Open Access Article
Open Access ArticleNeuroprotective thiazole sulfonamides against 6-OHDA-induced Parkinsonian model: in vitro biological and in silico pharmacokinetic assessments†
Waralee Ruankham ab,
Ratchanok Pingaew
ab,
Ratchanok Pingaew *c,
Veda Prachayasittikula,
Apilak Worachartcheewand,
Suphissara Sathuphonga,
Setthawut Apiraksattayakula,
Tanawut Tantimongcolwata,
Virapong Prachayasittikule,
Supaluk Prachayasittikula and
Kamonrat Phopin
*c,
Veda Prachayasittikula,
Apilak Worachartcheewand,
Suphissara Sathuphonga,
Setthawut Apiraksattayakula,
Tanawut Tantimongcolwata,
Virapong Prachayasittikule,
Supaluk Prachayasittikula and
Kamonrat Phopin *ae
*ae
aCenter for Research Innovation and Biomedical Informatics, Faculty of Medical Technology, Mahidol University, Bangkok 10700, Thailand. E-mail: kamonrat.php@mahidol.ac.th; kamonrat.php@mahidol.edu; Fax: +66 2 441 4380; Tel: +66 2 441 4376
bDepartment of Clinical Chemistry, Faculty of Medical Technology, Mahidol University, Bangkok 10700, Thailand
cDepartment of Chemistry, Faculty of Science, Srinakharinwirot University, Bangkok 10110, Thailand. E-mail: ratchanok@g.swu.ac.th
dDepartment of Community Medical Technology, Faculty of Medical Technology, Mahidol University, Bangkok 10700, Thailand
eDepartment of Clinical Microbiology and Applied Technology, Faculty of Medical Technology, Mahidol University, Bangkok 10700, Thailand
First published on 10th February 2025
Abstract
The limitations of currently existing medications in delaying or halting the development of Parkinson's disease (PD) remain dramatically problematic, making it the second most prevalent neurodegenerative disorder. Moreover, it is expected that the number of PD cases will double within the next 30 years. Herein, to discover a novel neuroprotective therapeutic strategy, a series of multifunctional thiazole sulfonamides underwent preliminary assessment owing to their neuroprotective capabilities against 6-hydroxydopamine (6-OHDA)-induced damage in human neuronal SH-SY5Y cells. Pretreatment with novel synthetic hybrids, including 1, 2, and 8, significantly improved cell viability, reduced lactate dehydrogenase (LDH) leakage, prevented mitochondrial dysfunction, and mitigated intracellular oxidative stress. Insight molecular mechanisms and potential targets of these compounds were elucidated through their activation and binding interaction with sirtuin 1 (SIRT1), suggesting their influencing roles on relevant downstream cascades of PD. Furthermore, in silico pharmacokinetic analysis revealed the drug-likeness of these three hybrids, which are capable of being distributed into the central nervous system (CNS) with slight toxicity. Therefore, these novel neuroprotective thiazole sulfonamides are promising candidates for further development (i.e., in vivo and clinical trials) of effective PD therapy.
Introduction
The entire aging population (over 65 years) has currently exceeded that of the children under five years old and is forecasted to double in 2050 worldwide.1 Accordingly, the gradually increasing prevalence of age-related neurodegenerative disorders (i.e., Alzheimer's disease (AD), PD, and moderate cognitive impairment) significantly contributes to the national and international socioeconomic impacts.2 The World Health Organization (WHO) states that PD is an impaired neurological condition with the second highest rate of global death cases. PD can be caused by both hereditary and non-genetic factors affecting uneven walking, tremors, muscular rigidity, and imbalanced physical movements.3,4 Although the exact pathological causes of PD are still unknown, the aggregation of alpha-synuclein has been hypothesized as a major hallmark of PD,5 incorporated with external risk factors, including unhealthy lifestyle habits and environmental pollutant exposure.6,7 To date, levodopa, dopamine agonists, anticholinergics as well as monoamine oxidase B (MAO B) and catechol-O-methyl transferase (COMT) inhibitors have been the only PD medications approved by the Food and Drug Administration (FDA). These medications are symptomatic agents, which are only capable of relieving the symptoms but incapable of delaying the progression of the disease or its adverse effects.8,9 Hence, the discovery of alternative neuroprotective agents that could slow down the progression of PD in the early stages is an urgent issue.Attention has been paid to sulfonamide as an initial core scaffold in medicinal chemistry because of its pivotal therapeutic applications, i.e., antibacterial,10 antiviral,11 antimalarial,11 and anticancer11 activities. Owing to their broad-ranging bioactivities, the sulfonamides are current candidates for drug repurposing as well as novel designs of multi-target-based drugs according to the expanding discipline of polypharmacology.10,12 Several studies have reported thiazole as another key pharmacophore for treating various disorders with strong therapeutic effects, including antibacterial13,14 and anti-inflammatory15 activities. Particularly, both sulfonamide and thiazole derivatives exert antioxidant16,17 and neuroprotective18 effects. Additionally, dopamine analogs bearing sulfonamide acted as anti-AD agents by inhibiting acetylcholinesterase (AChE) and butyrylcholinesterase (BChE).19 Bis-sulfonamides are also highlighted as potential PD agents through the activity of NAD-dependent deacetylase SIRT1.20 The current representative of sulfonamide and thiazole moieties has been illustrated in Scheme 1.
Specifically, drug development failures mostly occur in clinical trials owing to unfavorable pharmacokinetics and toxicity, leading to a low success rate for developing effective PD therapeutics.21,22 Therefore, computational (in silico) approaches have been successfully proven in drug discovery and development to increase the success rate as well as reduce the time, expenses, and labor.23 Herein, a combination of in vitro and in silico methodologies was employed to investigate the neuroprotective effects and potential mechanisms of twelve thiazole sulfonamides against the 6-OHDA-induced human neuroblastoma SH-SY5Y cell death by mimicking the Parkinsonian model. The protective efficiency in terms of cell viability, morphological changes, LDH leakage, intracellular reactive oxygen species (ROS) generation, mitochondrial membrane potential (MMP), and SIRT1 activation was explored. Molecular docking was further conducted to clarify the possible binding modes and key interactions against the SIRT1 target protein. In silico pharmacokinetics and target predictions were also performed to ensure their promising prospects for further PD advancement.
