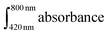Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Photoactive methylene blue-functionalized polymer for antimicrobial activation under red light†
Zeyu
Shao
a,
Huanli
Sun
 b and
Edgar H. H.
Wong
b and
Edgar H. H.
Wong
 *a
*a
aSchool of Chemical Engineering, University of New South Wales (UNSW), Sydney, NSW 2052, Australia. E-mail: edgar.wong@unsw.edu.au
bBiomedical Polymers Laboratory, College of Chemistry, Chemical Engineering and Materials Science, and State Key Laboratory of Radiation Medicine and Protection, Soochow University, Suzhou 215123, P. R. China
First published on 13th February 2025
Abstract
This study presents the synthesis of a novel methylene blue acrylamide monomer and its incorporation into a diblock copolymer, PolyMB, which exhibits potent antimicrobial activity against Gram-negative (Escherichia coli, Pseudomonas aeruginosa) and Gram-positive (Staphylococcus aureus) bacteria when photoirradiated under red light (λ = 630 nm). Mechanistic investigations revealed that singlet oxygen species, and not superoxides, are responsible for the antimicrobial activity, most likely by damaging cellular components such as proteins and DNA. The advantage of using red light as an external trigger because of its ability to penetrate skin and tissue is demonstrated here, where PolyMB is still active against E. coli when irradiated through a cover of chicken skin. In terms of biocompatibility, PolyMB is, significantly, 130 times more biocompatible than the original methylene blue dye. Overall, this study demonstrates the efficient modification of a red light-active photosensitiser into an antimicrobial macromolecule with improved biological properties for potential photodynamic applications in healthcare.
Introduction
The rise in the number of infections caused by multidrug-resistant (MDR) bacteria in recent years is a cause for concern for human health. To put this global healthcare issue called antimicrobial resistance (AMR) into perspective, about 1.14 million deaths in 2021 were directly attributed to MDR bacteria and it is forecasted that this figure could nearly double to 1.91 million deaths by 2050.1 AMR not only causes deaths but also disrupts the global economy and could result in costing the European economy €11.7 billion each year because of increased hospitalizations if no new effective solutions are found.2To combat AMR, researchers have been looking at natural immune system responses for inspiration, especially at host defense peptides (also known as antimicrobial peptides (AMPs)). AMPs are defined as sequence-specific polymers typically composed of cationic and hydrophobic groups that exert their antimicrobial mechanism by mainly causing perturbation of bacterial cell membranes.3–6 Given that this particular mode of mechanism hinders resistance development in bacteria, it is not surprising that many synthetic polymer mimics have been developed that follow the general compositional rules of AMPs.7–16 While AMPs and mimics thereof are excellent at killing bacteria, they are generally plagued by toxicity issues when administered as therapeutics and therefore have found limited application in clinical settings. Several recent strategies are being explored to overcome this toxicity barrier,17–22 including those developed by us that entail the use of self-immolative chemistry and combination therapy.23–27
Besides AMPs, the natural immune system is also capable of producing reactive oxygen species (ROS) that exert antimicrobial activity against extracellular bacteria. An attractive way to mimic this capability is via the use of photosensitisers as this class of compound not only enables external spatiotemporal activation, but also the efficient generation of singlet oxygen species to attack bacterial cells. This approach has been elegantly demonstrated by the groups of Bazan and Wang, for instance, where water-soluble cationic conjugated polymers were employed as photosensitisers to kill bacteria under light irradiation.28–30 In addition, the incorporation of small-molecule photosensitisers, either covalently linked to hydrophilic polymers or in nanoparticle encapsulations, has also been shown to be effective when triggered photochemically. Regardless of the different methodologies employed, they all share a common theme in improving the aqueous solubility of photosensitisers for bioapplications, which is essential considering that the general chemical structure of photosensitisers is often composed of hydrophobic aromatic groups.31–34
The ability to phototrigger the generation of ROS at higher wavelengths, for example, using red light (at λ ≥ 630 nm), is another key parameter to consider especially when light penetration across skin and tissue is desired. Several photosensitisers with this characteristic have been investigated for antimicrobial photodynamic therapy, including porphyrins,35,36 chlorins,37,38 and phthalocyanines.39,40 Additionally, methylene blue (MB) has emerged as a useful photosensitiser for antimicrobial applications given that it is a simple, low-cost dye with strong absorption at red light wavelengths that could generate ROS effectively.41,42 Furthermore, the ability to perform facile chemical modifications of MB for further functionalization43,44 makes this photosensitiser an attractive candidate for photoinduced antimicrobial applications.45–47
In this study, we describe the synthesis of a novel copolymer containing methylene blue moieties (PolyMB, Fig. 1) that could self-assemble in aqueous environments and generate ROS to kill bacteria under red light irradiation. This photoactive block copolymer when compared to the more hydrophobic methylene blue molecule has similar antimicrobial activity against Pseudomonas aeruginosa, Escherichia coli and Staphylococcus aureus, and inflicts damage to bacteria by generating singlet oxygen that targets proteins. More importantly, we demonstrate that PolyMB exhibits antimicrobial activity even when irradiated through a barrier of chicken skin and has superior biocompatibility (ca. 130 times improvement) compared to methylene blue.
