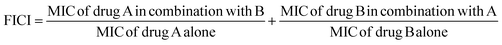Open Access Article
Open Access ArticleSynthesis and antibacterial evaluation of quinoline–sulfonamide hybrid compounds: a promising strategy against bacterial resistance†
Zohaib Saifia,
Asghar Ali bc,
Afreen Inama,
Amir Azam
bc,
Afreen Inama,
Amir Azam a,
Mohan Kamthanc,
Mohammad Abid
a,
Mohan Kamthanc,
Mohammad Abid *b and
Imran Ali*a
*b and
Imran Ali*a
aDepartment of Chemistry, Jamia Millia Islamia, Jamia Nagar, New Delhi-110025, India. E-mail: iali2@jmi.ac.in
bDepartment of Biosciences, Jamia Millia Islamia, New Delhi-110025, India. E-mail: mabid@jmi.ac.in
cDepartment of Biochemistry, School of Chemical and Life Sciences, Jamia Hamdard, New Delhi-110062, India
First published on 17th January 2025
Abstract
Antibiotic-resistant bacteria are a serious global health threat, making infections harder to treat and increasing medical costs and mortality rates. To combat resistant bacterial strains, a series of compounds (QS1–12) were synthesized with an excellent yield of 85–92%. Initial assessments of these analogues as potential antibacterial agents were conducted through a preliminary screening against a panel of diverse bacterial strains. The results identified compound QS-3 as the most effective antibacterial candidate, exhibiting exceptional inhibitory activity against P. aeruginosa with a minimum inhibitory concentration (MIC) of 64 μg mL−1. Furthermore, QS-3 demonstrated a favorable synergistic effect when combined with ciprofloxacin. Notably, the compound displayed minimal cytotoxicity, inducing less than 5% lysis of red blood cells (RBCs). Significantly, QS-3 exhibited enhanced inhibitory activity, particularly against the antibiotic-resistant strains AA202 and AA290. In silico predictions of physicochemical properties underscored the drug-like qualities of the designed compounds. Additionally, molecular docking poses, ligPlot images, and a binding affinity of −8.0 kcal mol−1 further reinforced their potential as promising antibacterial agents. Briefly, the reported compound QS3 may be a future broad-range antibacterial agent.
1. Introduction
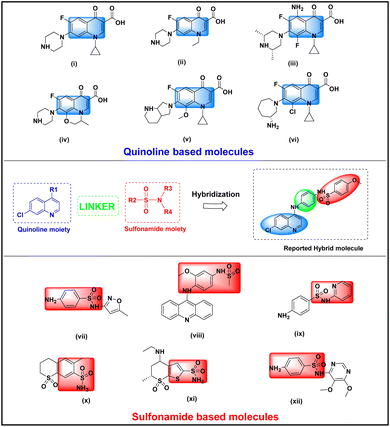
The improper use of limited antibiotics has made the problem much worse, rendering previously curable infections incurable, and the effectiveness of the currently available antibiotics is declining more quickly. As a result, bacterial infections are worsened by rising antibacterial resistance, especially in developing nations with widespread antibiotic access.1,2 Antibiotic-resistant bacteria contributed to around five million deaths in 2019, with projected mortality expected to rise by 2050. Recognized as a major global threat by the WHO, experts warn we may be entering a “post-antibiotic age”. This crisis highlights the urgent need for new, innovative antibacterials to combat infections.3–5 The combination therapy using multiple antibiotics simultaneously has been a key approach to combat bacterial infections and prevent resistance.6,7 Augmentin, a combination of amoxicillin and clavulanic acid, uses amoxicillin to disrupt growth while clavulanic acid inhibits β-lactamase enzymes that resist antibiotics.8,9 Hybrid drugs improve upon combination therapies by merging multiple elements into a single molecule, simplifying administration, reducing side effects, and avoiding drug interactions. This targeted approach enhances efficacy and safety, making hybrid drugs a promising solution for complex conditions.10,11 For this purpose, we selected two core moieties quinoline and sulfonamides, both have good antibacterial properties. Quinolones, the first class of antibiotics known to inhibit bacterial DNA synthesis, have revolutionized the treatment of bacterial infections. By targeting nucleic acid synthesis and disrupting key enzymes such as DNA gyrase and topoisomerase IV, these compounds effectively hinder bacterial growth.12 Beyond their antibacterial properties, quinoline derivatives have been extensively explored for diverse pharmacological applications, including cardiovascular, central nervous system (CNS),13 analgesic,14 antimalarial,15 anticancer,16 antibacterial,17 antiviral,18 antifungal,19 anti-inflammatory, and more20 Recognizing their critical role in drug development, this study focuses on the gyrase protein (PDB: 3qtd) and employs molecular docking analysis to further