Experimental
Chemicals and reagents
Human neuroblastoma (SH-SY5Y; ATCC CRL-2266) and normal embryonic lung (MRC-5; ATCC CCL-171) were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). Dulbecco's Modified Eagle Medium (DMEM), fetal bovine serum (FBS), 0.25% trypsin–ethylenediaminetetraacetic acid (EDTA), and 1% penicillin–streptomycin were purchased from Gibco BRL (Gaithersburg, MD, USA). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and 2′,7′-dichlorodihydrofluorescein diacetate (DCFDA) were obtained from Molecular Probes (Eugene, OR, USA). A 10× RIPA buffer and protease inhibitor cocktail were obtained from Merck Millipore (Darmstadt, Germany). The Bradford protein assay was purchased from Bio-Rad Laboratories (Hercules, CA, USA). LDH-activity assay kit (cat. no. MAK066), SIRT1 activity assay kit (cat. no. CS1040), mitochondrial-specific fluorescent rhodamine-123 (cat. no. R8004), 6-OHDA (cat. no. H4381), resveratrol (RSV), and reagent grade chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA).Chemistry
Tested thiazole sulfonamides 1–12 were synthesized by N-sulfonylation of 2-aminothiazole A with the corresponding benzenesulfonyl chloride B in the presence of sodium carbonate in dichloromethane, as shown in Fig. 1. Their chemical structures were confirmed by spectral data (1H, 13C, and mass spectra).24 Compound purities were determined by High Performance Liquid Chromatography (HPLC). HPLC was carried out using Waters; 600 pump and controller; 717 autosampler, equipped with 996 PDA detector at 254 nm; column (Nova-Pak, C18, 4 μm, 60 Å, 150 mm × 3.9 mm); mobile phases: CH3CN (A) and H2O (W); and condition: isocratic at 60% A and 40% W, flow rate 1 mL min−1, running time 8 min, and injection volume 2 μL. Each sample was prepared in acetonitrile/water (9/1). All biological tested compounds (1–12) were >95% purity as determined by HPLC and summarized in Table 1. | ||
| Fig. 1 Chemical structures of thiazole sulfonamides (1–12), 2-aminothiazole (parent compound A), and RSV. | ||
| Compound | Purity (%) | Cytotoxic activity (IC50, μM) |
|---|---|---|
| a Doxorubicin hydrochloride was used as a reference drug. | ||
| 1 | 100.00 | Non-cytotoxic |
| 2 | 100.00 | Non-cytotoxic |
| 3 | 100.00 | Non-cytotoxic |
| 4 | 98.87 | Non-cytotoxic |
| 5 | 100.00 | Non-cytotoxic |
| 6 | 95.85 | Non-cytotoxic |
| 7 | 99.61 | Non-cytotoxic |
| 8 | 100.00 | Non-cytotoxic |
| 9 | 96.44 | Non-cytotoxic |
| 10 | 99.11 | Non-cytotoxic |
| 11 | 98.94 | Non-cytotoxic |
| 12 | 97.16 | Non-cytotoxic |
| Doxorubicin hydrochloridea | — | 2.43 |
Cell culture
The SH-SY5Y and MRC-5 cells were grown in 75 cm2 culture flasks containing DMEM supplemented with 10% heat-inactivated FBS and 1% penicillin–streptomycin. The cultivated cells were maintained at a temperature of 37 °C in an incubator filled with a humidified atmosphere consisting of 95% air and 5% CO2. Every three days, the medium used for cultivation was replaced while passing the cells when they approached an overall confluence of around 80%.Measurement of cytotoxicity and cell viability by MTT assay
The MRC-5 cells suspended in the corresponding culture medium were seeded in 96 well plates at a density of 1–2 × 104 cells per well and were incubated for 24 h at 37 °C in the abovementioned humidified atmosphere with 95% air and 5% CO2. Serial dilutions of the thiazole sulfonamides (1–12), positive doxorubicin, or negative DMSO were added to the plates, followed by an additional 48 h of incubation. The number of surviving cells in each well was determined using an MTT assay. Briefly, MTT solution was introduced to each well and incubated for 2–4 h. Subsequently, DMSO was added to solubilize the purple formazan crystals by sonication. The absorbance of formazan was measured at a test wavelength of 550 nm and a reference wavelength of 650 nm using a microplate reader (Molecular Devices, CA, USA). The IC50 value is defined as the concentration of a compound that inhibits cell growth by 50%.25–27The SH-SY5Y cell line was also grown in 96 well plates for 24 h before being pretreated with various doses of thiazole sulfonamides (1–12) or RSV ranging from 0.1–100 μM for 3 h and then exposed to 100 μM of 6-OHDA for an additional 24 h. After treatment, MTT solution was applied to each well and further incubated in the dark at 37 °C for 3 h. The formazan was dissolved with 0.04 N HCl in isopropanol buffer, followed by optical density quantification at 570 nm using a microplate reader (Thermo Fisher Scientific, MA, USA).20
Assessment of cell morphology
The SH-SY5Y cells were cultured at a density of 1 × 105 cells per mL in 6 well plates and allowed to adhere overnight. After seeding, the cells were pretreated with thiazole sulfonamides (1–12) for 3 h, followed by an additional 100 μM 6-OHDA exposure for 24 h. Upon completion of the incubation period, an inverted light microscope (Olympus Corporation, Tokyo, Japan) was used to observe the morphology of the cells at a magnification of 20×.28Measurement of LDH leakage
The neuroprotective effect of thiazole sulfonamides (1–12) against 6-OHDA-induced cytotoxicity was evaluated by the amount of LDH that leaked into the culture medium.20 Briefly, 1 × 105 cells per mL of SH-SY5Y cells were grown in 6 well plates. Following a 3 h pretreatment with 1 μM thiazole sulfonamides, the pretreated cells were exposed to 100 μM 6-OHDA for a further 24 h. After that, the culture media was removed, and LDH activity was measured using the colorimetric LDH assay kit in accordance with the manufacturer's instructions. The LDH assay relies on the production of nicotinamide adenine dinucleotide (NADH) by the conversion of lactate to pyruvate, which can detect absorbance at 450 nm by applying a microplate reader.Determination of intracellular ROS production by DCFDA assay
Intracellular ROS generation was determined using the ROS-sensitive fluorescent DCFDA probe.28 After the mentioned treatment, the SH-SY5Y cells were incubated with a final concentration of 10 μM of DCFDA for 30 min in the dark. A microplate reader was used to measure the amounts of fluorescent ROS at excitation and emission wavelengths of 495 and 527 nm, respectively.Determination of catalase activity
Catalase antioxidant enzyme activity is traditionally quantified by the decomposition of hydrogen peroxide (H2O2) with potassium permanganate (KMnO4)-based catalysts.29,30 Briefly, the mixture of the tested compound and 0.059 M H2O2 was preincubated at room temperature for 30 min. Afterward, 0.2 M sulfuric acid and 0.1 M KMnO4 were added. The transition from purple KMnO4 to a colorless product was measured by absorbance at 525 nm to indicate the absence and presence of H2O2 quantity. Moreover, the cellular catalase was also determined using the lysate cell as in the above treatment.Assessment of MMP by rhodamine-123 staining
Fluorescent rhodamine-123 was used to measure the MMP.31 SH-SY5Y cells were seeded onto 96 well plates and treated as described above. At the end of the treatment, the cells were introduced to rhodamine-123 at a final concentration of 10 μM for 30 min in the dark. After the incubation period, the supernatant was removed, and the treated cells were cleaned twice with phosphate buffered saline (PBS). A microplate reader was employed to evaluate the MMP levels at excitation and emission spectra of 488 and 525 nm, respectively.Measurement of SIRT1 activity
Following the end of the treatment, the SH-SY5Y cells were washed with ice-cold PBS and extracted using 1× RIPA buffer containing protease inhibitors at 4 °C for 20 min. After collecting the lysate, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g centrifugation was performed for 20 min at 4 °C, followed by transferring the protein supernatant into the microcentrifuge tubes. The Bradford protein assay was performed to determine the protein content in the samples, adhering to the manufacturer's instructions. SIRT1 deacetylase activity was examined at an excitation wavelength of 340 nm and an emission wavelength of 445 nm.20