 | ||
| Fig. 1 Chemical structures of the methylene blue-functionalized polymer (PolyMB) and methylene blue (MB). | ||
Results and discussion
To furnish the diblock copolymer PolyMB, an acrylamide-functionalized methylene blue monomer was first synthesized and subsequently copolymerized with N-butyl acrylamide using a poly(N-hydroxyethyl acrylamide) macro-chain transfer agent via reversible addition fragmentation chain transfer (RAFT) polymerization. The design rationale behind PolyMB was to create two distinct blocks with one block being completely hydrophilic (i.e., the hydroxyl-rich polymer segment) and another consisting of the more hydrophobic methylene blue side chains such that PolyMB will undergo self-assembly to form nanoparticles in water. Furthermore, the introduction of the butyl groups was to ensure that there is sufficient hydrophobicity in the second block to induce self-assembly and to relieve potential steric hindrance between the methylene blue monomer units for the RAFT polymerization to proceed smoothly.The chemical structures of the synthesized final and intermediary compounds were confirmed by NMR analysis (Fig. S1–S3, ESI†). Meanwhile, GPC analysis revealed the successful chain extension of the methylene blue and N-butyl acrylamide copolymer from the poly(N-hydroxyethyl acrylamide) macro-chain RAFT agent, as evidenced by the evolution of the number-averaged molecular weight (Mn) from 6400 g mol−1 to 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 g mol−1 (Fig. 2a). Both the macro-chain RAFT agent and polyMB showed low dispersity (Đ) values of 1.17 and 1.21, respectively, thus indicating that the polymerization steps were well controlled. The ability of PolyMB to self-assemble in water was assessed using DLS where the polymer was initially dissolved in DMSO, which is a good solvent for both blocks, at high concentration (20 mg mL−1) before further dilution into water. The DLS trace in Fig. 2b confirmed the formation of self-assembled nanoparticles with an average hydrodynamic diameter (DH-avg) of 99 nm and polydispersity index value of 0.16, which indicated narrow particle size distribution. The critical micelle concentration (CMC) of PolyMB in water was determined to be 22 μg mL−1 based on the DLS count rate method48,49 (Fig. 2c), which was comparable to other polymer self-assemblies.50
100 g mol−1 (Fig. 2a). Both the macro-chain RAFT agent and polyMB showed low dispersity (Đ) values of 1.17 and 1.21, respectively, thus indicating that the polymerization steps were well controlled. The ability of PolyMB to self-assemble in water was assessed using DLS where the polymer was initially dissolved in DMSO, which is a good solvent for both blocks, at high concentration (20 mg mL−1) before further dilution into water. The DLS trace in Fig. 2b confirmed the formation of self-assembled nanoparticles with an average hydrodynamic diameter (DH-avg) of 99 nm and polydispersity index value of 0.16, which indicated narrow particle size distribution. The critical micelle concentration (CMC) of PolyMB in water was determined to be 22 μg mL−1 based on the DLS count rate method48,49 (Fig. 2c), which was comparable to other polymer self-assemblies.50
The antimicrobial performance of PolyMB was evaluated by first determining its bacteriostatic activity against Gram-negative (P. aeruginosa, E. coli) and Gram-positive (S. aureus) bacteria, in comparison with methylene blue. For this, the polymer or methylene blue at various concentrations was mixed with bacterial cells (ca. 5 × 105 cells per mL) in cell culture media and irradiated with red light (λmax = 630 nm, 18 mW cm−2) for 2 h (see Fig. S4, ESI,† for the photoreactor setup), before further incubating the samples at 37 °C for 20 h in the dark to determine the lowest concentration cut-off point, if any, that will inhibit bacterial growth by at least 90%. This concentration cut-off point is known as the minimum inhibitory concentration (MIC). Parallel control samples were also included where the same conditions were applied except that the compounds plus bacteria were not irradiated with red light but were instead kept in the dark during the initial 2 h. As observed in Fig. 3a, all samples that were not photoirradiated did not show any MIC values even at the highest tested concentration of 256 μg mL−1. In other words, neither PolyMB nor methylene blue produced any antimicrobial effects in the absence of light. On the other hand, both PolyMB and methylene blue inhibited the growth of all three tested bacterial species when red light was shone onto the samples. PolyMB yielded MIC values of 4, 4 and 1 μg mL−1 against P. aeruginosa, E. coli and S. aureus, respectively, whereas methylene blue registered MIC values of 1.5, 1 and 0.25 μg mL−1 against the same panel of bacteria. Although methylene blue appeared to be more antimicrobial than PolyMB, both compounds have similar potency when compared in terms of the number of moles of photoactive units due to the difference in molecular weight between the two (PolyMB is twice as heavy as methylene blue per photoactive group). When contrasting the activity between Gram-negative and Gram-positive bacteria, both PolyMB and methylene blue performed better against Gram-positive bacteria, which corroborates earlier studies that showed Gram-negative bacteria are less susceptible to ROS because of the additional lipid bilayer membrane in the cell wall.51–53 It is worth noting that bacterial growth was not affected by red light irradiation in the absence of either compound, thus validating that the observed bacteriostatic effect must have been derived from the photoirradiation of PolyMB and methylene blue.
Next, the bactericidal kinetics of PolyMB and methylene blue was assessed by irradiating the compounds under red light in the presence of E. coli in phosphate buffered saline (PBS) solution (Fig. 3b). Two different initial bacterial loadings were employed, one at a standard concentration of ca. 107 colony forming units (CFU) per mL, and another at a higher concentration of ca. 109 CFU per mL to challenge the limits of the system. In both cases, almost complete eradication of E. coli cells was achieved with a 6-log10 reduction compared to the bacteria only control sample (i.e., 99.9999% killing efficiency) after 1 h of irradiation, though a faster killing rate was observed in samples with a lower starting bacterial concentration, as expected. For example, PolyMB deactivated almost all bacterial cells within 30 min when the bacterial load was lower compared to taking 1 h when the bacteria content was higher by 2 orders of magnitude. Besides bacterial load, the duration of irradiation also had an important effect on the antimicrobial performance of both PolyMB and methylene blue as the photosensitisers need to be irradiated for a minimum number of minutes to achieve maximum bactericidal effects, most likely due to the necessity of generating sufficient ROS. Based on the bacteriostatic and bactericidal data, PolyMB demonstrated excellent antimicrobial activity.