understand its interactions with quinolone derivatives. The quinolone moiety forms the backbone of several widely recognized antibiotic drugs, such as ciprofloxacin (i), norfloxacin (ii), sparfloxacin (iii), levofloxacin (iv), moxifloxacin (v), and besifloxacin (vi) (Fig. 1).21 These medications represent successive generations of fluoroquinolones, each iteration designed to improve potency and therapeutic effectiveness. Over time, these advancements have significantly enhanced the clinical utility of quinolones, solidifying their role as critical agents in combating bacterial infections.22 In recent years, quinolones have been at the forefront of research as powerful antibacterial agents, garnering considerable attention for their potential in addressing infectious diseases. Efforts to develop and optimize these compounds have led to the discovery and synthesis of novel derivatives, showcasing a wide range of promising biological activities. These advancements not only underscore the evolving capabilities of fluoroquinolones but also highlight their crucial role in combating infectious diseases and tackling the growing challenge of antibacterial resistance.23On the other hand, sulfa drugs, renowned in the pharmaceutical industry for their sulfonamide moiety, are widely used to treat various bacterial infections. These compounds exert their antibacterial effects by inhibiting dihydropteroate synthase (DHPS), an essential enzyme in the bacterial folate biosynthesis pathway. This inhibition disrupts bacterial growth and cell division. Notably, this pathway is absent in higher organisms, making it an ideal target for developing effective antibacterial therapies.24,25 Additionally, they demonstrate antibacterial properties,26 antifungal capabilities,27 anti-inflammatory actions,28 antioxidant qualities,29 diuretic effects,30 anticancer properties,31 interactions with carbonic anhydrases,32 antitumor potentials,33 Alzheimer diseases,34 anti-tubercular activities,35 anti-diabetic effects,36 HIV inhibition,37 antiviral actions,38 and anti-malarial.39 Several sulfonamide derivatives are commercially available and widely used as antibacterial drugs. Notable examples include (vii), amsacrine (viii), sulfapyridine (ix), meticrane (x), dorzolamide (xi), sulfadoxine (xii), etc. (Fig. 1).21
To highlight the importance of this advancement, we explore several examples of quinoline and sulfonamide-based drugs that have demonstrated significant antibacterial properties. This paper presents the synthesis, spectroscopic characterization, and antibacterial activity of novel quinoline–sulfonamide hybrids.
2. Results and discussion
2.1 Chemistry
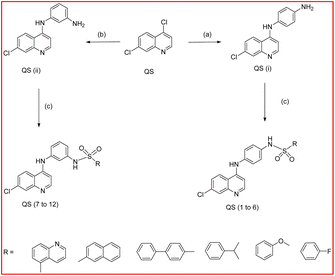
The availability of access to the title compounds via widely available starting materials is a significant advantage of our simple and fast synthesis process. The synthesis of title compounds QS(1–12) is achieved by a synthetic protocol as shown in Scheme 1. The 4,7-dichloroquinoline was condensed with p-phenylenediamine/m-phenylenediamine in presence of catalyst p-TSA to afford the N-(7-chloro-quinolinyl-4-yl)-benzene-1,4-diamine QS-(i) or N-(7-chloro-quinolinyl-4-yl)-benzene-1,3-diamine QS-(ii).40 Both these compounds QS(i–ii) were reacted with different substituted benzene sulfonyl chloride at room temperature to 60 °C for 12–16 hours using DMF and TEA as solvent and base, respectively, and produced the title compounds QS(1–12)41 with excellent percentage yield. All intermediates and title compounds have more than 90% yield. All Intermediates and final products were well purified using column chromatography with a mixer of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 methanol
9 methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dichloromethane/chloroform and characterized using multi-spectroscopic techniques like 1H NMR, 13C NMR, and FT-IR spectroscopy. The verification of purity was established through liquid chromatography-mass spectrometry (LC-MS).
dichloromethane/chloroform and characterized using multi-spectroscopic techniques like 1H NMR, 13C NMR, and FT-IR spectroscopy. The verification of purity was established through liquid chromatography-mass spectrometry (LC-MS).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations of the sulfonamide moiety, provided conclusive evidence of the formation of the sulfonamide derivatives.