000 × g centrifugation was performed for 20 min at 4 °C, followed by transferring the protein supernatant into the microcentrifuge tubes. The Bradford protein assay was performed to determine the protein content in the samples, adhering to the manufacturer's instructions. SIRT1 deacetylase activity was examined at an excitation wavelength of 340 nm and an emission wavelength of 445 nm.20
Prediction of physicochemical and pharmacokinetic properties
Physicochemical, pharmacokinetic (i.e., absorption, distribution, metabolism, and elimination; ADME), and toxicity characteristics of the thiazole sulfonamides (1–12) and the parent 2-aminothiazole (A) were predicted using web-based servers, including ProTox-II (https://tox-new.charite.de/protox_II/),32 pkCSM (https://biosig.unimelb.edu.au/pkcsm/),33 and SwissADME (https://www.swissadme.ch/).34 To estimate the above features, their chemical structures in Simplified Molecular Input Line Entry System (SMILES) format were submitted to the mentioned servers. Finally, Lipinski's, Ghose's, Veber's, Egan's, and Muegge's rules were employed to assess drug-likeness properties.35Molecular docking
To investigate the molecular interaction of thiazole sulfonamides and the SIRT1 protein target of interest, a free accessible SwissDock server (https://www.swissdock.ch/)36 was employed. The 2-dimensional (2D) structure of the ligands was drawn and converted to a 3D structure by applying ChemOffice 2018, while the crystallized protein structure was retrieved from the Protein Data Bank database (PDB 5BTR).37 Before the docking process, the co-crystallized ligands, including 3 subunits of RSV, were removed from the SIRT1 target protein, and only chain A was chosen for the docking process through the server with default parameters. The illustration of binding poses and estimated binding energy was analyzed using Discovery Studio Visualizer 2021 (BIOVIA, Dassault Systèmes).Target prediction
Possible protein target-related compounds were identified by uploading the SMILES format of the compounds through several web-based servers, i.e., SwissTargetPrediction (https://www.swisstargetprediction.ch/) with the inclusion of a probability more than 0 score38 and SuperPRED (https://prediction.charite.de/subpages/target_prediction.php).39 Additionally, potential targets associated with PD were screened using the keywords of human and Parkinson's diseases from DisGeNET (https://www.disgenet.org/)40 and GeneCards database (https://www.genecards.org/).41 The criterion for screening target-related disease genes was selected as greater than the average scores. After retrieving the target of interest, the compounds and protein-associated diseases were visualized as a Venn diagram by online Venny 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/).42 The overlapping target proteins for thiazole sulfonamides and PD were carried out from STRING database version 12.0 (https://string-db.org/),43 in which the highest confidence level of 0.9 was the minimum required interaction score. Subsequently, compound-target-disease (CTD) and protein–protein interaction (PPI) networks were constructed using open-source Cytoscape software 3.10.1 (https://cytoscape.org/).44 To comprehensively understand the possible molecular mechanistic relevance, centralities of degree (DC), closeness (CC), and betweenness (BC) were used as the reference standard to evaluate the essentiality of each target and compound.Statistical analysis
One-way analysis of variance (ANOVA) and a Tukey–Kramer post hoc test were used to express statistical comparisons across groups as mean ± SEM of three independent experiments (GraphPad Software Inc., CA, USA). A statistically significant value of probability (P) was defined as one that was less than 0.05.Results and discussion
Cytotoxicity and viability of cells affected by thiazole sulfonamides (1–12)
Researchers all over the world have greatly attempted to discover alternative therapeutics that significantly reduce motor symptoms or prevent neuronal damage in PD patients. Despite numerous potential therapeutics that have been shown in preclinical trials, the complex destructive nature of the disease along with the low success rate in drug discovery renders the search for novel PD-modifying agents a great challenge.21,22 2-Aminothiazole and sulfonamide derivatives exhibit broad biological activities and have been used as pharmaceutical cores to develop therapeutic agents for treating various non-communicable and infectious diseases.12,45 Particularly, 2-aminothiazole (parent A) is the sixth most known pharmacophore frequently found in FDA-approved drugs with crossover in multiple therapeutic indications. To enhance the synergistic effects of both key therapeutic scaffolds, several synthesized thiazole sulfonamides (1–12, Fig. 1) were explored. The designed compounds (1–12) were achieved by N-sulfonylation of the key compound A with benzene/aryl sulfonyl chlorides B bearing electron withdrawing/donating/hydrophobic effects (R substituent) at various positions, mostly at p-position, on the phenyl ring. Such properties of R groups and substitution patterns may provide an appropriate size/shape for interacting with the target site of action. Previously, it was noted that bioactivity can be increased by hydrogen bond donor substitution on the benzene ring at para substitution.46 Herein, the chemical synthesis of hybrids (1–12) was favorably modified relying on the efficacy of p-position with the satisfied purity as the percentage, as illustrated in Table 1 and ESI.†The cytotoxicity of such thiazole sulfonamides (1–12) against the normal lung MRC-5 cell line showed that all of the synthetic compounds had no cytotoxicity with IC50 > 50 μg mL−1 (non-cytotoxic), as summarized in Table 1.
Pretreatment of the thiazole sulfonamides (1–12) on SH-SY5Y cells at different doses (0.1–100 μM) was performed for 24 h. The results showed that no changes in cell viability were observed. Moreover, derivatives 1, 4, and the reference RSV significantly decreased cell viability at high concentrations of 10 and 100 μM, while the viability of parent compound A was predominantly enhanced at concentrations of 5, 10, and 100 μM compared with the untreated cells. To simulate the typical dopaminergic event in PD, 6-OHDA is a selective catecholaminergic neurotoxin commonly employed in in vitro and in vivo investigations to demonstrate preclinical PD models. Various concentrations of neurotoxic 6-OHDA have been applied to stimulate the SH-SY5Y cell line.20 At the concentration of 100 μM 6-OHDA, it showed significantly lower cell viability of the SH-SY5Y cells to 77% compared with the untreated cells. Additionally, it was clear that pretreatment with the most active derivatives 1, 2, and 8 (R = F, Br, and Cl) at 0.1–1 μM revealed a considerable recovery in cell viability against 100 μM 6-OHDA-mediated SH-SY5Y cell death in comparison to 6-OHDA alone (Fig. 2), which supported the enhanced activity resulting from the molecules with p-halogen substitutions. It was observed that most electron-withdrawing groups (i.e., haloalkyl (3), cyano (4), e− donating (5), and acetyl (9)) at the p-position on the benzene ring showed a decrease in activity. The nitro electron-withdrawing group at meta position (10) was less potent than p-position (6) at the highest dose. Interestingly, pretreatment with 1 μM of the thiazole sulfonamides (1, 2, and 8) against 6-OHDA exposure enhanced neuronal survival rates along with RSV pretreatment. The previous study demonstrates that the structural features of RSV or isatin-linked halogenated compounds enhance therapeutic efficacy compared with their parent moieties, thereby contributing to their potential PD treatments as MAO-A and MAO-B inhibitors.47,48 Therefore, the neuroprotective thiazole sulfonamides bearing halogen substituents at p-position (1, 2, and 8) exhibited protective effects due to their electron-withdrawing/negative ionic effects and were selected at the concentration of 1 μM for subsequent investigation through several in vitro-based methodologies.