Given that methylene blue is known to generate ROS, in particular singlet oxygen (1O2), the efficiency of PolyMB in doing this under red light irradiation was assessed using 9,10-dimethylanthracene (DMA) as a singlet oxygen scavenger. If singlet oxygen was generated, it would be trapped within the DMA ring, thereby causing a decrease in UV-vis absorbance. This was indeed the case as the UV-vis absorbance peaks corresponding to DMA decreased over time and nearly disappeared after 20 min of irradiation (Fig. 4a). Apart from singlet oxygen, the ability of PolyMB to generate other ROS such as superoxide was also checked using nitrotetrazolium blue chloride (NBT) as a superoxide trap. As seen in Fig. 4b, there were no changes to the UV-vis absorbance spectra of NBT even after 30 min of irradiation in the presence of PolyMB, hence demonstrating that PolyMB predominantly follows the type II photochemical pathway where its triplet state transfers energy to molecular oxygen, producing 1O2, without generating superoxide. This behavior mirrors that of MB and is strongly influenced by the experimental conditions employed in this study—relatively low dye concentrations, the presence of oxygen, and the absence of reducing agents—all of which favor the type II pathway.54,55 To gain an understanding of the antimicrobial mechanism, we investigated the ability of PolyMB to cause protein damage since the singlet oxygen species is known to result in undesirable oxidation and alteration of biomolecules such as proteins and DNA. Using bovine serum albumin (BSA) as a protein model, the fluorescence intensity of the protein decreased substantially over time as it was irradiated together with PolyMB, which most likely signified the modification of tryptophan and tyrosine residues on BSA because of singlet oxygen attack (Fig. 4c). Further mechanistic experiments were also conducted to check if PolyMB causes any perturbation to the bacterial cell membrane. Specifically, we investigated the ability of PolyMB to perturb the inner membrane of E. coli cells using propidium iodide (PI) as a fluorescent probe. Should PolyMB cause the disruption of the inner membrane, PI will permeate across the compromised cell wall and bind with the intracellular components of the bacteria, yielding red fluorescence. This change in fluorescence fold with respect to the negative control group (i.e., bacteria and PI only), herein defined as the dimensionless constant PI index, was used to estimate the extent of membrane perturbation. In essence, more PI permeation results in higher fluorescence fold change and PI index, and therefore the greater is the extent of membrane perturbation. Typical AMPs like polymyxin B showed a gradual increase in PI index over time since this is a dominant antimicrobial mechanism for such membrane-active compounds (Fig. 4d). However, unlike in the case of the polymyxin B AMP, the PI index of PolyMB (and methylene blue) remained constant over the entire course of irradiation, thus strongly suggesting that PolyMB does not act on the bacterial cell wall. Taken together, it can be postulated that PolyMB exerts its antimicrobial activity via the generation of singlet oxygen species, and not superoxides, which damage the cellular components such as proteins and DNA without affecting the cell membrane.
It is worthwhile noting that the methylene blue moiety on PolyMB could be uncaged under red light as demonstrated previously,43,56 though this is subject to the conditions employed (e.g., type of solvent, irradiation time). To investigate whether a similar phenomenon is occurring in this study, a detailed cleavage rate analysis was performed on both PolyMB and its monomer (MB-Am) using UV-vis spectroscopy (Fig. 5 and Fig. S5†). Since the self-aggregation of MB in water affects its visible light absorption spectrum due to the coexistence of monomer and dimer forms,57,58 the calibration of MB in water was conducted by integrating the entire absorption range from 420 to 800 nm to encompass the monomer (A664 nm) and dimer (A610 nm) peaks (Fig. 5a and b). After 3 h of irradiation, a noticeable color change was observed; however, the cleavage rate remained low where only 12.6 mol% of the photoactive group was cleaved from the polymer backbone (Fig. 5d). On the other hand for the monomer MB-Am, a higher cleavage rate of 32 mol% (more than double that of PolyMB) was achieved, though the cleavage experiment was performed in acetonitrile due to its poor solubility in water. The relatively low cleavage rates for both PolyMB and MB-Am suggest that the chemical structural integrity of PolyMB remained largely unaffected by the photoirradiation process.
One of the advantages of using red light as an external trigger for biomedical applications is that it can penetrate through skin and tissue to photoinduce a desired chemical process. We emulated this scenario by shining red light onto a vial that contained a solution of E. coli bacteria and PolyMB wrapped with chicken skin (ca. 1 mm thick) (Fig. 6a). The initial bacterial loading was ca. 107 CFU mL−1 and the killing kinetics of PolyMB in terms of the reduction of viable cells was monitored over 1 h of irradiation (Fig. 6b). No detectable bacteria were observed after 1 h of irradiation, with complete eradication being achieved within the detection limit, although the efficiency was about half that compared to the case above without any chicken skin cover where it took only 30 minutes to achieve the same antimicrobial effect (vide supra). However, this is not entirely surprising and in fact correlates with other examples in the literature where the efficiency of red light irradiation is typically reduced by ca. 50% when encountering a skin barrier.59,60 Though beyond the scope of this study, one potential solution to this is to fundamentally study the system with photochemical action plots by following wavelength-dependent ROS generation, as this will enable the determination of the most optimal irradiation wavelength for maximum reactivity, which may not necessarily be at the currently employed 630 nm.61,62 Regardless, the current data suggest that PolyMB could potentially be used in vivo in the future for photodynamic applications.
