxicity and reduced cellular internalization capacity.26
Recent studies demonstrated the potential of cell-penetrating peptides (CPPs) for nucleic acid delivery.27 However, the high cationic nature of the peptides often leads to increased toxicity, limiting their clinical utility.28–30 To address these limitations, gold nanoparticles (AuNPs) have emerged as promising carriers in nucleic acid delivery, owing to their large surface area, high drug-loading capacity, and biocompatibility.31–35 By functionalizing AuNPs with polymers and cationic peptides, a positively charged surface is created, enabling the formation of stable complexes with nucleic acids through electrostatic interactions, thereby protecting siRNAs from enzymatic degradation.20,33 AuNPs functionalized with linear or cyclic peptides have been demonstrated as efficient carriers for siRNA delivery. For instance, Panigrahi et al. successfully synthesized gold nanoparticles using peptides containing tryptophan and arginine, achieving effective siRNA delivery into cells.25 Similarly, Shirazi et al. demonstrated an effective cellular internalization of siRNA using cyclic peptide-capped AuNPs.24 Additionally, another study reported that borohydride-reduced Au3+ ions capped with the CPIR28 peptide facilitated efficient gene delivery.36 Moreover, these nanoparticles were synthesized through a green-synthesis process, ensuring both sustainability and scalability. This innovative approach makes pep-AuNPs highly feasible for clinical applications, bridging key gaps in siRNA-based cancer therapies.
Developing efficient and targeted delivery systems for siRNA is essential for advancement in cancer therapeutics, particularly for breast cancer, which affects millions globally and remains a leading cause of death among women. This study addresses this challenge by introducing peptide-based gold nanoparticles (pep-AuNPs) as a promising siRNA delivery platform targeting Src, a proto-oncogene heavily involved in cancer progression. The involvement of peptides as delivery agents marks a significant improvement over conventional carriers like cationic lipids and polymers, which are often associated with cytotoxicity. By minimizing the toxicity, pep-AuNPs enhance the safety and efficacy of the siRNA delivery system. This innovative approach makes pep-AuNPs highly feasible for clinical applications, bridging key gaps in siRNA-based cancer therapies.
Herein, we have synthesized peptide-capped gold nanoparticles using newly developed peptide sequences under sunlight. The nanoparticles were characterized using analytical tools and subsequently explored for their efficacy in siRNA delivery within mammalian cells.
Materials and methods
Materials
Chloroauric acid (HAuCl4) was purchased from MERCK, MA, USA. Milli-Q water was used for all preparations of the nanoparticles. Silencer™ Select Negative Control No. 1 siRNA was purchased from ThermoFisher scientific. FAM-GAPDH siRNA was purchased from Ambion, Waltham, Massachusetts, USA and Hs_SRC_1 siRNA was purchased from Qaigen. Monoclonal GAPDH anti-mouse antibody was purchased from Abgenex, India and Src anti-rabbit antibody (#2108) was bought from Cell Signalling Technology (Boston, MA, USA). Anti-rabbit & anti-mouse IgG AP conjugated secondary antibodies were purchased from Abgenex, India. Acetic acid, methanol, ethanol and other chemicals were purchased from SRL Chemicals, India. Glycine, Tris, agarose, Coomassie Brilliant Blue R250, and SDS were purchased form MP Biomedicals, USA. MTT dye was purchased from HiMedia Lab. Mumbai India. The BCIP®/NBT liquid substrate system was purchased from Sigma Aldrich. The NextGen HM protease inhibitor was purchased from BIOPNEER Pvt. Ltd.Peptide synthesis
All the peptides were resynthesized using protocols reported in our previous literature, molecular weights of the peptides were confirmed by MALDI-TOF mass spectroscopy.27Sequences of the peptides are given below.
Series 1:
1. W1R4-(Ac-WGGGRRRR-Am): HR-MS (ESI-TOF) (m/z): [M]+ calcd for C43H72N22O9, 1040.59; found 521.5 [M + 2H]2+.
2. W2R4-(Ac-WWGGGRRRR-Am): HR-MS (MALDI-TOF) (m/z): [M]+ calcd for C54H82N24O10, 1226.66; found 1227.7628.
3. W3R3-(Ac-WWWGGGRRR-Am): HR-MS (MALDI-TOF) (m/z): [M]+ calcd for C59H80N22O10, 1256.64; found 1257.6398.
4. W4R4-(Ac-WWWWGGRRRR-Am): HR-MS (MALDI-TOF) (m/z): [M]+ calcd for C74H99N27O11, 1541.80; found 1542.7693.
Series 2:
1. W1H4-(Ac-WGGHHHH-Am): HR-MS (MALDI-TOF) (m/z): [M]+ calcd for C43H52N18O9, 964.42; found 966.3987.
2. W2H4-(Ac-WWGGGHHHH-Am): HR-MS (MALDI-TOF) (m/z): [M]+ calcd for C54H63N20O10, 1150.50; found 1151.5424.
3. W3H4-(Ac-WWWGGHHHH-Am): HR-MS (MALDI-TOF) (m/z): [M]+ calcd for C65H72N22O11, 1336.58; found 1337.5797.
4. W4H4-(Ac-WWWWGGHHHH-Am): HR-MS (MALDI-TOF) (m/z): [M]+ calcd for C74H79N23O11, 1465.63; found 1466.6166.
Synthesis of peptide generated gold nanoparticles
For the synthesis of peptide-based gold nanoparticles (AuNPs), fresh stock solutions of 2 mM peptides (W1H4, W2H4, W3H4, W4H4, W1R4, W2R4, W3R3, and W4R4) and 2 mM HAuCl4 were prepared using Milli-Q water. To synthesize the nanoparticles, 200 μL of each peptide (2 mM) solution was mixed with 200 μL of HAuCl4 (2 mM) solution in a microcentrifuge tube, maintaining a final concentration ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The mixture was then exposed to sunlight during noon for 1 hour, without any stirring. Control experiments were conducted under two additional conditions: (1) under dark conditions for 6 hours, and (2) at 80 °C under ambient light, to assess the role of sunlight in the synthesis process. No chemical reducing agents (e.g., NaBH4) or stabilizing agents (e.g., sodium citrate) were used in the synthesis. The resulting nanoparticles were subsequently utilized for in vitro assays.
1. The mixture was then exposed to sunlight during noon for 1 hour, without any stirring. Control experiments were conducted under two additional conditions: (1) under dark conditions for 6 hours, and (2) at 80 °C under ambient light, to assess the role of sunlight in the synthesis process. No chemical reducing agents (e.g., NaBH4) or stabilizing agents (e.g., sodium citrate) were used in the synthesis. The resulting nanoparticles were subsequently utilized for in vitro assays.
Characterization studies of nanoparticles
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (siRNA
100 (siRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AuNP or peptide) molar ratios in serum-free DMEM cell culture media. The ratio mentioned here indicates the molar ratio. That means 1 molar concentration of siRNA binds to 100 molar concentrations of AuNPs. [For example, when 100 nM siRNA is used, 10 μM peptides or pep-AuNPs was added to achieve a 1
AuNP or peptide) molar ratios in serum-free DMEM cell culture media. The ratio mentioned here indicates the molar ratio. That means 1 molar concentration of siRNA binds to 100 molar concentrations of AuNPs. [For example, when 100 nM siRNA is used, 10 μM peptides or pep-AuNPs was added to achieve a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (siRNA
100 (siRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AuNP) molar ratio. Similarly, for a 1
AuNP) molar ratio. Similarly, for a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 (siRNA
80 (siRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AuNP) ratio, 8 μM peptides or pep-AuNPs was used with 100 nM siRNA, and so on.] The mixtures were incubated at room temperature for 30 minutes. Subsequently, the complexes were mixed with a gel loading dye and loaded onto a 2% agarose gel containing ethidium bromide (1 μg mL−1). The gel was electrophoresed at 80 V for 30 minutes and then visualized using a ImageQuant LAS 500 GE Health care, USA. The band intensity of siRNA was quantified using ImageJ software. To optimize the AuNP–siRNA molar ratio, different molar ratios of AuNPs and siRNA ranging from 1
AuNP) ratio, 8 μM peptides or pep-AuNPs was used with 100 nM siRNA, and so on.] The mixtures were incubated at room temperature for 30 minutes. Subsequently, the complexes were mixed with a gel loading dye and loaded onto a 2% agarose gel containing ethidium bromide (1 μg mL−1). The gel was electrophoresed at 80 V for 30 minutes and then visualized using a ImageQuant LAS 500 GE Health care, USA. The band intensity of siRNA was quantified using ImageJ software. To optimize the AuNP–siRNA molar ratio, different molar ratios of AuNPs and siRNA ranging from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; 1
20; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40; 1
40; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 and 1
80 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (siRNA
100 (siRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AuNP) were screened using gel retardation assay, where 100 nM of siRNA was taken constantly in all ratios and the delivery agent concentration was adjusted according to different ratios.