Impact of thiazole sulfonamides on biological cell alterations
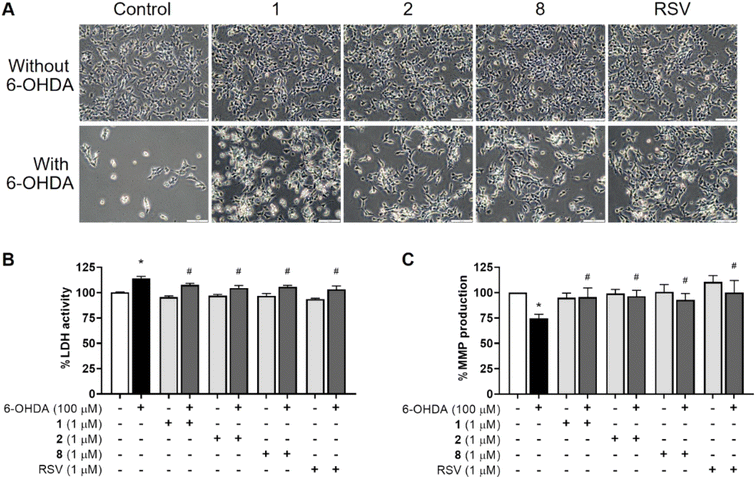
The potential thiazole sulfonamide hybrids, including 1, 2, and 8, against 6-OHDA-treated SH-SY5Y cells were morphologically observed using the light microscope at a magnification of 20×. Comparing the pretreated cells to the control, the toxic 100 μM 6-OHDA treatment caused morphological changes, as observed by a small amount of viable cells due to the shrinking cells into spherical shapes and detaching, while none of the abovementioned was observed by the exposure of studied compounds. Before being exposed to the 6-OHDA, the cells pretreated with derivatives 1, 2, 8, or positive RSV at 1 μM showed slight aberrant morphological changes, less destruction, and a more intact appearance of the cell growth with sufficient confluence than that of the 6-OHDA-exposed cells. This suggests that the aberrant morphological alterations in the cells may be avoided by pretreating with the potential compounds 1, 2, and 8 (Fig. 3A).Owing to its high binding affinity to the dopamine transporter, the neurotoxicity of 6-OHDA disrupts the activities of mitochondrial complexes I and IV by ROS oxidization, leading to the sequelae of insufficient energy and finally neuronal death.49 LDH is an enzymatic indicator elevated when the cell membrane is ruptured. The effects of thiazole sulfonamides 1, 2, and 8 on LDH activity in the culture medium were then assessed. As shown in Fig. 3B, there was no LDH change by 1, 2, and 8 pretreatments. In contrast, the cells exposed to neurotoxic 6-OHDA significantly increased LDH leakage by 114% compared with the control group. Interestingly, the pretreated cells with the indicated sulfonamides and RSV at a concentration of 1 μM showed lower LDH leakage compared with the 6-OHDA exposure, indicating the protective properties of compounds 1, 2, and 8 in maintaining the viable status of the SH-SY5Y cells against 6-OHDA-mediated cell death. These results were concurrent with previous studies, including bis-sulfonamides.20 Similarly, the thiazole-based scaffolds further extended their intensive LDH inhibitory activity.50
Metabolic energy production depends heavily on the mitochondrial powerhouse, and the pathophysiology of PD has been linked to the malfunction of mitochondria. As an indicator of the early stage of apoptosis and mitochondrial malfunction, MMP has been utilized to reflect the mitochondrial activities using the fluorescent rhodamine-123 staining.51,52 Herein, MMP analysis was carried out to evaluate the neuroprotective effects of hybrids 1, 2, and 8. Comparing the affected cells to the control group, the hybrids did not influence MMP activity, while the percentage of MMP was dramatically reduced to 75% for those exposed to 100 μM of the toxic 6-OHDA. Conversely, the cells were pretreated with 1 μM of the synthetic hybrids (1, 2, and 8) or RSV, and an increased percentage of MMP was observed against 6-OHDA-exposed cells. This suggests that pretreatment with the investigated derivatives protected the neurons against mitochondrial dysfunction using 6-ODHA (Fig. 3C).
Impact of thiazole sulfonamides on antioxidant activity
Oxidative stress is one of the most common underlying processes behind cellular dysfunction in neurodegenerative diseases, particularly PD. Thus, the antioxidative strategy has attracted considerable interest as a potential strategy for neuroprotection.53,54 To determine their antioxidant potentials, the fluorescent DCFDA probe was used to examine the levels of intracellular ROS production affected by hybrids 1, 2, and 8. Following the treatment, the cells treated with 100 μM 6-OHDA showed a notable increase in fluorescence intensity of up to 138.6%, while the hybrids-treated cells did not alter the production of ROS compared with the control group (Fig. 4A). Interestingly, intracellular ROS accumulation was significantly reduced by pretreating the cells with 1 μM of the investigated compounds or RSV, down to the range of 120.50–128.52% compared with the 6-OHDA group, suggesting that the intracellular ROS would be sustained after the pretreatment of these novel candidates.Catalase is one of the key antioxidant enzymes that responsibly catalyzes non-radical H2O2 to water and oxygen. This decomposition of H2O2 can neutralize intracellular ROS overaccumulation and maintain the optimum level of antioxidant defense-associated cellular signaling processes. By utilizing H2O2 reduction, the activity of catalase can be referred. It was found that the pure synthetic thiazole sulfonamide substances (1, 2, and 8) and parent A were responsible for reducing the H2O2 formation similar to the antioxidant RSV compared with the H2O2 control (Fig. 4B). As expected, thiazole sulfonamides-pretreated neurons significantly reduced the presence of intracellular H2O2, which was observed in the same level of RSV treatment as in the 6-OHDA group (Fig. 4C). The reduction of H2O2 percentage could be due to their antioxidant properties, particularly in catalase activation, indicating considerable prevention of mitochondrial dysfunction in SH-SY5Y cells by the antioxidant properties of thiazole sulfonamide hybrids.
Effects of thiazole sulfonamides on SIRT1 activation
Proteins of the SIRT family serve a dynamic role in cellular processes to genomic, metabolic, inflammatory, and oxidative stresses.55 Because activation of SIRT1 by a well-known RSV activator outstandingly represents a promise for slowing PD and neurological progression, the involvement of SIRT1 pathways in neurotoxin-regulated PD has been previously reported.56,57 Therefore, the cellular mechanisms of SIRT1 underlying the neuroprotective actions of thiazole sulfonamides in SH-SY5Y cells undergoing 6-OHDA-induced oxidative damage were investigated. Following the exposure to 100 μM 6-OHDA, the results revealed a considerable reduction in SIRT1 activity of the 6-OHDA-treated cells, which was 65% compared with the untreated cells (Fig. 4D). In contrast, no SIRT1 change was observed by the hybrid treatment, which further predominantly maintained the SIRT1 activity within the high range of 99–111% against 6-OHDA exposure. These were consistent with the positive RSV-activated SIRT1 activity, exhibiting notable SIRT1 regulation by the neuroprotective effects of 1, 2, and 8.Additionally, in silico molecular docking simulations are one of the powerful computational methods to reveal the ligand binding interactions and affinities against the target proteins of interest,58 confirming the effectiveness of the investigated compounds (i.e., 1, 2, and 8) as the SIRT1 activating agents. Possible binding modes of the investigated ligands against the SIRT1 target protein were exploited. The docking system was ensured owing to its reliability and accuracy as shown by an acceptable root mean square deviation (RMSD < 2.0) value obtained from the redocking of co-crystallized RSV. The docking simulations revealed that