Lastly, the biocompatibility of PolyMB and methylene blue was assessed by determining the mammalian cell cytotoxicity on murine embryonic fibroblast (MEF) cells. Specifically, different concentrations of each compound were screened with MEF cells to determine the half-maximal inhibitory concentration value (IC50), which is defined as the concentration that reduces the cell viability by half. The cell viability curves of MEF cells after incubation with the compounds for 24 h are shown in Fig. 7a. Evidently, PolyMB exhibited a significantly higher IC50 value compared to methylene blue (387 vs. 3 μg mL−1), which suggested it was nearly 130 times less toxic. Closer inspection of the MEF cell morphology after exposure to the compounds showed that PolyMB did not alter the cell morphology, whereas methylene blue caused the cells to shrivel and shrink (Fig. 7b). In addition, rod-shaped crystals/aggregates of methylene blue were also observed, indicating its poor solubility in water. The superior biocompatibility shown by PolyMB relative to methylene blue could be attributed to the functionalization of the photoactive unit with hydrophilic polymers to improve water solubility, which ultimately prevents the crystallization of poorly soluble aggregates that are responsible for cytotoxicity.
Conclusion
In conclusion, we developed a new antimicrobial block copolymer, PolyMB, comprising photoactive methylene blue side chains that could undergo self-assembly in water to form nanoparticles. When irradiated under red light (λ = 630 nm), PolyMB exhibited strong antimicrobial activity against Gram-negative and Gram-positive pathogens via the generation of singlet oxygen species. Further mechanistic studies revealed that PolyMB does not affect the bacterial cell membrane, and the antimicrobial activity stems from singlet oxygen attack on cellular components such as proteins and DNA. Additionally, the use of red light in combination with PolyMB enabled the eradication of bacteria even when illuminated through a chicken skin barrier, thus demonstrating the potential for biomedical applications where spatiotemporal activation across skin and tissue is desired. Importantly, PolyMB showed excellent biocompatibility and was about 130 times less toxic than the original methylene blue molecule. This study thus demonstrated the strategic design and transformation of small molecule dyes/photosensitisers into advanced functional antimicrobial macromolecules with improved biological properties for potential photodynamic therapy applications. Work is currently underway to expand the scope of this study to include testing of MDR bacterial strains in vitro and in vivo.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the Australian Research Council and UNSW via the Future Fellowship (FT210100150, E. H. H. W.) and Scientia Fellowship (E. H. H. W.) schemes, respectively. We acknowledge the facilities and technical assistance provided by the NMR Facility and Cell Culture Facility within the Mark Wainwright Analytical Centre at UNSW. We also acknowledge Prof. Christopher Barner-Kowollik and A/Prof. Hendrik Frisch from Queensland University of Technology, Australia, for helpful discussions.References
- M. Naghavi, S. E. Vollset, K. S. Ikuta, L. R. Swetschinski, A. P. Gray, E. E. Wool, G. R. Aguilar, T. Mestrovic, G. Smith, C. Han, R. L. Hsu, J. Chalek, D. T. Araki, E. Chung, C. Raggi, A. G. Hayoon, N. D. Weaver, P. A. Lindstedt, A. E. Smith, U. Altay, N. V. Bhattacharjee, K. Giannakis, F. Fell, B. McManigal, N. Ekapirat, J. A. Mendes, T. Runghien, O. Srimokla, A. Abdelkader, S. Abd-Elsalam, R. G. Aboagye, H. Abolhassani, H. Abualruz, U. Abubakar, H. J. Abukhadijah, S. Aburuz, A. Abu-Zaid, S. Achalapong, I. Y. Addo, V. Adekanmbi, T. E. Adeyeoluwa, Q. E. S. Adnani, L. A. Adzigbli, M. S. Afzal, S. Afzal, A. Agodi, A. J. Ahlstrom, A. Ahmad, S. Ahmad, T. Ahmad, A. Ahmadi, A. Ahmed, H. Ahmed, I