AuNP) were screened using gel retardation assay, where 100 nM of siRNA was taken constantly in all ratios and the delivery agent concentration was adjusted according to different ratios.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 (siRNA
80 (siRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AuNPs) in DMEM media containing either 10% or 25% FBS, in two separate experimental setups. The mixtures were incubated at room temperature for 30 minutes. Additionally, serum stability studies were conducted at different time intervals of 4 hours and 24 hours under the 25% FBS condition to evaluate stability under harsher conditions over an extended duration. The complex samples were then run in 2% agarose gel, and imaged under ImageQuant LAS 500 from GE Health care. The bands were quantitatively analysed using Image J software.
AuNPs) in DMEM media containing either 10% or 25% FBS, in two separate experimental setups. The mixtures were incubated at room temperature for 30 minutes. Additionally, serum stability studies were conducted at different time intervals of 4 hours and 24 hours under the 25% FBS condition to evaluate stability under harsher conditions over an extended duration. The complex samples were then run in 2% agarose gel, and imaged under ImageQuant LAS 500 from GE Health care. The bands were quantitatively analysed using Image J software.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well were seeded in 6-well plates 24 hours prior to the addition of the AuNP/siRNA complexes. These complexes were prepared in serum containing DMEM media at molar ratios of 1
000 cells per well were seeded in 6-well plates 24 hours prior to the addition of the AuNP/siRNA complexes. These complexes were prepared in serum containing DMEM media at molar ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 (siRNA/AuNPs). Lipofectamine 2000 was used as a positive control for the cellular uptake study. The final concentration of fluorescence labeled siRNA (FAM-siRNA) used for this experiment was 200 nM. The cells were then treated with the siRNA/AnNPs complexes and incubated under standard growth conditions at 37 °C for 24 hours. After the incubation period, the media was removed and the cells were washed twice with 1× PBS. To detach the cells from the plate trypsinization was conducted. Furthermore, the trypsinized cells were washed twice with 1XPBS and the cells were collected by centrifugation at 700 rpm for 5 min. Then the cells were suspended in 1× PBS and analysed using FITC channel to measure cell-associated fluorescence. Cellular internalization was quantified by gating the response against a non-treated cell population. The percentage of cells exhibiting FITC fluorescence was determined by normalizing with the total cell count.
80 (siRNA/AuNPs). Lipofectamine 2000 was used as a positive control for the cellular uptake study. The final concentration of fluorescence labeled siRNA (FAM-siRNA) used for this experiment was 200 nM. The cells were then treated with the siRNA/AnNPs complexes and incubated under standard growth conditions at 37 °C for 24 hours. After the incubation period, the media was removed and the cells were washed twice with 1× PBS. To detach the cells from the plate trypsinization was conducted. Furthermore, the trypsinized cells were washed twice with 1XPBS and the cells were collected by centrifugation at 700 rpm for 5 min. Then the cells were suspended in 1× PBS and analysed using FITC channel to measure cell-associated fluorescence. Cellular internalization was quantified by gating the response against a non-treated cell population. The percentage of cells exhibiting FITC fluorescence was determined by normalizing with the total cell count.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 molar ratios with AuNPs. The complexes were first made in 100 μl solution followed by incubation for 30 min and then volume was adjusted up to 800 μl. The studies were conducted in 3 runs and each run was consisting of 11 scans. The values were then obtained by calculating average of triplicate readings. For zeta potential measurement 500 μl of the sample was loaded on to capillaries of the cuvette. Each sample was was subjected to 3 runs consisting of 15 scans. The values were obtained by calculating the average of triplicate readings.
80 molar ratios with AuNPs. The complexes were first made in 100 μl solution followed by incubation for 30 min and then volume was adjusted up to 800 μl. The studies were conducted in 3 runs and each run was consisting of 11 scans. The values were then obtained by calculating average of triplicate readings. For zeta potential measurement 500 μl of the sample was loaded on to capillaries of the cuvette. Each sample was was subjected to 3 runs consisting of 15 scans. The values were obtained by calculating the average of triplicate readings.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells in 200 μL of complete media were seeded into 96-well plates and allowed to attach for 24 hours at 37 °C under a humidified atmosphere with 5% CO2. The cells were then treated with different AuNPs at various concentrations (10 μM and 20 μM) for 24 hours under the same conditions. After the incubation period, 100 μL of 0.1 mg mL−1 MTT solution was added to each well and incubated for an additional 4 hours. The MTT solution was then carefully removed, and 100 μL of DMSO was added to each well to dissolve the formazan crystals formed. The resulting purple-blue color was quantified using an Epoch Biotek microplate ELISA reader at 570 nm. Untreated cells were served as the negative control for the experiment, and all the experiments were performed in triplicate. The cell viability was calculated using the following formula in MS Excel.
000 cells in 200 μL of complete media were seeded into 96-well plates and allowed to attach for 24 hours at 37 °C under a humidified atmosphere with 5% CO2. The cells were then treated with different AuNPs at various concentrations (10 μM and 20 μM) for 24 hours under the same conditions. After the incubation period, 100 μL of 0.1 mg mL−1 MTT solution was added to each well and incubated for an additional 4 hours. The MTT solution was then carefully removed, and 100 μL of DMSO was added to each well to dissolve the formazan crystals formed. The resulting purple-blue color was quantified using an Epoch Biotek microplate ELISA reader at 570 nm. Untreated cells were served as the negative control for the experiment, and all the experiments were performed in triplicate. The cell viability was calculated using the following formula in MS Excel.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 (siRNA
80 (siRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AuNPs) molar ratios in complete media for 4 hours. The complexes were then replaced with growth media and incubated at 37 °C and 5% CO2 48 hours. After the incubation period, media was removed from cells and the cells were washed three times with ice-cold PBS. Then RIPA lysis buffer containing protease inhibitor was used to generate the whole-cell lysate. The cell lysates were then centrifuged at 12
AuNPs) molar ratios in complete media for 4 hours. The complexes were then replaced with growth media and incubated at 37 °C and 5% CO2 48 hours. After the incubation period, media was removed from cells and the cells were washed three times with ice-cold PBS. Then RIPA lysis buffer containing protease inhibitor was used to generate the whole-cell lysate. The cell lysates were then centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 minutes at 4 °C. The supernatant was transferred to precooled microtubes, and the total protein concentration was determined using the Bradford reagent.
000 rpm for 15 minutes at 4 °C. The supernatant was transferred to precooled microtubes, and the total protein concentration was determined using the Bradford reagent.
Proteins (20 μg per well) were loaded onto 10% SDS PAGE gel and electrophoresed in TRIS buffer for 1.5 h at 120 V. The protein bands determined by gel electrophoresis were subsequently transferred onto a polyvinylidene fluoride (PVDF) membrane via western blotting. Briefly, membranes were blocked with 5% skim milk (SRL) in TBS-T (1× Tris-buffered saline, 0.1% Tween 20) for 1 hour and then incubated overnight at 4 °C with Src antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in TBS-T) and GAPDH primary antibody (1
1000 in TBS-T) and GAPDH primary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 in TBS-T) in 2 different setups. The membrane was washed three times with TBS-T (5 minutes each) and the membrane was probed with secondary antibodies conjugated to alkaline phosphatase (anti-mouse for GAPDH and anti-rabbit for Src) at a 1
5000 in TBS-T) in 2 different setups. The membrane was washed three times with TBS-T (5 minutes each) and the membrane was probed with secondary antibodies conjugated to alkaline phosphatase (anti-mouse for GAPDH and anti-rabbit for Src) at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 dilution in TBS-T for 2 hours, followed by additional washings. The blots were then developed using AP buffer along with 16 μl of BCIP and 32 μl of NBT. The bands were scanned and visualised using with a GE Health Care ImageQuant LAS 500 scanner and further quantified using Image J software by normalizing against the GAPDH endogenous expression.