all studied compounds (1, 2, and 8) could be occupied within the same binding region of the RSV activator on the SIRT1 protein (Fig. 5) and provided the estimated lowest binding energies of −7.35, −7.58, and −7.83 kcal mol−1, respectively (Table 2). All three compounds mimicked the ability of RSV binding to the SIRT1 active site with a similar estimated binding energy, supported by shared key residues, such as pi-alkyl/alkyl hydrophobic interactions (i.e., ARG446, PRO447, and ILE223) and hydrogen bond interactions (i.e., ASN226, LYS3, and HSD2), as summarized in Table 2. Considerably, the hybrids displayed a binding affinity comparable to that of the RSV (−7.20 kcal mol−1). This could be because the presence of the sulfonamide group of these hybrid molecules played essential roles in hydrogen bonding formation with ASN226 via one of the sulfonyl oxygens. The terminal benzene and thiazole rings of compounds (1, 2, and 8) can mimic two terminal benzene rings of the RSV to form pi-alkyl/alkyl hydrophobic interactions with ARG446 and PRO447 of SIRT1. This is in concordance with a previous report, which suggested that the sulfonamide moiety, particularly the sulfonyl oxygen interacting with ASN226, played a crucial deacetylated role under the RSV treatment, highlighting the significance of this interaction in SIRT1 activation20 and corresponding to the abovementioned in vitro model. Additionally, substituted halogen atoms (i.e., Cl and Br) at the p-position on the benzene ring of compounds 2 and 8 could display hydrophobic interactions with PRO212 and LEU215 residues, which may be attributed to their better binding energies compared with the compound 1. In summary, the neuroprotective effects of thiazole sulfonamides bearing halogen atoms (i.e., F, Br, and Cl) may be partly attributed to their SIRT1 activations, enhancing downstream pathways linked to cell longevity. Additionally, aminothiazole analogues have been reported as selective SIRT2 inhibitors, making them potential therapeutic targets for PD via the SIRT family.59,60
| Ligand | Binding free energy (kcal mol−1) | Interacting type | Bonding interaction | Interacting amino acids | Bond distance (Å) |
|---|---|---|---|---|---|
| 1 | −7.35 | Hydrogen bonding | Conventional | ASN226 | 2.31 |
| LYS3 | 2.38 | ||||
| Carbon | ILE223 | 2.84 | |||
| HSD2 | 2.65 | ||||
| Hydrophobic bonding | Pi-alkyl | ILE223 | 4.10 | ||
| ARG446 | 4.47 | ||||
| PRO447 | 5.03 | ||||
| 2 | −7.58 | Hydrogen bonding | Conventional | ASN226 | 2.26 |
| LYS3 | 2.41 | ||||
| Carbon | ILE223 | 2.83 | |||
| HSD2 | 2.61 | ||||
| Hydrophobic bonding | Alkyl | PRO212 | 4.74 | ||
| LEU215 | 5.50 | ||||
| Pi-alkyl | ILE223 | 4.10 | |||
| ARG446 | 4.55 | ||||
| PRO447 | 5.07 | ||||
| 8 | −7.83 | Hydrogen bonding | Conventional | ASN226 | 2.27 |
| LYS3 | 2.40 | ||||
| Carbon | ILE223 | 2.84 | |||
| HSD2 | 2.64 | ||||
| Hydrophobic bonding | Alkyl | PRO212 | 4.81 | ||
| LEU215 | 5.35 | ||||
| Pi-alkyl | ILE223 | 4.08 | |||
| ARG446 | 4.50 | ||||
| PRO447 | 5.04 | ||||
| RSV | −7.20 | Hydrogen bonding | Conventional | GLU230 | 1.81 |
| LYS3 | 1.72 | ||||
| Carbon | HSD2 | 2.40 | |||
| Pi-donor | ASN226 | 3.08 | |||
| Hydrophobic bonding | Pi-alkyl | ARG446 | 4.05 | ||
| PRO447 | 5.31 | ||||
| LEU206 | 5.47 | ||||
| ILE223 | 5.21 |
In silico pharmacokinetics and target prediction of thiazole sulfonamides on PD
Finally, the majority of failures during the late phases of the drug development pipeline are due to their undesirable pharmacokinetic and toxicological characteristics22 resulting from sulfonamide-based compounds concerning toxicities (especially hepatotoxicity) and drug allergy issues.10 Conventional methods, such as in vitro high-throughput and in vivo models, are available for screening pharmacokinetic profiles in the early-stage processes of drug development. In silico approaches have been developed to aid the design and structural optimization of lead compounds in the early stages of drug development processes and have gained popularity owing to their low cost and rapid prediction capabilities.23 To avoid undesirable ADMET properties and unattractive underlying targets, in silico drug-likeness and target prediction of the three promising hybrids (i.e., 1, 2, and 8) were carried out to ensure their potentials for further successful development as anti-PD agents.The in silico physicochemical and pharmacokinetic characteristics of the candidates (i.e., 1, 2, 8, and parent A) were predicted using web-based tools, including SwissADME, pkCSM, and ProTox-II. All the investigated compounds meet the drug-like criteria of Lipinski, Ghose, Veber, Egan, and Muegge, which provide great drug-likeness over the parent A, showing one violation of the mentioned Ghose's and Muegge's rules (Table 3). According to the predicted pharmacokinetic profiles of thiazole sulfonamides (Table 4), all selected compounds display optimal lipophilicity, water solubility, and absorption-related parameters (i.e., GI absorption, Caco2 permeability, and skin permeation). Most of the investigated compounds (1 and 8) demonstrate moderate blood–brain barrier (BBB) and CNS permeabilities, while 2 exhibited significant BBB permeability in comparison to the parent A, suggesting its ability to reach the target site in the brain. None of the studied compounds are predicted to be non-acting substrates or inhibitors of the main metabolizing enzymes, particularly CYP2D6 and CYP3A4 isoforms. Moreover, hybrids 1, 2, and 8 are not likely to be inhibitors of CYP450 enzymes, suggesting their low chances of inducing drug–drug or food–drug interactions. The hybrids displayed total clearance ranging from −0.012 to −0.03 log mL min−1 kg−1, while that of the parent A was higher up to 0.235 log mL min−1 kg−1. Additionally, all three thiazole sulfonamides (1, 2, and 8) were not substrates for renal organic cation transporter 2 (ROCT2). According to the Globally Harmonized System of Classification and Labeling of Chemicals (GHS), the LD50 values of hybrids 1, 2, and 8 were in the range of 2000–5000 mg kg−1, which is classified as slightly hazardous, while parent A represents moderately hazardous in the range of 50–2000 mg kg−1. All selected hybrids are unlikely to induce carcinogenicity, immunotoxicity, mutagenicity, and cytotoxicity but potentially induce hepatotoxicity. However, parent A could enhance carcinogenicity and mutagenicity. Based on these predictions, the candidates (i.e., 1, 2, and 8) showed good absorption ability, preferable lipophilicity, and acceptable CNS penetration without inducing carcinogenicity, immunotoxicity, mutagenicity, and cytotoxicity. This was consistent with previously reported the halogenated isatin upon binding to SIRT2 protein, which demonstrated that its high BBB permeability could be particularly due to the presence of a substituted Cl group.48 However, slight hepatotoxicity should be carefully investigated for further appropriate usage in subsequent clinical trials.