. Ahmed, M. Ahmed, S. Ahmed, S. A. Ahmed, M. A. Akkaif, S. A. Awaidy, Y. A. Thaher, S. O. Alalalmeh, M. T. AlBataineh, W. A. Aldhaleei, A. A. S. Al-Gheethi, N. B. Alhaji, A. Ali, L. Ali, S. S. Ali, W. Ali, K. Allel, S. Al-Marwani, A. Alrawashdeh, A. Altaf, A. B. Al-Tammemi, J. A. Al-Tawfiq, K. H. Alzoubi, W. A. Al-Zyoud, B. Amos, J. H. Amuasi, R. Ancuceanu, J. R. Andrews, A. Anil, I. A. Anuoluwa, S. Anvari, A. E. Anyasodor, G. L. C. Apostol, J. Arabloo, M. Arafat, A. Y. Aravkin, D. Areda, A. Aremu, A. A. Artamonov, E. A. Ashley, M. O. Asika, S. S. Athari, M. M. D. W. Atout, T. Awoke, S. Azadnajafabad, J. M. Azam, S. Aziz, A. Y. Azzam, M. Babaei, F.-X. Babin, M. Badar, A. A. Baig, M. Bajcetic, S. Baker, M. Bardhan, H. J. Barqawi, Z. Basharat, A. Basiru, M. Bastard, S. Basu, N. S. Bayleyegn, M. A. Belete, O. O. Bello, A. Beloukas, J. A. Berkley, A. S. Bhagavathula, S. Bhaskar, S. S. Bhuyan, J. A. Bielicki, N. I. Briko, C. S. Brown, A. J. Browne, D. Buonsenso, Y. Bustanji, C. G. Carvalheiro, C. A. Castañeda-Orjuela, M. Cenderadewi, J. Chadwick, S. Chakraborty, R. M. Chandika, S. Chandy, V. Chansamouth, V. K. Chattu, A. A. Chaudhary, P. R. Ching, H. Chopra, F. R. Chowdhury, D.-T. Chu, M. Chutiyami, N. Cruz-Martins, A. G. da Silva, O. Dadras, X. Dai, S. D. Darcho, S. Das, F. P. De la Hoz, D. M. Dekker, K. Dhama, D. Diaz, B. F. R. Dickson, S. G. Djorie, M. Dodangeh, S. Dohare, K. G. Dokova, O. P. Doshi, R. K. Dowou, H. L. Dsouza, S. J. Dunachie, A. M. Dziedzic, T. Eckmanns, A. Ed-Dra, A. Eftekharimehrabad, T. C. Ekundayo, I. El Sayed, M. Elhadi, W. El-Huneidi, C. Elias, S. J. Ellis, R. Elsheikh, I. Elsohaby, C. Eltaha, B. Eshrati, M. Eslami, D. W. Eyre, A. O. Fadaka, A. F. Fagbamigbe, A. Fahim, A. Fakhri-Demeshghieh, F. O. Fasina, M. M. Fasina, A. Fatehizadeh, N. A. Feasey, A. Feizkhah, G. Fekadu, F. Fischer, I. Fitriana, K. M. Forrest, C. F. Rodrigues, J. E. Fuller, M. A. Gadanya, M. Gajdács, A. P. Gandhi, E. E. Garcia-Gallo, D. O. Garrett, R. K. Gautam, M. W. Gebregergis, M. Gebrehiwot, T. G. Gebremeskel, C. Geffers, L. Georgalis, R. M. Ghazy, M. Golechha, D. Golinelli, M. Gordon, S. Gulati, R. D. Gupta, S. Gupta, V. K. Gupta, A. D. Habteyohannes, S. Haller, H. Harapan, M. L. Harrison, A. I. Hasaballah, I. Hasan, R. S. Hasan, H. Hasani, A. H. Haselbeck, M. S. Hasnain, I. I. Hassan, S. Hassan, M. S. H. Z. Tabatabaei, K. Hayat, J. He, O. E. Hegazi, M. Heidari, K. Hezam, R. Holla, M. Holm, H. Hopkins, M. M. Hossain, M. Hosseinzadeh, S. Hostiuc, N. R. Hussein, L. D. Huy, E. D. Ibáñez-Prada, A. Ikiroma, I. M. Ilic, S. M. S. Islam, F. Ismail, N. E. Ismail, C. D. Iwu, C. J. Iwu-Jaja, A. Jafarzadeh, F. Jaiteh, R. J. Yengejeh, R. D. G. Jamora, J. Javidnia, T. Jawaid, A. W. J. Jenney, H. J. Jeon, M. Jokar, N. Jomehzadeh, T. Joo, N. Joseph, Z. Kamal, K. K. Kanmodi, R. S. Kantar, J. A. Kapisi, I. M. Karaye, Y. S. Khader, H. Khajuria, N. Khalid, F. Khamesipour, A. Khan, M. J. Khan, M. T. Khan, V. Khanal, F. F. Khidri, J. Khubchandani, S. Khusuwan, M. S. Kim, A. Kisa, V. A. Korshunov, F. Krapp, R. Krumkamp, M. Kuddus, M. Kulimbet, D. Kumar, E. A. P. Kumaran, A. Kuttikkattu, H. H. Kyu, I. Landires, B. K. Lawal, T. T. T. Le, I. M. Lederer, M. Lee, S. W. Lee, A. Lepape, T. L. Lerango, V. S. Ligade, C. Lim, S. S. Lim, L. W. Limenh, C. Liu, X. Liu, X. Liu, M. J. Loftus, H. I. M. Amin, K. L. Maass, S. B. Maharaj, M. A. Mahmoud, P. Maikanti-Charalampous, O. M. Makram, K. Malhotra, A. A. Malik, G. D. Mandilara, F. Marks, B. A. Martinez-Guerra, M. Martorell, H. Masoumi-Asl, A. G. Mathioudakis, J. May, T. A. McHugh, J. Meiring, H. N. Meles, A. Melese, E. B. Melese, G. Minervini, N. S. Mohamed, S. Mohammed, S. Mohan, A. H. Mokdad, L. Monasta, A. M. Ghalibaf, C. E. Moore, Y. Moradi, E. Mossialos, V. Mougin, G. D. Mukoro, F. Mulita, B. Muller-Pebody, E. Murillo-Zamora, S. Musa, P. Musicha, L. A. Musila, S. Muthupandian, A. J. Nagarajan, P. Naghavi, F. Nainu, T. S. Nair, H. H. R. Najmuldeen, Z. S. Natto, J. Nauman, B. P. Nayak, G. T. Nchanji, P. Ndishimye, I. Negoi, R. I. Negoi, S. A. Nejadghaderi, Q. P. Nguyen, E. A. Noman, D. C. Nwakanma, S. O’Brien, T. J. Ochoa, I. A. Odetokun, O. A. Ogundijo, T. R. Ojo-Akosile, S. R. Okeke, O. C. Okonji, A. T. Olagunju, A. Olivas-Martinez, A. A. Olorukooba, P. Olwoch, K. I. Onyedibe, E. Ortiz-Brizuela, O. Osuolale, P. Ounchanum, O. T. Oyeyemi, P. A. Mahesh, J. L. Paredes, R. R. Parikh, J. Patel, S. Patil, S. Pawar, A. Y. Peleg, P. Peprah, J. Perdigão, C. Perrone, I.