5000 dilution in TBS-T for 2 hours, followed by additional washings. The blots were then developed using AP buffer along with 16 μl of BCIP and 32 μl of NBT. The bands were scanned and visualised using with a GE Health Care ImageQuant LAS 500 scanner and further quantified using Image J software by normalizing against the GAPDH endogenous expression.
Statistical analysis
Data from the experiments have been presented as mean ± SD from three independent trials, unless stated otherwise. Western blotting data were analyzed using one-way ANOVA with Bartlett's test for multiple comparisons. Pairwise comparisons between two groups were made using the Student's t-test. A p-value of <0.05 was considered statistically significant. Statistical significance displayed was *p < 0.05, **p < 0.01, ***p < 0.001.Results and discussion
Conceptualization of peptides and synthesis of peptide-derived gold nanoparticles
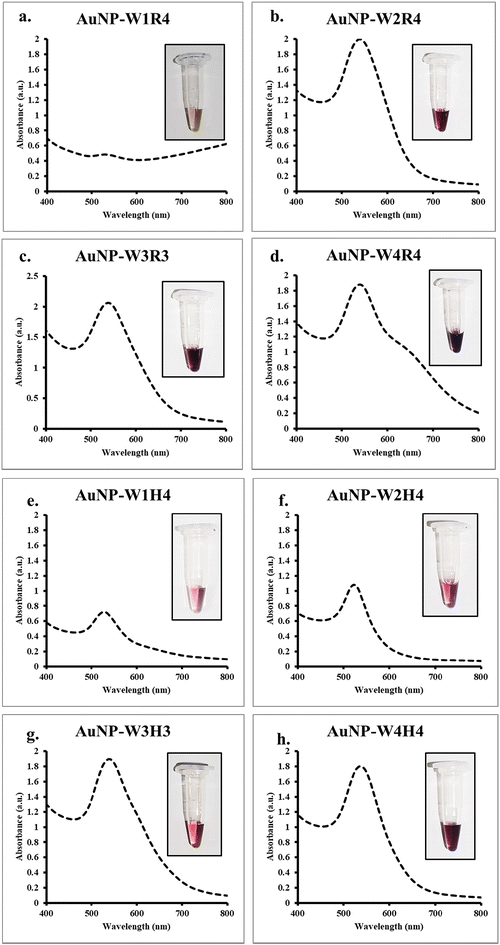
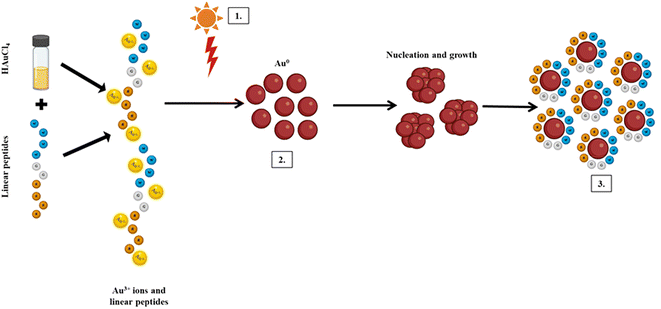
A set of linear peptide sequences have been previously reported containing hydrophobic amino acids (tryptophan) and hydrophilic amino acids (arginine or histidine) that are able to deliver nucleic acid inside the cells efficiently and silence the target proteins.27 Herein, our objective was to investigate the capability of the peptides toward nanoparticle synthesis and compare the protein silencing efficiency of the nanoparticles with a native peptide sequence. Previous studies by Lévy et al. have demonstrated that cationic peptides play a crucial role in nanoparticle stabilization.37 Recently, we also demonstrated that short hexapeptides containing tryptophan and arginine residues could be used to synthesize gold nanoparticles.25 To accomplish the objective, previously developed peptides (series 1 and series 2) (Fig. S1†) were exposed to gold salts in the presence of sunlight. The gradual development of a reddish-pink color indicated the formation of gold nanoparticles.The efficiency of nanoparticle synthesis varied among the peptides. Peptides with fewer tryptophan residues were found to be less effective in reducing Au3+ ions to Au0, thus failing to form nanoparticles. Conversely, peptides with more tryptophan residues successfully reduced Au3+ and formed stable nanoparticles. UV-Vis spectroscopy revealed a characteristic peak at 525 nm, indicating the formation of gold nanoparticles. Peptides such as W2R4, W3R3, W4R4, W2H4, W3H4, and W4H4 exhibited a narrow surface plasmon resonance (SPR) band, indicating the formation of monodispersed nanoparticles (Fig. 1). To understand the role of sunlight, the same reaction was conducted in the dark. However, no color change of the solution occurred even after 6 hours, further emphasizing the necessity of sunlight for nanoparticle formation. Furthermore, the stability of the nanoparticles (AuNP-W2R4; AuNP-W3R3 and AuNP-W4R4) was assessed for 20 days by analyzing the UV-vis spectra which displayed negligible changes in the intensity as well as the position of the peaks, ascertaining the stability of the synthesized nanoparticles (Fig. S3†).
Previous studies have demonstrated that light or heat is a crucial factor in peptide-based metal nanoparticle synthesis.25,38,39 In the current study, a similar effect was investigated, which revealed negligible formation of nanoparticles when the synthesis was performed under ambient light and temperature conditions (Fig. S2†). Among the amino acids, tryptophan, histidine, and arginine residues are known to interact with Au3+ ions, through the indole group, imidazole group and guanidine group, respectively.40–42 Sunlight irradiation increases the reaction temperature, enhancing the polarizability of amino acids and their oxidation potential. The reduction of ions is facilitated by the photoexcitation of tryptophan under sunlight, while other peptides stabilize the growth of Au0 to prevent aggregation. This process highlights the simplicity and efficiency of using sunlight and peptides for the green synthesis of stable gold nanoparticles (Scheme 1). For the purpose of molecular transport, the positively charged surface of the nanoparticles facilitates ionic interactions with the negatively charged cell membrane, while the hydrophobic residues enhance interactions with the phospholipid bilayer, promoting effective cellular internalization of cargo.