| Compound | 1 | 2 | 8 | A |
|---|---|---|---|---|
| Formula | C9H7FN2O2S2 | C9H7BrN2O2S2 | C9H7ClN2O2S2 | C3H4N2S |
| Molecular weight | 258.29 | 319.20 | 274.75 | 100.14 |
| Rotatable bonds | 3 | 3 | 3 | 0 |
| H-bond acceptors | 4 | 4 | 4 | 3 |
| H-bond donors | 1 | 1 | 1 | 1 |
| Polar surface area | 95.68 | 95.68 | 95.68 | 67.15 |
| Molar refractivity | 59.19 | 66.93 | 64.24 | 26.52 |
| Lipinski's rule | Yes | Yes | Yes | Yes |
| Ghose's rule | Yes | Yes | Yes | No |
| Veber's rule | Yes | Yes | Yes | Yes |
| Egan's rule | Yes | Yes | Yes | Yes |
| Muegge's rule | Yes | Yes | Yes | No |
| Compound | 1 | 2 | 8 | A |
|---|---|---|---|---|
| Absorption | ||||
Lipophilicity (i![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) P) |
Soluble | Soluble | Soluble | Soluble |
| Water solubility | Soluble | Soluble | Soluble | Soluble |
| GI absorption | High | High | High | High |
| Caco2 permeability | High | High | High | High |
| Skin permeation | Yes | Yes | Yes | Yes |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Distribution | ||||
| BBB permeability | Adequate | High | Adequate | Adequate |
| CNS permeability | Adequate | Adequate | Adequate | Adequate |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Metabolism | ||||
| CYP2D6 substrate | No | No | No | No |
| CYP3A4 substrate | No | No | No | No |
| CYP1A2 inhibitor | No | Yes | Yes | Yes |
| CYP2C19 inhibitor | No | Yes | Yes | No |
| CYP2C9 inhibitor | No | No | No | Yes |
| CYP2D6 inhibitor | No | No | No | No |
| CYP3A4 inhibitor | No | No | No | Yes |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Excretion | ||||
| Total clearance (log mL min−1 kg−1) | −0.029 | −0.03 | −0.012 | 0.235 |
| Renal OCT2 substrate | No | No | No | No |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Toxicity | ||||
| Predicted LD50 (mg kg−1) | 4500 | 4500 | 4500 | 500 |
| Predicted toxicity class | 5 | 5 | 5 | 4 |
| Hepatotoxicity | Active | Active | Active | Active |
| Carcinogenicity | Inactive | Inactive | Inactive | Active |
| Immunotoxicity | Inactive | Inactive | Inactive | Inactive |
| Mutagenicity | Inactive | Inactive | Inactive | Active |
| Cytotoxicity | Inactive | Inactive | Inactive | Inactive |
Therapeutic molecules can interact with several biological targets to exhibit multiple pharmacological effects. The multilayer computational networks of CTD and PPI are well-known to robustly offer data integration contributing to PD or several age-associated neurological diseases.44,61 Herein, the PD-expressed proteins possibly interacting with the thiazole sulfonamides were predicted using in silico data resources. The Venn diagram showed the overlap of differentially PD-expressed proteins between the various thiazole sulfonamide hybrids (1, 2, and 8) (Fig. 6A). A total of 4726 proteins were screened as predicted targets for modulating PD. The hybrids (1, 2, and 8) have 100, 85, and 95 anticipated targets, respectively. The core relationship between key active PD and key candidate targets was identified as the CTD network (Fig. 6B). To better comprehend the intricate interplay between thiazole sulfonamide protein targets and PD, the 280 target genes of the overlapping targets were employed to construct the PPI network by introducing them into the STRING database. As shown in Fig. 6C, 122 targets of the effective proteins in PD based on a confidence score above 0.9 were obtained, which included 126 nodes and 283 edges. Based on the degree value of the PPI network, the top 10 core target proteins were PRKACA, NFKB1, GSK3B, FYN, PIK3CB, CDK2, PIK3CA, STAT3, SIRT1, and PPARG (Table 5). The highest degree of thiazole sulfonamides in PD management is related to PRKACA, which is downregulated in PD patients compared to the normal healthy controls.61 Following the PRKACA connectivity, NFKB1 and GSK3B demonstrated in the top 3 ranks of the network, mainly responding to inflammation, cell growth, apoptosis, proliferation, and survival.62,63 Because PD is a devastating multifactorial disorder, several cellular and molecular underlying mechanisms interplay in PD initiation and progression. The key regulator such as SIRT1 is also among the top 10 active target proteins of thiazole sulfonamides (1, 2, and 8), supporting the roles of these compounds in modulating the vital cellular processes frequently disrupted in age-related disorders.56,57,64 SIRT1 and other SIRT members could be involved in all downstream signaling cascades, as shown by the connected possible node and edge illustrations. These perspectives will be extensively robust for further in vitro, in vivo experimental, and functional validations in the clinical stages.
| Uniprot ID | Protein | Gene | Pathway | DC | CC | BC | Ref. |
|---|---|---|---|---|---|---|---|
| P17612 | Catalytic subunit α of protein kinase A | PRKACA | Differentiation, proliferation, and apoptosis | 11 | 776.33 | 0.04 | 61 |
| P19838 | Nuclear factor NF-kappa-B p105 subunit | NFKB1 | Inflammation, differentiation, cell growth, and apoptosis | 6 | 715.67 | 0.04 | 63 |
| P49841 | Glycogen synthase kinase-3 beta | GSK3B | Cell division, proliferation, motility, and survival | 6 | 157.67 | 0.03 | 62 |
| P06241 | Tyrosine–protein kinase Fyn | FYN | Cell growth, survival, motility, and immune response | 4 | 140.00 | 0.03 | 65 |
| P42338 | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform | PIK3CB | Cell growth, survival, proliferation, and motility | 4 | 48.33 | 0.03 | 66 |
| P24941 | Cyclin-dependent kinase 2 | CDK2 | Cell cycle and apoptosis | 4 | 266.00 | 0.03 | 67 |
| P42336 | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | PIK3CA | Cell growth, proliferation, metabolism, and survival | 4 | 48.33 | 0.03 | 68 |
| P40763 | Signal transducer and activator of transcription 3 | STAT3 | Cell growth, immune response, and apoptosis | 4 | 246.67 | 0.03 | 69 |
| Q96EB6 | NAD-dependent protein deacetylase sirtuin-1 | SIRT1 | Cell cycle, DNA regulation, metabolism, apoptosis, and autophagy | 4 | 101.67 | 0.03 | 57 |
| P37231 | Peroxisome proliferator-activated receptor gamma | PPARG | Redox balance, immune response, and mitochondrial function | 4 | 453.33 | 0.03 | 70 |
Conclusions
The synergistic neuroprotective efficiency of twelve thiazole sulfonamides against 6-OHDA-induced Parkinsonian characteristics was highlighted to enlighten the success rate in drug discovery and development. From this study, the substitution of thiazole sulfonamides at the p-position of the benzene ring enhanced the pharmaceutical properties with halogen atoms (i.e., 1, 2, and 8) in human neuroblastoma SH-SY5Y cells, exhibiting potent neuroprotective effects with promising potential to regulate the common mechanisms underlying the pathogenesis of PD by improving cell survival, intracellular antioxidants, and mitochondrial function. Molecular docking demonstrated that these hybrids acted as SIRT1 modulators capable of mimicking the binding mode of the well-known SIRT1 activator, RSV, with favorable binding energies. Thiazole ring, sulfonyl oxygen, and substituted halogen moieties presented in the molecules were highlighted as key chemical features essential for preferable ligand–protein binding interactions with key amino acid residues. All the three candidates display preferable drug-like properties with less toxicity and moderate BBB and CNS permeabilities targeting the brain. Additionally, the integrated CTD and PPI network-based computational analyses pointed out the possible relationship of these hybrids with PD-related pathways. In summary, these thiazole sulfonamides are promising candidates that are potentially further developed for all stages of PD management.Data availability
All data generated or analyzed during this study are included in this published article.Author contributions
WR, SA, SS, KP: investigation; SA, WR, VeP, RP, KP: formal analysis; WR, SS: writing – original draft preparation; RP, TT, AW, VP, SP, KP: conceptualization; AW, VeP, WR, TT, SP, KP: methodology; RP, SP, KP: resources; AW, RP, SP, KP: funding acquisition; AW, RP, VeP, TT, VP, SP, KP: writing – reviewing and editing; RP, KP: supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research project is supported by Mahidol University (Fundamental Funds: fiscal year 2023 by National Science Research and Innovation Fund (NSRF)), Office of the Permanent Secretary, Ministry of Higher Education, Science, Research and Innovation (OPS MHESI) (Grant No. RGNS 63-155), Thailand Science Research and Innovation (TSRI), and Mahidol University. We are also indebted to Chulabhorn Research Institute for bioactivity testing.References
- World social report 2023: leaving no one behind in an aging world, United Nations Publication, 2023, pp. 1–162 Search PubMed.