-R. Petcu, K. Phommasone, Z. Z. Piracha, D. Poddighe, A. J. Pollard, R. Poluru, A. Ponce-De-Leon, J. Puvvula, F. N. Qamar, N. H. Qasim, C. D. Rafai, P. Raghav, L. Rahbarnia, F. Rahim, V. Rahimi-Movaghar, M. Rahman, M. A. Rahman, H. Ramadan, S. K. Ramasamy, P. S. Ramesh, P. W. Ramteke, R. K. Rana, U. Rani, M.-M. Rashidi, D. Rathish, S. Rattanavong, S. Rawaf, E. M. M. Redwan, L. F. Reyes, T. Roberts, J. V. Robotham, V. D. Rosenthal, A. G. Ross, N. Roy, K. E. Rudd, C. J. Sabet, B. A. Saddik, M. R. Saeb, U. Saeed, S. S. Moghaddam, W. Saengchan, M. Safaei, A. Saghazadeh, N. S. Sharif-Askari, A. Sahebkar, S. S. Sahoo, M. Sahu, M. Saki, N. Salam, Z. Saleem, M. A. Saleh, Y. L. Samodra, A. M. Samy, A. Saravanan, M. Satpathy, A. E. Schumacher, M. Sedighi, S. Seekaew, M. Shafie, P. A. Shah, S. Shahid, M. J. Shahwan, S. Shakoor, N. Shalev, M. A. Shamim, M. A. Shamshirgaran, A. Shamsi, A. Sharifan, R. P. Shastry, M. Shetty, A. Shittu, S. Shrestha, E. E. Siddig, T. Sideroglou, J. Sifuentes-Osornio, L. M. L. R. Silva, E. A. F. Simões, A. J. H. Simpson, A. Singh, S. Singh, R. Sinto, S. S. M. Soliman, S. Soraneh, N. Stoesser, T. Z. Stoeva, C. K. Swain, L. Szarpak, S. S. T. Yella, S. Tabatabai, C. Tabche, Z. M.-A. Taha, K.-K. Tan, N. Tasak, N. Y. Tat, A. Thaiprakong, P. Thangaraju, C. C. Tigoi, K. Tiwari, M. R. Tovani-Palone, T. H. Tran, M. Tumurkhuu, P. Turner, A. J. Udoakang, A. Udoh, N. Ullah, S. Ullah, A. G. Vaithinathan, M. Valenti, T. Vos, H. T. L. Vu, Y. Waheed, A. S. Walker, J. L. Walson, T. Wangrangsimakul, K. G. Weerakoon, H. F. L. Wertheim, P. C. M. Williams, A. A. Wolde, T. M. Wozniak, F. Wu, Z. Wu, M. K. K. Yadav, S. Yaghoubi, Z. S. Yahaya, A. Yarahmadi, S. Yezli, Y. E. Yismaw, D. K. Yon, C.-W. Yuan, H. Yusuf, F. Zakham, G. Zamagni, H. Zhang, Z.-J. Zhang, M. Zielińska, A. Zumla, S. E. H. H. Zyoud, S. H. Zyoud, S. I. Hay, A. Stergachis, B. Sartorius, B. S. Cooper, C. Dolecek and C. J. L. Murray, Lancet, 2024, 404, 1199–1226 CrossRef PubMed.
- Antimicrobial resistance, https://www.who.int/europe/news-room/fact-sheets/item/antimicrobial-resistance, (accessed September 19, 2024).
- Y. Wu, K. Chen, J. Wang, M. Chen, Y. Chen, Y. She, Z. Yan and R. Liu, Prog. Polym. Sci., 2023, 141, 101679 CrossRef CAS.
- H. Zhang, Q. Chen, J. Xie, Z. Cong, C. Cao, W. Zhang, D. Zhang, S. Chen, J. Gu and S. Deng, Sci. Adv., 2023, 9, eabn0771 CrossRef CAS PubMed.
- M. Magana, M. Pushpanathan, A. L. Santos, L. Leanse, M. Fernandez, A. Ioannidis, M. A. Giulianotti, Y. Apidianakis, S. Bradfute and A. L. Ferguson, Lancet Infect. Dis., 2020, 20, e216–e230 CrossRef CAS PubMed.
- B. H. Gan, J. Gaynord, S. M. Rowe, T. Deingruber and D. R. Spring, Chem. Soc. Rev., 2021, 50, 7820–7880 RSC.
- P. R. Judzewitsch, T. K. Nguyen, S. Shanmugam, E. H. H. Wong and C. Boyer, Angew. Chem., 2018, 130, 4649–4654 CrossRef.
- S. J. Lam, E. H. H. Wong, N. M. O’Brien-Simpson, N. Pantarat, A. Blencowe, E. C. Reynolds and G. G. Qiao, ACS Appl. Mater. Interfaces, 2016, 8, 33446–33456 CrossRef CAS PubMed.
- T.-K. Nguyen, S. J. Lam, K. K. Ho, N. Kumar, G. G. Qiao, S. Egan, C. Boyer and E. H. H. Wong, ACS Infect. Dis., 2017, 3, 237–248 CrossRef CAS PubMed.
- P. R. Judzewitsch, N. Corrigan, F. Trujillo, J. Xu, G. Moad, C. J. Hawker, E. H. H. Wong and C. Boyer, Macromolecules, 2020, 53, 631–639 CrossRef CAS.
- P. Pham, S. Oliver, E. H. H. Wong and C. Boyer, Polym. Chem., 2021, 12, 5689–5703 RSC.