Binding affinity and serum stability of the siRNA/AuNP complex
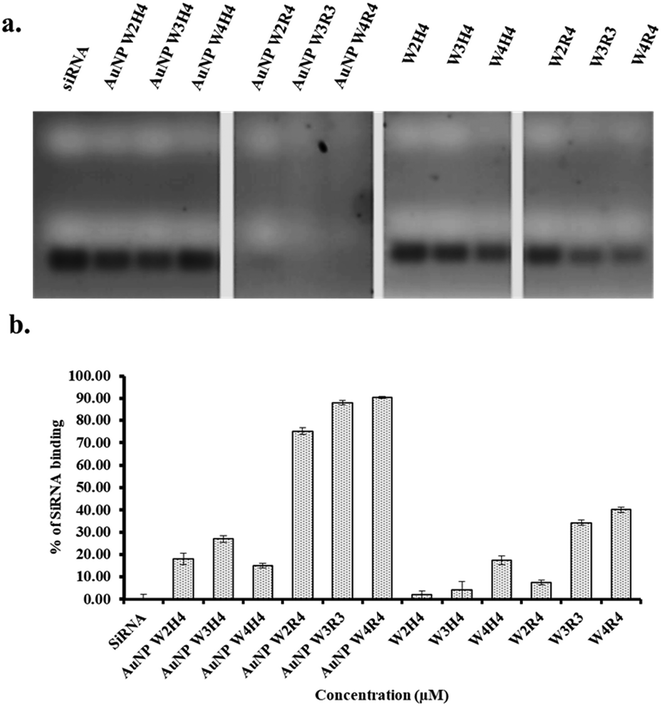
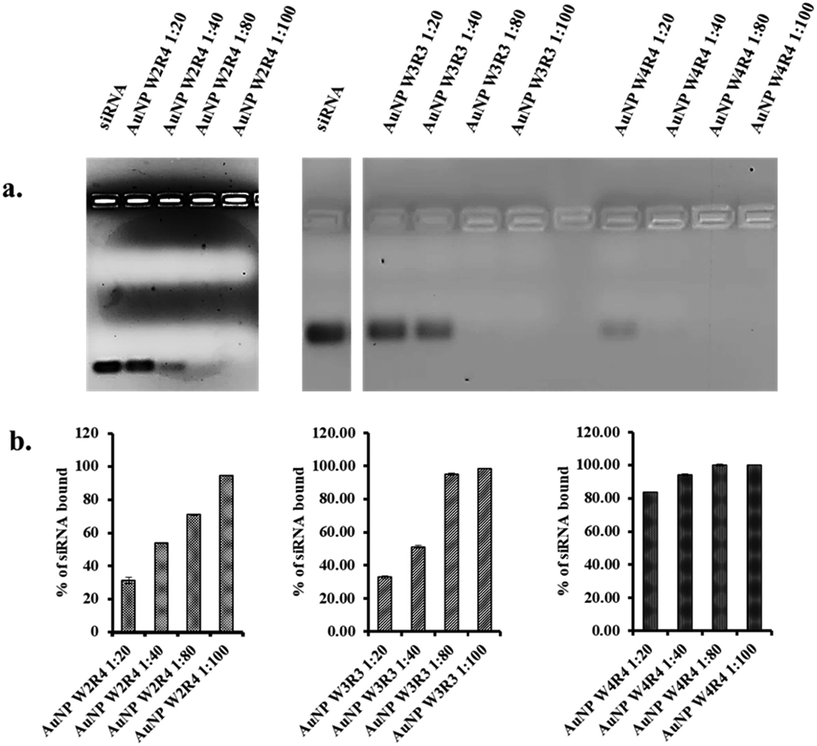
The binding study between siRNA and Pep-AuNPs are depicted in Fig. 2, where the free siRNA band indicates the unbound or released siRNA from the complex. Among the nanoparticles, AuNPs synthesized using peptides containing tryptophan and arginine exhibited strong interactions with siRNA. Furthermore, among all AuNPs, AuNP-W4R4 and AuNP-W3R3 showed nearly 90% siRNA binding and AuNP-W2R4 demonstrated 75% binding as very faint bands of siRNA were observed compared to the control, indicating efficient complex formation in the presence of higher number of arginine residues. In contrast, nanoparticles synthesized with peptides containing histidine did not exhibit significant affinity toward siRNA, as strong bands of siRNA were found in the gel, which were comparable to the control.To find the answer whether peptides alone exhibit similar binding affinity toward siRNA, only peptide samples were used to synthesize complexes with siRNA and analyzed in agarose gel. WH peptides did not show promising results, where the siRNA band intensity was found to be similar to the control. However, WR peptides showed minimal siRNA binding (W2R4: 7%, W3R3: 34%, and W4R4: 40%) which was significantly lower than that of AuNPs, ascertaining the role of peptide derived AuNPs for increased siRNA binding.
In a nut shell, AuNPs outperformed native peptides in complex formation due to their size, shape, and cationic surface. Arginine-based gold nanoparticles demonstrated superior siRNA binding efficiency compared to other materials. Based on this result, AuNP-W2R4, AuNP-W3R3, and AuNP-W4R4 were selected for further studies of cellular uptake and protein silencing and AuNP-W1R4 or AuNP-W1H4 were not used further for other experiments as these peptides failed to synthesize gold nanoparticles successfully.
To optimize the binding ratio, the selected nanoparticles were further analyzed at various molar ratios to determine the most effective ratio for efficient siRNA binding. Silencer™ Select Negative Control No. 1 siRNA was mixed with AuNPs (AuNP-W2R4, AuNP-W3R3, and AuNP-W4R4) at different molar ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, 1
20, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40, 1
40, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80, and 1
80, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) and incubated for 30 minutes to allow the complex formation before running them on an agarose gel. The efficiency of the complexes was determined by measuring the bands of free siRNA for each molar ratio and comparing them with the control siRNA band. Among them, AuNP-W4R4 was found to be highly efficient for complex formation, with 80% of siRNA bound even at the lower molar ratio of 1
100) and incubated for 30 minutes to allow the complex formation before running them on an agarose gel. The efficiency of the complexes was determined by measuring the bands of free siRNA for each molar ratio and comparing them with the control siRNA band. Among them, AuNP-W4R4 was found to be highly efficient for complex formation, with 80% of siRNA bound even at the lower molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20. Similarly, AuNP-W3R3 showed effective binding, achieving 100% siRNA binding at a 1
20. Similarly, AuNP-W3R3 showed effective binding, achieving 100% siRNA binding at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 molar ratio. In contrast, AuNP-W2R4 exhibited less capability in siRNA binding (70%), even at higher molar ratios of 1
80 molar ratio. In contrast, AuNP-W2R4 exhibited less capability in siRNA binding (70%), even at higher molar ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 (Fig. 3). The results of this study indicate that materials with a higher number of hydrophobic amino acids such as tryptophan residues and hydrophilic amino acids such as arginine residues together enhance the packaging capacity and complex formation. For subsequent assays, an optimal molar ratio of 1
80 (Fig. 3). The results of this study indicate that materials with a higher number of hydrophobic amino acids such as tryptophan residues and hydrophilic amino acids such as arginine residues together enhance the packaging capacity and complex formation. For subsequent assays, an optimal molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 was used.
80 was used.
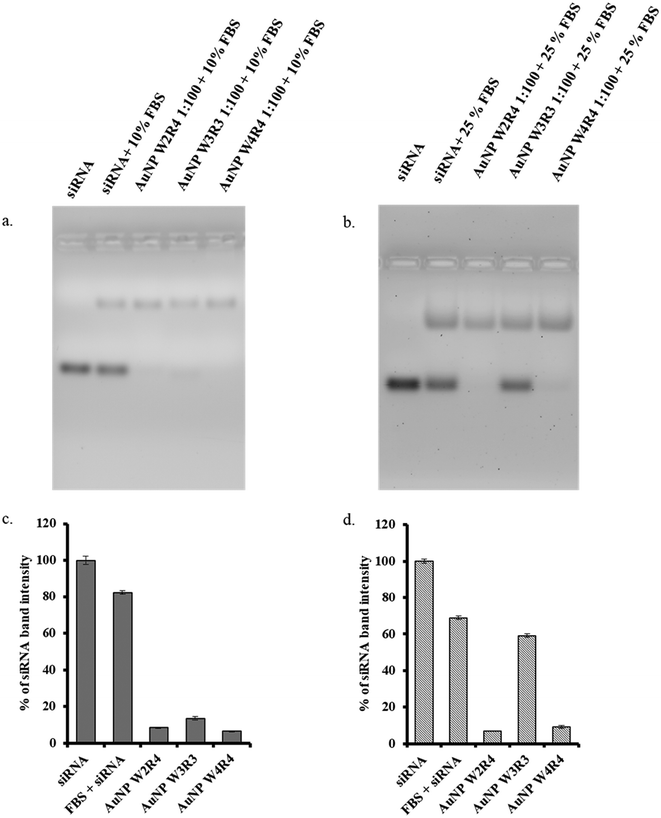
One of the most significant challenges in siRNA delivery is the susceptibility of nucleic acids to enzymatic degradation under in vivo conditions, which hampers the effective internalization of the complex into cells. Previous work by Mandal et al. demonstrated that peptide sequences with a high content of hydrophobic and cationic amino acids in a linear configuration could successfully facilitate nucleic acid delivery in vitro, even in the presence of serum.20 To make it consistent, in this study all in vitro assays have been conducted in the presence of 10% FBS.
Furthermore, to check the efficiency of the nanoparticles toward the protection of siRNA in the presence of an enzymatic environment, we assessed the serum stability of the siRNA/AuNP complex in a time dependent manner. The experiment was conducted in two sets: one set contains 10% FBS and another set contains 25% FBS, to evaluate the impact of varying serum concentrations. siRNA and AuNPs were mixed at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 molar ratio and incubated under these different FBS conditions for 30 min at room temperature; other time dependent serum stability assays were performed for 4 hours and 24 hours at a serum concentration of 25% FBS. A faint band of unbound siRNA was found in the presence of 10% FBS in the case of AuNP-W3R3, whereas others did not show any significant band, showing good stability of the complex in serum. Upon the increase in the concentration of FBS to 25%, the siRNA band was clearly visible in the case of AuNP-W3R3, indicating the instability of the complex under higher serum conditions as the total siRNA was released from the complex (Fig. 4). The results for other time dependent experiments at 4 hours and 24 hours also displayed similar results where AuNP-W3R3 was not stable and an siRNA band for free siRNA was present whereas other two materials AuNP-W2R4 and AuNP-W4R4 displayed promising results of complex formation even at 4 hours and 24 hours (Fig. S6†).