- E. Kesidou, P. Theotokis, O. Damianidou, M. Boziki, N. Konstantinidou, C. Taloumtzis, S. A. Sintila, P. Grigoriadis, M. E. Evangelopoulos, C. Bakirtzis and C. Simeonidou, J. Clin. Med., 2023, 12(6), 2255 CrossRef PubMed.
- E. R. Dorsey, T. Sherer, M. S. Okun and B. R. Bloem, J. Parkinsons Dis., 2018, 8, S3–S8 CrossRef PubMed.
- A. Kouli, K. M. Torsney and W. L. Kuan, Parkinson’s Disease: Pathogenesis and Clinical Aspects, Codon Publications, 2018 Search PubMed.
- A. Wood-Kaczmar, S. Gandhi and N. W. Wood, Trends Mol. Med., 2006, 12, 521–528 CrossRef CAS PubMed.
- P. Wal, H. Vig, A. Wal, S. Rathore, S. S. Pandey, N. K. Jain and A. Srivastava, Curr. Aging Sci., 2023, 16, 170–187 CrossRef CAS PubMed.
- A. Priyadarshi, S. A. Khuder, E. A. Schaub and S. S. Priyadarshi, Environ. Res., 2001, 86, 122–127 CrossRef CAS PubMed.
- M. Regensburger, C. W. Ip, Z. Kohl, C. Schrader, P. P. Urban, J. Kassubek and W. H. Jost, J. Neural Transm., 2023, 130, 847–861 CrossRef CAS PubMed.
- L. Dezsi and L. Vecsei, CNS Neurol. Disord.: Drug Targets, 2017, 16, 425–439 CrossRef CAS PubMed.
- A. Ovung and J. Bhattacharyya, Biophys. Rev., 2021, 13, 259–272 CrossRef CAS PubMed.
- C. Zhao, K. P. Rakesh, L. Ravidar, W. Y. Fang and H. L. Qin, Eur. J. Med. Chem., 2019, 162, 679–734 CrossRef CAS PubMed.
- A. Kołaczek, I. Fusiarz, J. Lawecka and D. Branowska, Chemik, 2014, 68, 620–628 Search PubMed.
- S. H. Sumrra, U. Habiba, W. Zafar, M. Imran and Z. H. Chohan, J. Coord. Chem., 2020, 73, 2838–2877 CrossRef CAS.
- S. H. Sumrra, Z. Arshad, W. Zafar, K. Mahmood, M. Ashfaq, A. U. Hassan, E. U. Mughal, A. Irfan and M. Imran, R. Soc. Open Sci., 2021, 8, 210910 CrossRef CAS PubMed.
- K. H. Narasimhamurthy, T. R. Swaroop and K. S. Rangappa, Eur. J. Med. Chem. Rep., 2024, 12, 100225 CAS.
- S. A. Hassan, D. M. Aziz, M. N. Abdullah, A. R. Bhat, R. S. Dongre, T. B. Hadda, F. A. Almalki, S. M. A. Kawsar, A. K. Rahiman, S. Ahmed, M. H. Abdellattif, M. Berredjem, S. A. Sheikh and J. Jamalis, J. Biomol. Struct. Dyn., 2024, 42, 3747–3763 CrossRef CAS PubMed.
- B. Z. Kurt, F. Sonmez, C. Bilen, A. Ergun, N. Gencer, O. Arslan and M. Kucukislamoglu, J. Enzyme Inhib. Med. Chem., 2016, 31, 991–998 CrossRef CAS PubMed.
- Y. Hasegawa, E. Sugimoto, T. Endo, K. Ogawa, H. Aratake, A. Morikawa and N. Kitaguchi, Biol. Pharm. Bull., 1995, 18, 1750–1754 CrossRef CAS PubMed.
- N. Gok, A. Akincioglu, E. E. Binici, H. Akincioglu, N. Kilinc and S. Goksu, Arch. Pharm., 2021, 354, e2000496 CrossRef PubMed.
- S. Apiraksattayakul, R. Pingaew, V. Prachayasittikul, W. Ruankham, P. Jongwachirachai, N. Songtawee, W. Suwanjang, T. Tantimongcolwat, S. Prachayasittikul, V. Prachayasittikul and K. Phopin, Front. Mol. Neurosci., 2022, 15, 890838 CrossRef CAS PubMed.
- R. N. L. Lamptey, B. Chaulagain, R. Trivedi, A. Gothwal, B. Layek and J. Singh, Int. J. Mol. Sci., 2022, 23, 1851 CrossRef CAS PubMed.
- D. Sun, W. Gao, H. Hu and S. Zhou, Acta Pharm. Sin. B, 2022, 12, 3049–3062 CrossRef CAS PubMed.
- B. Shaker, S. Ahmad, J. Lee, C. Jung and D. Na, Comput. Biol. Med., 2021, 137, 104851 CrossRef PubMed.
- A. Worachartcheewan, R. Pingaew, V. Prachayasittikul, A. Apiraksattayakul, S. Prachayasittikul, S. Ruchirawat and V. Prachayasittikul, EXCLI J., 2025, 24, 60–81 Search PubMed.
- A. R. Alzahrani, N. Hosny, D. I. Mohamed, H. H. A. Nahas, A. Albogami, T. M. I. Al-Hazani, I. A. A. Ibrahim, A. H. Falemban, G. A. Bamagous and E. M. Saied, RSC Adv., 2024, 14, 19400–19427 RSC.
- V. Prachayasittikul, R. Pingaew, C. Nantasenamat, S. Prachayasittikul, S. Ruchirawat and V. Prachayasittikul, Drug Des., Dev. Ther., 2014, 8, 1089–1096 CrossRef CAS PubMed.
- A. Alzamami, E. M. Radwan, E. Abo-Elabass, M. E. Behery, H. A. Alshwyeh, E. Al-Olayan, A. S. Altamimi, N. G. M. Attallah, N. Altwaijry, M. Jaremko and E. M. Saied, BMC Chem., 2023, 17, 174 CrossRef CAS PubMed.
- N. H. Gay, K. Phopin, W. Suwanjang, N. Songtawee, W. Ruankham, P. Wongchitrat, S. Prachayasittikul and V. Prachayasittikul, Neurochem. Res., 2018, 43, 619–636 CrossRef CAS PubMed.
- R. T. Rasheed, H. S. Mansoor and A. S. Mansoor, Mater. Res. Express, 2020, 7, 025405 CrossRef CAS.
- R. O. Costa, S. S. Ferreira, C. A. Pereira, J. R. Harmer, C. J. Noble, G. Schenk, R. W. A. Franco, J. Resende, P. Comba, A. E. Roberts, C. Fernandes and A. Horn Jr, Front. Chem., 2018, 6, 491 CrossRef CAS PubMed.
- P. Sooknual, R. Pingaew, K. Phopin, W. Ruankham, S. Prachayasittikul, S. Ruchirawat and V. Prachayasittikul, Bioorg. Chem., 2020, 105, 104384 CrossRef CAS PubMed.
- P. Banerjee, A. O. Eckert, A. K. Schrey and R. Preissner, Nucleic Acids Res., 2018, 46, W257–W263 CrossRef CAS PubMed.
- D. E. Pires, T. L. Blundell and D. B. Ascher, J. Med. Chem., 2015, 58, 4066–4072 CrossRef CAS PubMed.
- A. Daina, O. Michielin and V. Zoete, Sci. Rep., 2017, 7, 42717 CrossRef PubMed.
- E. M. Radwan, E. Abo-Elabass, A. E. Abd El-Baky, H. A. Alshwyeh, R. A. Almaimani, G. Almaimani, I. A. A. Ibrahim, A. Albogami, M. Jaremko, S. Z. Alshawwa and E. M. Saied, Front. Chem., 2023, 11, 1231030 CrossRef CAS PubMed.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- D. Cao, M. Wang, X. Qiu, D. Liu, H. Jiang, N. Yang and R. M. Xu, Genes Dev., 2015, 29, 1316–1325 CrossRef CAS PubMed.