- S. Shabani, S. Hadjigol, W. Li, Z. Si, D. Pranantyo, M. B. Chan-Park, N. M. O'Brien-Simpson and G. G. Qiao, Nat. Rev. Bioeng., 2024, 2, 343–361 CrossRef CAS.
- Z. Shao, H. Luo, T. H. Q. Nguyen and E. H. H. Wong, Biomacromolecules, 2024, 25, 6899–6912 CrossRef CAS PubMed.
- D. Wyers, T. Jirapanjawat, J. F. Quinn, M. R. Whittaker, C. Greening and T. Junkers, Polym. Chem., 2023, 14, 2126–2134 RSC.
- M. Aquib, W. Yang, L. Yu, V. K. Kannaujiya, Y. Zhang, P. Li, A. Whittaker, C. Fu and C. Boyer, Chem. Sci., 2024, 15, 19057–19069 RSC.
- A. M. Bapolisi, A. C. Lehnen, R. Machatschek, G. Mangiapia, E. Mark, J. F. Moulin, P. Wendler, S. C. Hall and M. Hartlieb, Small, 2024, 2406534 CrossRef PubMed.
- B. Zhang, D. Lu, D. B. R. Wang, Z. Y. Kok, M. B. Chan-Park and H. Duan, Adv. Funct. Mater., 2024, 34, 2407869 CrossRef CAS.
- S. Barman, L. B. Kurnaz, X. Yang, M. Nagarkatti, P. Nagarkatti, A. W. Decho and C. Tang, ACS Infect. Dis., 2023, 9, 1769–1782 CrossRef CAS PubMed.
- L. B. Kurnaz, Y. Luo, X. Yang, A. Alabresm, R. Leighton, R. Kumar, J. Hwang, A. W. Decho, P. Nagarkatti and M. Nagarkatti, Bioact. Mater., 2023, 20, 519–527 Search PubMed.
- Q. Zhou, Z. Si, K. Wang, K. Li, W. Hong, Y. Zhang and P. Li, J. Controlled Release, 2022, 352, 507–526 CrossRef CAS PubMed.
- A. C. Weiss, S. J. Shirbin, H. G. Kelly, Q. A. Besford, S. J. Kent and G. G. Qiao, ACS Appl. Mater. Interfaces, 2021, 13, 33821–33829 CrossRef CAS PubMed.
- J. Tan, Y. Fang, C. Yang, J. Tay, N. Tan, N. D. O. B. Krishnan, B. L. Chua, Y. Zhao, Y. Chen and J. L. Hedrick, Biomacromolecules, 2023, 24, 5551–5562 CrossRef CAS PubMed.
- Z. Shao, Y. D. Xu, H. Luo, K. Hakobyan, M. Zhang, J. Xu, M. H. Stenzel and E. H. H. Wong, Macromol. Rapid Commun., 2024, 2400350 CrossRef CAS PubMed.
- Z. Shao, K. Hakobyan, J. Xu, R. Chen, N. Kumar, M. Willcox and E. H. H. Wong, ACS Appl. Polym. Mater., 2023, 5, 8735–8743 CrossRef CAS.
- R. Namivandi-Zangeneh, Z. Sadrearhami, D. Dutta, M. Willcox, E. H. H. Wong and C. Boyer, ACS Infect. Dis., 2019, 5, 1357–1365 CrossRef CAS PubMed.
- R. Namivandi-Zangeneh, Z. Sadrearhami, A. Bagheri, M. Sauvage-Nguyen, K. K. K. Ho, N. Kumar, E. H. H. Wong and C. Boyer, ACS Macro Lett., 2018, 7, 592–597 CrossRef CAS PubMed.
- Z. Shao, E. Wulandari, R. C. Lin, J. Xu, K. Liang and E. H. H. Wong, ACS Infect. Dis., 2022, 8, 1480–1490 CrossRef CAS PubMed.
- B. Wang, M. Wang, A. Mikhailovsky, S. Wang and G. C. Bazan, Angew. Chem., 2017, 129, 5113–5116 CrossRef.
- B. Wang, G. Feng, M. Seifrid, M. Wang, B. Liu and G. C. Bazan, Angew. Chem., Int. Ed., 2017, 56, 16063–16066 CrossRef CAS PubMed.
- C. Zhu, Q. Yang, L. Liu, F. Lv, S. Li, G. Yang and S. Wang, Adv. Mater., 2011, 41, 4805–4810 CrossRef PubMed.
- M. T. Yaraki, B. Liu and Y. N. Tan, Nanomicro Lett., 2022, 14, 123 CAS.
- P. R. Judzewitsch, N. Corrigan, E. H. H. Wong and C. Boyer, Angew. Chem., 2021, 133, 24450–24458 CrossRef.
- A. Escudero, C. Carrillo-Carrión, M. C. Castillejos, E. Romero-Ben, C. Rosales-Barrios and N. Khiar, Mater. Chem. Front., 2021, 5, 3788–3812 RSC.
- X. Zhao, J. Liu, J. Fan, H. Chao and X. Peng, Chem. Soc. Rev., 2021, 50, 4185–4219 RSC.
- L. Jiang, C. R. R. Gan, J. Gao and X. J. Loh, Small, 2016, 12, 3609–3644 CrossRef CAS PubMed.
- H. H. Buzzá, F. Alves, A. J. B. Tomé, J. Chen, G. Kassab, J. Bu, V. S. Bagnato, G. Zheng and C. Kurachi, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2216239119 CrossRef PubMed.