80 molar ratio and incubated under these different FBS conditions for 30 min at room temperature; other time dependent serum stability assays were performed for 4 hours and 24 hours at a serum concentration of 25% FBS. A faint band of unbound siRNA was found in the presence of 10% FBS in the case of AuNP-W3R3, whereas others did not show any significant band, showing good stability of the complex in serum. Upon the increase in the concentration of FBS to 25%, the siRNA band was clearly visible in the case of AuNP-W3R3, indicating the instability of the complex under higher serum conditions as the total siRNA was released from the complex (Fig. 4). The results for other time dependent experiments at 4 hours and 24 hours also displayed similar results where AuNP-W3R3 was not stable and an siRNA band for free siRNA was present whereas other two materials AuNP-W2R4 and AuNP-W4R4 displayed promising results of complex formation even at 4 hours and 24 hours (Fig. S6†).
Conversely, the complexes formed by the other two materials remained intact, indicating that peptide sequences with a higher number of cationic residues (Arg) were able to protect the siRNA complex under harsh serum conditions, independent of the number of hydrophobic residues in the peptide sequence, as the in vitro assays require only up to 4 hours for the uptake of siRNA and under 10% FBS in DMEM conditions, whereas the complexes were found to be intact for 24 hours even under harsh serum conditions such as 25% in DMEM during binding study. Naked siRNA in the presence of FBS served as a negative control to observe the stability of free siRNA in serum.
Based on the binding studies, nanoparticles generated from W2R4, W3R3 and W4R4 only were characterized further. The synthesized nanoparticles were characterized by transmission electron microscopy (TEM) and their size distributions were analyzed. AuNP-W2R4 exhibited the largest average size of 35 nm and high polydispersity compared to the other materials. AuNP-W3R3 had the smallest average size of 14 nm, while AuNP-W4R4 displayed an average size of 19 nm, with the least polydispersity (Fig. 5). These findings suggest that polydispersity is influenced by the number of hydrophobic residues present in the peptide sequences (W2R4 > W3R3 > W4R4). Fig. S4† illustrates that the materials comprised a mix of spherical and triangular nanoparticles.
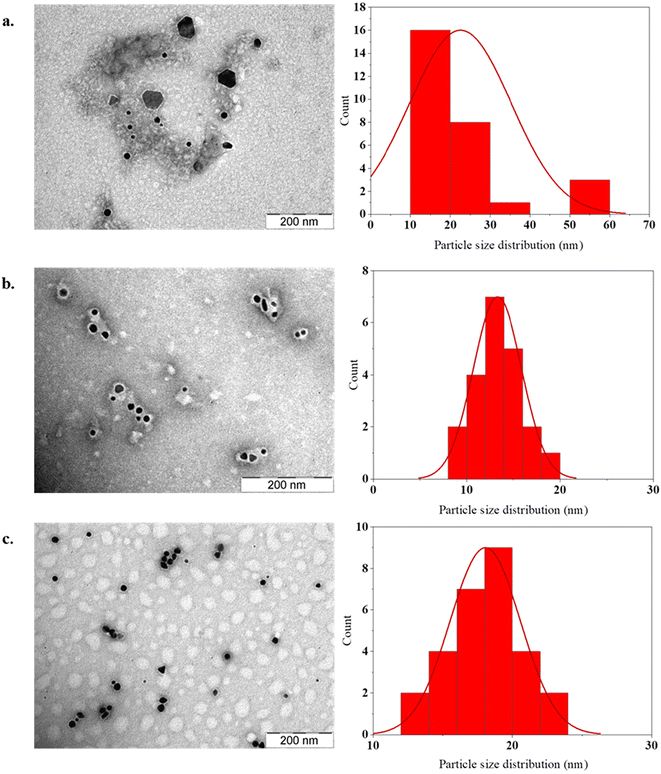 | ||
| Fig. 5 TEM images and size distributions of the selected AuNPs for uptake and silencing study: (a) AuNP-W2R4, (b) AuNP-W3R3 and (c) AuNP-W4R4 (scale = 200 nm). | ||
DLS analysis of the AuNP/siRNA complex
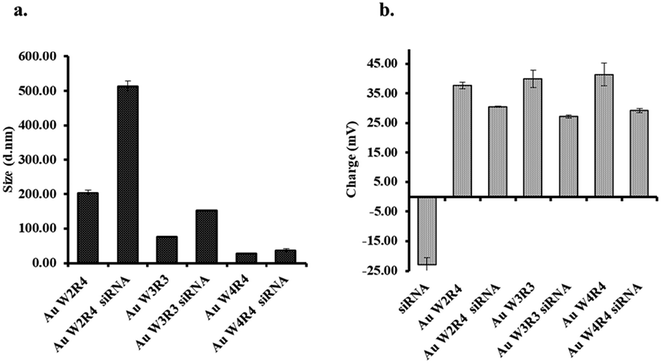
To evaluate the effective complex formation between AuNP and siRNA, the hydrodynamic size and zeta potential of control AuNPs and their corresponding complexes were determined. The hydrodynamic sizes of the AuNPs were found to be 203 nm, 76.11 nm, and 27.95 nm for AuNP-W2R4, AuNP-W3R3, and AuNP-W4R4 (Fig. 6). The relatively larger size of AuNP-W2R4 particles can be attributed to the lower number of hydrophobic amino acid residues and a higher proportion of charged residues, thus, behaving like amphiphilic molecules which may form micelle like structures. In contrast, AuNP-W3R3 and AuNP-W4R4, with a balanced composition of hydrophobic and charged residues exhibited stable and smaller particles. Similarly, in case of the complexes, AuNP-W2R4 formed a large complex with siRNA exhibiting a hydrodynamic size of 521 nm, which would likely impede the ability of the complex to cross the membrane barrier. Conversely, the sizes of complexes formed with AuNP-W3R3 (151 nm) and AuNP-W4R4 (36 nm) were found to be significantly smaller (Fig. S5†) which are favorable for cellular internalization. Another parameter for membrane permeability is the surface charge of the complex. To determine the surface charge of the nanoparticles as well as the complex, zeta potential analysis was performed. The zeta potentials were found to be −23.30 mV, +37.75 mV, +40 mV, and +37.75 mV for naked siRNA, AuNP-W2R4, AuNP-W3R3, and AuNP-W4R4, respectively. In comparison, the zeta potentials of the corresponding complexes with siRNA showed a significant decline. The observed decrease in the zeta potential for all complexes, compared to control nanoparticles suggests that the binding of negatively charged siRNA occurs with the positive charge of the AuNPs and hence, neutralizes the charge.Cytotoxicity assay of the AuNPs
In order to develop one drug delivery system one of the primary criteria is find a nontoxic carrier system. To evaluate the cytotoxicity of the synthesized AuNPs, an MTT assay was conducted on the MCF 7 cell line. The nanoparticles were tested at concentrations of 10 μM and 20 μM over a 48-hour incubation period. The results indicate that nanoparticle-treated cells exhibit more than 90% viability at the tested concentration which is higher than the used concentration in this work, as depicted in Fig. 7. These results suggest that newly generated nanoparticles can be used as an siRNA delivery system.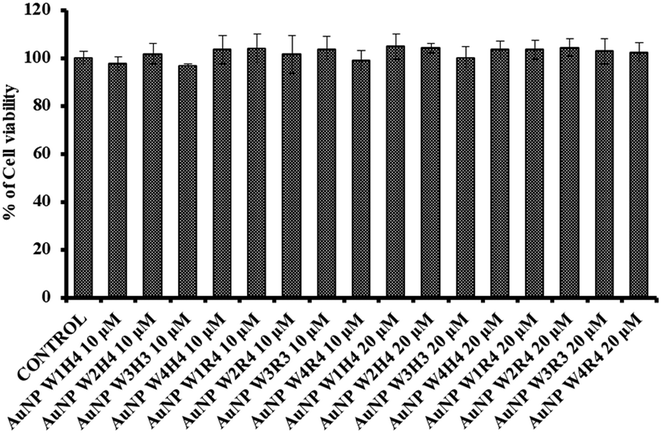 | ||
| Fig. 7 Cytotoxicity of the synthesised peptide derived AuNPs at 10 μM and 20 μM (incubation time = 48 hours) in the MCF 7 cell line. | ||