O stretching vibrations of the sulfonamide moiety, provided conclusive evidence of the formation of the sulfonamide derivatives.2.2 Chemical synthesis
The synthesis protocol of the reported compounds is shown in Scheme 1 as given below.2.3 Antibacterial studies
| Compound | E. coli | E. faecalis | P. aeruginosa | S. typhi. |
|---|---|---|---|---|
| a CIP: ciprofloxacin. | ||||
| QS1 | 06 | — | — | — |
| QS2 | — | — | — | — |
| QS3 | 12 | 07 | 09 | 11 |
| QS4 | 07 | 06 | — | 06 |
| QS5 | — | — | 06 | — |
| QS6 | — | — | — | — |
| QS7 | — | 06 | — | — |
| QS8 | — | — | — | — |
| QS9 | 06 | — | — | 07 |
| QS10 | — | — | — | — |
| QS11 | — | — | — | — |
| QS12 | — | — | — | — |
| Ciprofloxacin | 25 | 18 | 24 | 18 |
| Isolates | Zone of inhibition (in mm) at different concentrations of test compound | ||
|---|---|---|---|
| ½MIC | MIC | 2MIC | |
| E. coli | 06 | 07 | 07 |
| E. faecalis | 07 | 08 | 08 |
| P. aeruginosa | 06 | 08 | 10 |
| S. typhi. | 07 | 07 | 08 |
| Bacterial strain | MIC alone (μg mL−1) | MIC in combination (μg mL−1) | FICI | Mode of interaction | ||
|---|---|---|---|---|---|---|
| Comp | CIP | Comp | CIP | |||
| E. coli | 128 | 0.5 | 2 | 0.125 | 0.26 | Synergistic |
| E. faecalis | 128 | 0.5 | 2 | 0.125 | 0.26 | Synergistic |
| P. aeruginosa | 64 | 0.5 | 4 | 0.5 | 1 | Indifferent |
| S. typhi. | 512 | 0.5 | 2 | 0.25 | 0.5 | Synergistic |
2.4 In silico studies
The binding affinities of QS-3 were determined across all four chains (A, B, C, and D) of the target protein, with the best energy observed in chain C at −8.0 kcal mol−1—better than the reference drug ciprofloxacin, which had a binding energy of −7.6 kcal mol−1. The molecular docking poses (A) cartoon image and (B) solid surface images show the interactions of QS-3 with Ser264 and Ser332, and ciprofloxacin with Asn209, Ala214, and Asp207, respectively (chain C). The ligPlot images (C) highlight hydrophobic interactions for QS-3 with Gly262, Leu260, Gly257, Gly346, Val327, Gly329, Tyr331, Tyr269, Arg270, Gly266, and for ciprofloxacin with Leu260, Gly257, Gly261, Gly346, Val327, Gly266, Arg270, Gly329, Tyr331, and Tyr269. QS-3 forms two hydrogen bond interactions, while ciprofloxacin forms three, as shown in Fig. 3. Additional details are provided in Table 4.