- A. Daina, O. Michielin and V. Zoete, Nucleic Acids Res., 2019, 47, W357–W364 CrossRef CAS PubMed.
- K. Gallo, A. Goede, R. Preissner and B. O. Gohlke, Nucleic Acids Res., 2022, 50, W726–w731 CrossRef CAS PubMed.
- J. Piñero, J. M. Ramírez-Anguita, J. Saüch-Pitarch, F. Ronzano, E. Centeno, F. Sanz and L. I. Furlong, Nucleic Acids Res., 2020, 48, D845–D855 Search PubMed.
- G. Stelzer, N. Rosen, I. Plaschkes, S. Zimmerman, M. Twik, S. Fishilevich, T. I. Stein, R. Nudel, I. Lieder, Y. Mazor, S. Kaplan, D. Dahary, D. Warshawsky, Y. Guan-Golan, A. Kohn, N. Rappaport, M. Safran and D. Lancet, Curr. Protoc. Bioinf., 2016, 54, 1.30.1–1.30.33 Search PubMed.
- A. Jia, L. Xu and Y. Wang, Briefings Bioinf., 2021, 22, bbab108 CrossRef PubMed.
- D. Szklarczyk, A. L. Gable, K. C. Nastou, D. Lyon, R. Kirsch, S. Pyysalo, N. T. Doncheva, M. Legeay, T. Fang, P. Bork, L. J. Jensen and C. von Mering, Nucleic Acids Res., 2021, 49, D605–D612 CrossRef CAS PubMed.
- P. Shannon, A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski and T. Ideker, Genome Res., 2003, 13, 2498–2504 CrossRef CAS.
- M. F. Elsadek, B. M. Ahmed and M. F. Farahat, Molecules, 2021, 26, 6158 CrossRef PubMed.
- B. Soni, M. S. Ranawat, R. Sharma, A. Bhandari and S. Sharma, Eur. J. Med. Chem., 2010, 45, 2938–2942 CrossRef CAS PubMed.
- W. Nawaz, Z. Zhou, S. Deng, X. Ma, X. Ma, C. Li and X. Shu, Nutrients, 2017, 9, 1188 CrossRef PubMed.
- S. Kumar, J. M. Oh, P. Prabhakaran, A. Awasti, H. Kim and B. Mathew, Sci. Rep., 2024, 14, 1264 CrossRef CAS PubMed.
- D. Hernandez-Baltazar, L. M. Zavala-Flores and A. Villanueva-Olivo, Neurologia, 2017, 32, 533–539 CrossRef CAS PubMed.
- D. Sharma, M. Singh, J. Joshi, M. Garg, V. Chaudhary, D. Blankenberg, S. Chandna, V. Kumar and R. Rani, ACS Omega, 2023, 8, 17552–17562 CrossRef CAS PubMed.
- A. Bose and M. F. Beal, J. Neurochem., 2016, 139(suppl. 1), 216–231 CrossRef CAS PubMed.
- A. Baracca, G. Sgarbi, G. Solaini and G. Lenaz, Biochim. Biophys. Acta, Bioenerg., 2003, 1606, 137–146 CrossRef CAS PubMed.
- M. T. Islam, Neurol. Res., 2017, 39, 73–82 CrossRef CAS PubMed.
- R. I. Teleanu, C. Chircov, A. M. Grumezescu, A. Volceanov and D. M. Teleanu, J. Clin. Med., 2019, 8, 1659 CrossRef CAS PubMed.
- Q. J. Wu, T. N. Zhang, H. H. Chen, X. F. Yu, J. L. Lv, Y. Y. Liu, Y. S. Liu, G. Zheng, J. Q. Zhao, Y. F. Wei, J. Y. Guo, F. H. Liu, Q. Chang, Y. X. Zhang, C. G. Liu and Y. H. Zhao, Signal Transduct. Targeted Ther., 2022, 7, 402 CrossRef CAS PubMed.
- R. Manjula, K. Anuja and F. J. Alcain, Front. Pharmacol., 2020, 11, 585821 CrossRef CAS PubMed.
- X. Li, Y. Feng, X. X. Wang, D. Truong and Y. C. Wu, Aging Dis., 2020, 11, 1608–1622 CrossRef PubMed.
- P. C. Agu, C. A. Afiukwa, O. U. Orji, E. M. Ezeh, I. H. Ofoke, C. O. Ogbu, E. I. Ugwuja and P. M. Aja, Sci. Rep., 2023, 13, 13398 CrossRef CAS PubMed.
- S. G. Kaya and G. Eren, Bioorg. Chem., 2024, 143, 107038 CrossRef CAS PubMed.
- S. Sundriyal, S. Moniot, Z. Mahmud, S. Yao, P. Di Fruscia, C. R. Reynolds, D. T. Dexter, M. J. Sternberg, E. W. Lam, C. Steegborn and M. J. Fuchter, J. Med. Chem., 2017, 60, 1928–1945 CrossRef CAS PubMed.
- W. Quan, J. Li, X. Jin, L. Liu, Q. Zhang, Y. Qin, X. Pei and J. Chen, Parkinson's Dis., 2021, 2021, 1690341 Search PubMed.
- J. J. Credle, J. L. George, J. Wills, V. Duka, K. Shah, Y. C. Lee, O. Rodriguez, T. Simkins, M. Winter, D. Moechars, T. Steckler, J. Goudreau, D. I. Finkelstein and A. Sidhu, Cell Death Differ., 2015, 22, 838–851 CrossRef CAS.
- S. Perez-Oliveira, D. Vazquez-Coto, S. Pardo, M. Blazquez-Estrada, M. Menendez-Gonzalez, P. Siso, E. Suarez, C. Garcia-Fernandez, B. C. Fages, E. Coto and V. Alvarez, J. Neural Transm., 2024, 131, 773–779 CrossRef CAS.
- U. Kilic, O. Gok, U. Erenberk, M. R. Dundaroz, E. Torun, Y. Kucukardali, B. Elibol-Can, O. Uysal and T. Dundar, PLoS One, 2015, 10, e0117954 CrossRef PubMed.
- E. Angelopoulou, Y. N. Paudel, T. Julian, M. F. Shaikh and C. Piperi, Mol. Neurobiol., 2021, 58, 1372–1391 Search PubMed.
- J. Fores-Martos, C. Boullosa, D. Rodrigo-Dominguez, J. Sanchez-Valle, B. Suay-Garcia, J. Climent, A. Falco, A. Valencia, J. A. Puig-Butille, S. Puig and R. Tabares-Seisdedos, Cancers, 2021, 13, 2990 CrossRef CAS.
- O. A. Levy, C. Malagelada and L. A. Greene, Apoptosis, 2009, 14, 478–500 CrossRef PubMed.
- A. Goyal, A. Agrawal, A. Verma and N. Dubey, Exp. Mol. Pathol., 2023, 129, 104846 Search PubMed.
- X. Geng, Y. Zou, J. Li, S. Li, R. Qi, H. Yu and L. Zhong, Cell Tissue Res., 2023, 393, 455–470 CrossRef CAS PubMed.
- T. Behl, P. Madaan, A. Sehgal, S. Singh, N. Sharma, S. Bhatia, A. Al-Harrasi, S. Chigurupati, I. Alrashdi and S. G. Bungau, Int. J. Mol. Sci., 2021, 22, 7432 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra04941a |
| This journal is © The Royal Society of Chemistry 2025 |