Cellular internalization: flow cytometry analysis
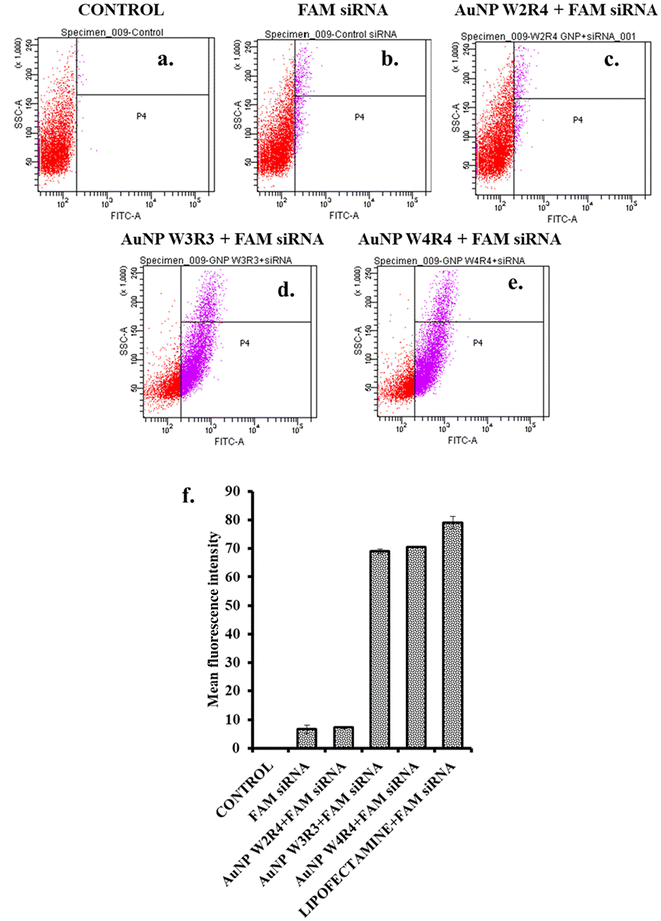
Although some of the nanoparticles exhibit strong binding affinity for siRNA, it is imperative to investigate the cellular internalization of the nanoparticle/siRNA complex. To obtain the quantitative idea about the cellular internalization efficiency of the nanoparticles, FAM-siRNA was used as a model drug. Upon the delivery of FAM-siRNA, fluorescence labeled cells were quantified for different delivery systems. The results indicated that AuNPs capped with peptides W3R3 and W4R4 effectively facilitated siRNA inside the cells, achieving fluorescence positive cells of 69% and 70.6%, respectively, which indicate a tenfold increase in the cellular uptake of siRNA compared with FAM-siRNA alone, highlighting their efficacy as delivery agents (Fig. 8). In contrast, AuNP-W2R4 showed only 7.2% positive cells, this indicates that it was the least efficient delivery agent among those tested. For cellular internalization study, Lipofectamine 2000 served as a positive control (Fig. S7†).This outcome underscores the importance of the balanced arrangement of tryptophan and arginine residues in the peptide sequences (W3R3 and W4R4), which effectively facilitates the cellular uptake.
Cellular uptake study: fluorescence microscopy
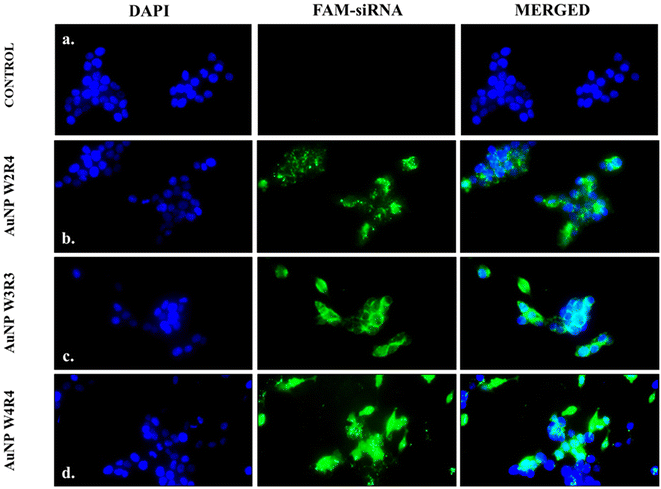
To determine the intracellular localization of nucleic acids, fluorescence microscopy was employed using FAM siRNA as a model cargo. MCF 7 cells were treated with AuNPs/FAM-siRNA complexes, and the cell nuclei were stained with DAPI. As a control, the cells were treated with naked FAM siRNA without any delivery agent. As illustrated in Fig. 9, the control cells did not exhibit any fluorescence. In contrast, cells treated with AuNP-W2R4, AuNP-W3R3, and AuNP-W4R4-FAM siRNA emit green fluorescence when exposed to the light of FITC wavelength, confirming the successful internalization of the complexes inside the cells.In order to gain a detailed insight into the specific localization of FAM-siRNA inside the cells, confocal microscopy studies were performed. The results revealed that in case of AuNP-W2R4, the complexes were found to be localized within the cytosol (Fig. S8†). AuNP-W3R3 and AuNP-W4R4 exhibited significant localization of FAM-siRNA in both the nucleus and cytoplasm confirming effective nuclear uptake in addition to cytoplasmic presence. These observations support the potential of AuNP-W3R3 and AuNP-W4R4 as effective nucleic acid delivery vectors. Previous studies have demonstrated that cyclic peptides can efficiently deliver antisense oligonucleotides and siRNA into both the cytoplasm and nucleus, where the complex was presumed to be dissociated in the cytosol and subsequently entered the nucleus.9,43,44
Protein silencing
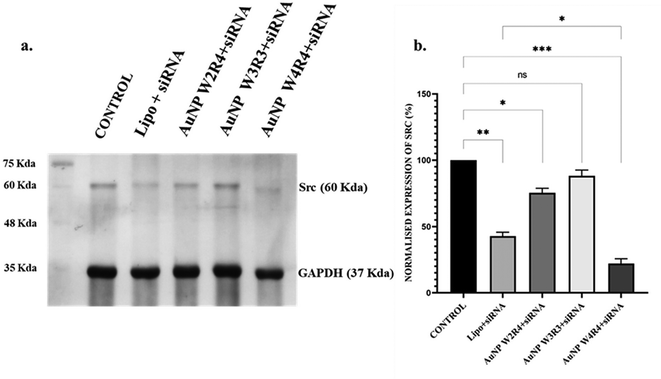
Src is a proto-oncogene frequently overexpressed in breast cancer, and its activation is associated with poor prognosis. The activation of Src contributes to several malignant phenotypes, including enhanced proliferation and survival of cancer cells, increased motility and invasiveness, and the promotion of metastasis to distant organs. Additionally, Src stimulates the production of angiogenic factors such as VEGF, which promotes the formation of new blood vessels, further supporting tumor growth and metastasis.45 Recent studies have focused on targeting Src kinase through gene therapies and targeted delivery systems for cancer theranostics. For instance, monodispersed gold nanoparticles conjugated with polyethylenimine have demonstrated the ability to knock down eEF-2K and deregulate genes in vitro, thereby inhibiting the Src and MAPK-ERK signalling pathways in tumor models.46 This prompted us to explore the potential of the synthesized AuNPs as siRNA delivery agents for silencing the Src protein, given their promising cellular uptake results. The efficiency of protein silencing was assessed using western blot analysis after transfecting MCF 7 cells with Src siRNA in the presence of AuNPs. The results (Fig. 10) revealed that treatment with Lipofectamine led to 60% reduction in Src protein levels, while AuNP W2R4, AuNP W3R3, and AuNP W4R4 vectors resulted in 25%, 12%, and 78% downregulation of the Src protein, respectively, compared to untreated cells. Notably, AuNP-W4R4 exhibited a highly significant downregulation of Src protein levels, outperforming the commercial standard Lipofectamine 2000.In our study we have utilized the linear peptide W4R4 for the synthesis of AuNPs as well as to facilitate Src protein silencing. To ascertain that the observed Src silencing was indeed attributable to the complex AuNP-W4R4/Src-siRNA rather than the linear peptide (W4R4) and AuNP-W4R4, control experiments were conducted, where cells were treated with similar concentrations of both W4R4 and AuNP-W4R4, respectively, and their Src protein expression levels were assessed. These control experiments showed no significant downregulation of the Src protein at this concentration (Fig. S9†); this confirms that the effective silencing of Src protein expression is due to the synergistic action of the AuNP-W4R4/siRNA complex only.