- A. S. Amorim, Z. A. Arnaut, A. I. Mata, B. Pucelik, A. Barzowska, G. J. da Silva, M. M. Pereira, J. M. Dąbrowski and L. G. Arnaut, ACS Infect. Dis., 2024, 10, 3368–3377 CrossRef CAS PubMed.
- Q. Gao, D. Huang, Y. Deng, W. Yu, Q. Jin, J. Ji and G. Fu, Chem. Eng. J., 2021, 417, 129334 CrossRef CAS.
- E. Baigorria, M. E. Milanesio and E. N. Durantini, Eur. Polym. J., 2020, 134, 109816 CrossRef CAS.
- S. N. Nyamu, L. Ombaka, E. Masika and M. Ng'ang'a, Adv. Chem., 2018, 2018, 2598062 Search PubMed.
- M. Wainwright and K. Crossley, J. Chemother., 2002, 14, 431–443 CrossRef CAS PubMed.
- A. P. M. Cardozo, D. D. F. T. da Silva, K. P. S. Fernandes, R. D. C. Ferreira, A. Lino-dos-Santos-Franco, M. F. S. D. Rodrigues, L. J. Motta and R. B. Cecatto, Photodermatol., Photoimmunol. Photomed., 2024, 40, e12978 CrossRef CAS PubMed.
- I. M. Irshadeen, V. X. Truong, H. Frisch and C. Barner-Kowollik, Chem. Commun., 2023, 59, 11959–11962 RSC.
- V. X. Truong and C. Barner-Kowollik, ACS Macro Lett., 2020, 10, 78–83 CrossRef PubMed.
- J. P. Tardivo, F. Adami, J. A. Correa, M. A. S. Pinhal and M. S. Baptista, Photodiagn. Photodyn. Ther., 2014, 11, 342–350 CrossRef PubMed.
- Y.-Y. Huang, A. Wintner, P. C. Seed, T. Brauns, J. A. Gelfand and M. R. Hamblin, Sci. Rep., 2018, 8, 7257 CrossRef PubMed.
- L. M. De Freitas, E. N. Lorenzón, N. A. Santos-Filho, L. H. D. P. Zago, M. P. Uliana, K. T. De Oliveira, E. M. Cilli and C. R. Fontana, Sci. Rep., 2018, 8, 4212 CrossRef PubMed.
- J.-M. Noy, F. Chen, D. T. Akhter, Z. H. Houston, N. L. Fletcher, K. J. Thurecht and M. H. Stenzel, Biomacromolecules, 2020, 21, 2320–2333 CrossRef CAS PubMed.
- M. Callari, P. L. De Souza, A. Rawal and M. H. Stenzel, Angew. Chem., 2017, 129, 8561–8565 CrossRef.
- L. Glavas, P. Olsén, K. Odelius and A.-C. Albertsson, Biomacromolecules, 2013, 14, 4150–4156 CrossRef CAS PubMed.
- A. Galstyan, D. Block, S. Niemann, M. C. Grüner, S. Abbruzzetti, M. Oneto, C. G. Daniliuc, S. Hermann, C. Viappiani and M. Schäfers, Chem. – Eur. J., 2016, 22, 5243–5252 CrossRef CAS PubMed.
- N. S. Soukos, L. A. Ximenez-Fyvie, M. R. Hamblin, S. S. Socransky and T. Hasan, Antimicrob. Agents Chemother., 1998, 42, 2595–2601 CrossRef CAS PubMed.
- Y. Nitzan, M. Gutterman, Z. Malik and B. Ehrenberg, Photochem. Photobiol., 1992, 55, 89–96 CrossRef CAS PubMed.
- J. P. Tardivo, A. Del Giglio, C. S. De Oliveira, D. S. Gabrielli, H. C. Junqueira, D. B. Tada, D. Severino, R. de Fátima Turchiello and M. S. Baptista, Photodiagn. Photodyn. Ther., 2005, 2, 175–191 CrossRef CAS PubMed.
- H. C. Junqueira, D. Severino, L. G. Dias, M. S. Gugliotti and M. S. Baptista, Phys. Chem. Chem. Phys., 2002, 4, 2320–2328 RSC.
- I. M. Irshadeen, V. X. Truong, H. Frisch and C. Barner-Kowollik, Chem. Commun., 2022, 58, 12975–12978 RSC.
- A. Fernandez-Perez and G. Marban, ACS Omega, 2020, 5, 29801–29815 CrossRef CAS PubMed.
- A. Fernández-Pérez, T. Valdés-Solís and G. Marbán, Dyes Pigm., 2019, 161, 448–456 CrossRef.
- G. Ng, Z. Wu, T. Zhang, A. P. Dang, Y. Yao, A. R. Nelson, S. W. Prescott, A. Postma, G. Moad and C. J. Hawker, Macromolecules, 2023, 56, 7898–7908 CrossRef CAS.
- Z. Wu, T. Zhang, X. Shi, N. Corrigan, G. Ng and C. Boyer, Angew. Chem., 2023, 135, e202302451 CrossRef.
- S. L. Walden, J. A. Carroll, A. N. Unterreiner and C. Barner-Kowollik, Adv. Sci., 2024, 11, 2306014 CrossRef CAS PubMed.
- I. M. Irshadeen, S. L. Walden, M. Wegener, V. X. Truong, H. Frisch, J. P. Blinco and C. Barner-Kowollik, J. Am. Chem. Soc., 2021, 143, 21113–21126 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Full experimental protocols, NMR spectra of compounds, picture of photoreactor setup, and data for the uncaging kinetics of the methylene blue monomer. See DOI: https://doi.org/10.1039/d5py00068h |
| This journal is © The Royal Society of Chemistry 2025 |