Conclusion
Stable gold nanoparticle solutions were successfully synthesized using linear peptides. It was anticipated that nanoparticles containing tryptophan and arginine residues on the surface would make a strong complex with siRNA and would be able to cross the cell membrane easily and consequently deliver siRNA inside the cells efficiently. Our experimental results were found to be in line with our anticipation. The newly developed nanoparticles containing arginine and tryptophan residues were found to be efficient in delivering siRNA inside the cells. Around 70% fluorescence-labeled cells were detected from FACS analysis upon the delivery of the AuNP/siRNA complex, which was comparable to Lipofectamine. Significant downregulation of the Src protein (around 80%) was determined while delivering Src siRNA using W4R4-AuNPs, which outperformed the efficiency of Lipofectamine. These results suggest that peptide-based gold nanoparticles offer a promising, less-toxic alternative for siRNA delivery, with enhanced protection from nuclease degradation and efficient protein silencing.This study establishes peptide-capped gold nanoparticles as a robust platform for siRNA delivery, paving the way for their application in advanced cancer therapeutics and beyond. A key future direction is to expand this delivery agent to deliver multiple therapeutic agents, such as a combination of siRNA and chemotherapeutic drugs, to achieve synergistic effects in tackling multi-drug-resistant cancers. The environment-friendly, sunlight-driven synthesis method aligns with the growing demand for sustainable nanotechnology in biomedical applications. Additionally, the biocompatibility and minimal toxicity of the nanoparticles make them strong candidates for preclinical and clinical studies. Future research could focus on evaluating their efficacy in vivo, particularly in animal models of cancer, to confirm their therapeutic potential under physiological conditions.
Data availability
Original data will be available upon request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
US acknowledges the financial support from the ICMR Senior Research Fellowship (SRF) award (2021-12330/GTGE-BMS). BP acknowledges the financial support from the National Post Doctoral Fellowship (N-PDF) award (PDF/2022/000419). We sincerely thank Dr. Suresh K. Verma for his support to obtain confocal microscopy images. We also acknowledge the facility provided through the DBT-BUILDER program (BT/INF/22/SP42155/2021) at KIIT University.References
- R. Hong and B. Xu, Breast cancer: an up–to–date review and future perspectives, Cancer Commun., 2022, 42(10), 913–936 CrossRef PubMed.
- S. J. Parsons and J. T. Parsons, Src family kinases, key regulators of signal transduction, Oncogene, 2004, 23(48), 7906–7909 CrossRef CAS PubMed.
- S. Martellucci, L. Clementi, S. Sabetta, V. Mattei, L. Botta and A. Angelucci, Src family kinases as therapeutic targets in advanced solid tumors: what we have learned so far, Cancers, 2020, 12(6), 1448 CrossRef CAS PubMed.
- J. Luo, H. Zou, Y. Guo, T. Tong, L. Ye, C. Zhu, L. Deng, B. Wang, Y. Pan and P. Li, SRC kinase-mediated signaling pathways and targeted therapies in breast cancer, Breast Cancer Res., 2022, 24(1), 99 CrossRef CAS PubMed.
- Z. Tian, G. Liang, K. Cui, Y. Liang, Q. Wang, S. Lv, X. Cheng and L. Zhang, Insight into the prospects for RNAi therapy of cancer, Front. Pharmacol., 2021, 16(12), 644718 CrossRef PubMed.
- X. Yang, Y. Zhang, C. Malichewe, Z. Shi, L. Wang, Z. Lu and X. Guo, Chitosan nanoparticle mediated upregulation of microRNA34a expression to suppress the proliferation, migration, invasion of MDA-MB-231 cells, J. Drug Delivery Sci. Technol., 2019, 52, 1061–1069 CrossRef CAS.
- X. Deng, M. Cao, J. Zhang, K. Hu, Z. Yin, Z. Zhou, X. Xiao, Y. Yang, W. Sheng, Y. Wu and Y. Zeng, Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer, Biomaterials, 2014, 35(14), 4333–4344 CrossRef CAS PubMed.
- B. Panigrahi, R. K. Singh, S. Mishra and D. Mandal, Cyclic peptide-based nanostructures as efficient siRNA carriers, Artif. Cells, Nanomed., Biotechnol., 2018, 46(sup3), 763–773 CrossRef PubMed.
- B. Panigrahi, R. K. Singh, U. Suryakant, S. Mishra, A. A. Potnis, A. B. Jena, R. G. Kerry, H. Rajaram, S. K. Ghosh and D. Mandal, Cyclic peptides nanospheres: A ‘2-in-1′ self-assembled delivery system for targeting nucleus and cytoplasm, Eur. J. Pharm. Sci., 2022, 171, 106125 CrossRef CAS PubMed.
- X. Xu, J. Wu, Y. Liu, P. E. Saw, W. Tao, M. Yu, H. Zope, M. Si, A. Victorious, J. Rasmussen, D. Ayyash, O. C. Farokhzad and J. Shi, Multifunctional envelope-type siRNA delivery nanoparticle platform for prostate cancer therapy, ACS Nano, 2017, 11(3), 2618–2627 CrossRef CAS PubMed.
- L. Zhang, Z. Dong, S. Yu, G. Li, W. Kong, W. Liu, H. He, Y. Lu, W. Wu and J. Qi, Ionic liquid-based in situ dynamically self-assembled cationic lipid nanocomplexes (CLNs) for enhanced intranasal siRNA delivery, Chin. Chem. Lett., 2024, 35(7), 109101 CrossRef CAS.
- X. Huang, N. Kong, X. Zhang, Y. Cao, R. Langer and W. Tao, The landscape of mRNA nanomedicine, Nat. Med., 2022, 28(11), 2273–2287 CrossRef CAS PubMed.
- G. Kara, G. A. Calin and B. Ozpolat, RNAi-based therapeutics and tumor targeted delivery in cancer, Adv. Drug Delivery Rev., 2022, 182, 114113 CrossRef CAS PubMed.
- K. A. Whitehead, R. Langer and D. G. Anderson, Knocking down barriers: advances in siRNA delivery, Nat. Rev. Drug Discovery, 2009, 8(2), 129–138 CrossRef CAS PubMed.
- R. Kanasty, J. R. Dorkin, A. Vegas and D. Anderson, Delivery materials for siRNA therapeutics, Nat. Mater., 2013, 12(11), 967–977 CrossRef CAS PubMed.
- S. H. El Moukhtari, E. Garbayo, A. Amundarain, S. Pascual-Gil, A. Carrasco-León, F. Prosper, X. Agirre and M. J. Blanco-Prieto, Lipid nanoparticles for siRNA delivery in cancer treatment, J. Controlled Release, 2023, 361, 130–146 CrossRef CAS PubMed.
- M. Lu, H. Xing, Z. Xun, T. Yang, P. Ding, C. Cai, D. Wang and X. Zhao, Exosome-based small RNA delivery: Progress and prospects, Asian J. Pharm. Sci., 2018, 13(1), 1–11 CrossRef PubMed.
- Y. Dong, T. Yu, L. Ding, E. Laurini, Y. Huang, M. Zhang, Y. Weng, S. Lin, P. Chen, D. Marson, Y. Jiang, S. Giorgio, S. Pricl, X. Liu, P. Rocchi and L. Peng, A dual targeting dendrimer-mediated siRNA delivery system for effective gene silencing in cancer therapy, J. Am. Chem. Soc., 2018, 140(47), 16264–16274 CrossRef CAS PubMed.
- F. Eweje, M. L. Walsh, K. Ahmad, V. Ibrahim, A. Alrefai, J. Chen and E. L. Chaikof, Protein-based nanoparticles for therapeutic nucleic acid delivery, Biomaterials, 2024, 305, 122464 CrossRef CAS PubMed.
- D. Mandal, E. H. M. Mohammed, S. Lohan, P. Mandipoor, D. Baradaran, R. K. Tiwari, K. Parang and H. M. Aliabadi, Redox-responsive disulfide cyclic peptides: a new strategy for siRNA delivery, Mol. Pharmaceutics, 2022, 19(5), 1338–1355 CrossRef CAS PubMed.
- D. Mandal, S. Lohan, M. I. Sajid, A. Alhazza, R. K. Tiwari, K. Parang and H. Montazeri Aliabadi, Modified Linear Peptides Effectively Silence STAT-3 in Breast Cancer and Ovarian Cancer Cell Lines, Pharmaceutics, 2023, 15(2), 666 CrossRef CAS PubMed.
- J. Guo, C. M. O'Driscoll, J. D. Holmes and K. Rahme, Bioconjugated gold nanoparticles enhance cellular uptake: A proof of concept study for siRNA delivery in prostate cancer cells, Int. J. Pharm., 2016, 509(1–2), 16–27 CrossRef CAS PubMed.
- W.-J. Song, J.-Z. Du, T.-M. Sun, P.-Z. Zhang and J. Wang, Gold nanoparticles capped with polyethyleneimine for enhanced siRNA delivery, Small, 2010, 6(2), 239–246 CrossRef CAS PubMed.
- A. N. Shirazi, K. L. Paquin, N. G. Howlett, D. Mandal and K. Parang, Cyclic peptide-capped gold nanoparticles for enhanced siRNA delivery, Molecules, 2014, 19(9), 13319–13331 CrossRef PubMed.
- B. Panigrahi, S. Mishra, R. K. Singh, N. Siddiqui, R. Bal and D. Mandal, Peptide generated anisotropic gold nanoparticles as efficient siRNA vectors, Int. J. Pharm., 2019, 563, 198–207 CrossRef CAS PubMed.
- Y. Xin, M. Huang, W. W. Guo, Q. Huang, L. Z. Zhang and G. Jiang, Nano-based delivery of RNAi in cancer therapy, Mol. Cancer, 2017, 16, 1–9 CrossRef PubMed.
- B. Baral, B. Panigrahi, A. Kar, K. D. Tulsiyan, U. Suryakant, D. Mandal and U. Subudhi, Peptide nanostructures-based delivery of DNA nanomaterial therapeutics for regulating gene expression, Mol. Ther. – Nucleic Acids, 2023, 33, 493–510 CrossRef CAS PubMed.
- A. M. Weiss, M. A. I. Lopez, B. W. Rawe, S. Manna, Q. Chen, E. J. Mulder, S. J. Rowan and A. P. Esser-Kahn, Understanding How Cationic Polymers’ Properties Inform Toxic or Immunogenic Responses via Parametric Analysis, Macromolecules, 2023, 56(18), 7286–7299 CrossRef CAS PubMed.
- D. G. Anderson, D. M. Lynn and R. Langer, Semi–automated synthesis and screening of a large library of degradable cationic polymers for gene delivery, Angew. Chem., Int. Ed., 2003, 42(27), 3153–3158 CrossRef CAS PubMed.
- C. E. Nelson, J. R. Kintzing, A. Hanna, J. M. Shannon, M. K. Gupta and C. L. Duvall, Balancing cationic and hydrophobic content of PEGylated siRNA polyplexes enhances endosome escape, stability, blood circulation time, and bioactivity in vivo, ACS Nano, 2013, 7(10), 8870–8880 CrossRef CAS PubMed.
- P. Ghosh, G. Han, M. De, C. K. Kim and V. M. Rotello, Gold nanoparticles in delivery applications, Adv. Drug Delivery Rev., 2008, 60(11), 1307–1315 CrossRef CAS PubMed.
- X. Qiu, X. You, X. Chen, H. Chen, A. Dhinakar, S. Liu, Z. Guo, J. Wu and Z. Liu, Development of graphene oxide-wrapped gold nanorods as robust nanoplatform for ultrafast near-infrared SERS bioimaging, Int. J. Nanomed., 2017, 12, 4349–4360 CrossRef CAS PubMed.
- J.-J. Liang, Y.-Y. Zhou, J. Wu and Y. Ding, Gold nanoparticle-based drug delivery platform for antineoplastic chemotherapy, Curr. Drug Metab., 2014, 15(6), 620–631 CrossRef CAS PubMed.
- H. Chen, Z. Liu, O. Jiang, J. Zhang, J. Huang, X. You, Z. Liang, W. Tao and J. Wu, Nanocomposite of Au and black phosphorus quantum dots as versatile probes for amphibious SERS spectroscopy, 3D photoacoustic imaging and cancer therapy, Giant, 2021, 8, 100073 CrossRef CAS.
- J. Guo, M. J. Armstrong, C. M. O'Driscoll, J. D. Holmes and K. Rahme, Positively charged, surfactant-free gold nanoparticles for nucleic acid delivery, RSC Adv., 2015, 5(23), 17862–17871 RSC.
- X. Xu, Y. Liu, Y. Yang, J. Wu, M. Cao and L. Sun, One-pot synthesis of functional peptide-modified gold nanoparticles for gene delivery, Colloids Surf., A, 2022, 640, 128491 CrossRef CAS.
- R. Lévy, N. T. K. Thanh, R. C. Doty, I. Hussain, R. J. Nichols, D. J. Schiffrin, M. Brust and D. G. Fernig, Rational and combinatorial design of peptide capping ligands for gold nanoparticles, J. Am. Chem. Soc., 2004, 126(32), 10076–10084 CrossRef PubMed.
- S. Dong, C. Tang, H. Zhou and H. Zhao, Photochemical synthesis of gold nanoparticles by the sunlight radiation using a seeding approach, Gold Bull., 2004, 37(3), 187–195 CrossRef CAS.
- X. Yan, J. Blacklock, J. Li and H. Möhwald, One-pot synthesis of polypeptide–gold nanoconjugates for in vitro gene transfection, ACS Nano, 2012, 6(1), 111–117 CrossRef CAS PubMed.
- X. Zhao, Y. Jia, J. Li, R. Dong, J. Zhang, C. Ma, H. Wang, Y. Rui and X. Jiang, Indole derivative-capped gold nanoparticles as an effective bactericide in vivo, ACS Appl. Mater. Interfaces, 2018, 10(35), 29398–29406 CrossRef CAS PubMed.
- L. Morillas-Becerril, S. Franco-Ulloa, I. Fortunati, R. Marotta, X. Sun, G. Zanoni, M. De Vivo and F. Mancin, Specific and nondisruptive interaction of guanidium-functionalized gold nanoparticles with neutral phospholipid bilayers, Commun. Chem., 2021, 4(1), 1–12 CrossRef PubMed.
- R. Imperatore, G. Carotenuto, M. A. Di Grazia, I. Ferrandino, L. Palomba, R. Mariotti, E. Vitale, S. De Nicola, A. Longo and L. Cristino, Imidazole–stabilized gold nanoparticles induce neuronal apoptosis: An in vitro and in vivo study, J. Biomed. Mater. Res., Part A, 2015, 103(4), 1436–1446 CrossRef PubMed.
- G. M. Cooper and R. O. Hausman, A molecular approach. The Cell, Sinauer Associates, 2nd edn, 2000, pp. 1394–1140 Search PubMed.
- S. D. Patil, D. G. Rhodes and D. J. Burgess, DNA-based therapeutics and DNA delivery systems: a comprehensive review, AAPS J., 2005, 7, E61–E77 CrossRef CAS PubMed.
- T. J. Boggon and M. J. Eck, Structure and regulation of Src family kinases, Oncogene, 2004, 23(48), 7918–7927 CrossRef CAS PubMed.
- R. Shahbazi, E. Asik, N. Kahraman, M. Turk, B. Ozpolat and K. Ulubayram, Modified gold-based siRNA nanotherapeutics for targeted therapy of triple-negative breast cancer, Nanomedicine, 2017, 12(16), 1961–1973 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4pm00249k |
| This journal is © The Royal Society of Chemistry 2025 |