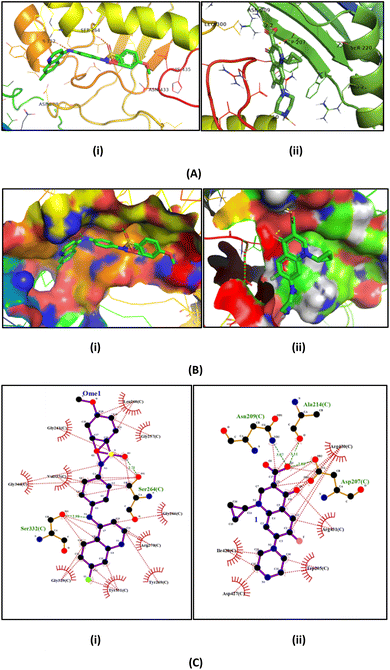 | ||
| Fig. 3 Molecular docking poses with gyrase enzyme (PDB: 3QTD): (A) cartoon images (B) solid surface images of (i) QS-3 and (ii) ciprofloxacin; showing interactions with protein residues; (C) ligPlot images highlighting hydrophobic interactions (orange) and hydrogen bonds (green). | ||
| Compound | Target (PDB code) | Binding affinities (kcal mol−1) | No. of H-bonds | H-bond length (Å) | Residue involved in H-bonds | Residue involved in hydrophobic interaction |
|---|---|---|---|---|---|---|
| QS3 | Putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa (PDB ID: 3QTD) | −8.00 | 2 | 2.71 | Ser 264 | Gly262, Leu260, Gly257, Gly346, Val327, Gly329, Tyr331, Tyr269, Arg270, Gly266 |
| 2.98 | Ser 332 | |||||
| Ciprofloxacin | −7.6 | 3 | 307 | Asn209 | Leu260, Gly257, Gly261, Gly346, Val327, Gly266, Arg270, Gly329, Tyr331, Try269 | |
| 311 | Ala214 | |||||
| 280 | Asp207 |
3. Experimental
The details of the experiment are given in the ESI file.† However, some important methodologies are described herein.3.1 Chemicals and reagents
All chemicals and solvents were of analytical grade and purchased from Sigma-Aldrich, USA. The chemicals used were 4,7-dichloroquinoline p-phenylenediamine/m-phenylenediamine, m-phenylenediamine, substituted sulfonyl chloride, catalyst para toluene sulfonic acid (p-TSA) or tosylic acid (TsOH), ethanol (EtOH), triethyle amine (TEA), dimethylformamide (DMF), potassium hydroxide, methanol, sulfonyl chloride, sodium bicarbonate, sodium sulfate, brine water, dichloromethane, ethyl acetate dimethylsulfoxide (DMSO), ciprofloxacin (CIP), ampicillin (AMP), triton-X, ethylenediaminetetraacetic acid (EDTA), sodium chloride, potassium chloride, disodium hydrogen phosphate, dipotassium hydrogen phosphate and ethanol.3.2 Instruments used
The instruments used were A digital Buchi melting point (MP) apparatus (M-560), TLC aluminum sheets of Merck silica gel 60 F254, FT-IR spectrometer of Agilent Cary 630, UV light chamber, NMR of Bruker Spectro Spin DPX-300, spectrometer using CDCl3/DMSO-d6 as a solvent Thermo Scientific Multiskan for optical density of the cultures, Laminar air flow for aseptic condition, Orbitek Incubator Shaker (Scigenics Biotech) and LC-MS of Agilent Quadrupole-6150 LC-MS.3.3 In vitro studies
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
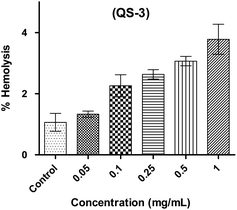
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10. From each diluted suspension, 100.0 μL was added in triplicate to 100.0 μL of various dilution series of test compounds present in micro-centrifuge tubes containing the same buffer solution. 1.0% Triton X-100 was used to attain complete haemolysis. The tubes were then placed in an incubator at 37 °C for duration of one hour, followed by centrifugation at 2000 rpm for 10.0 minutes at 20.0 °C. 150.0 μL of the resultant supernatant was transferred to a flat-bottomed Tarson microtiter plate, and the absorbance was measured. at a wavelength of 450 nm using a spectrophotometer.48
10. From each diluted suspension, 100.0 μL was added in triplicate to 100.0 μL of various dilution series of test compounds present in micro-centrifuge tubes containing the same buffer solution. 1.0% Triton X-100 was used to attain complete haemolysis. The tubes were then placed in an incubator at 37 °C for duration of one hour, followed by centrifugation at 2000 rpm for 10.0 minutes at 20.0 °C. 150.0 μL of the resultant supernatant was transferred to a flat-bottomed Tarson microtiter plate, and the absorbance was measured. at a wavelength of 450 nm using a spectrophotometer.48| % Hemolysis = [(A − B)/(C − B)] × 100 |
The molecular modeling studies followed a well-established protocol using various software tools, including ChemDraw Professional 16.0, AutoDock Tools 1.5.6, AutoDock Vina 4.0, Discovery Studio, PyMOL and Ligplot. The crystal structure of the putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa (PDB ID: 3QTD), obtained from the Protein Data Bank, served as the target protein. The PDB files for the selected X-ray structures were sourced from the protein database. The 2D structures were generated using ChemDraw software. The PDB file for Ligand preparation involved converting these structures into energy-minimized 3D models with Chem3D 16.0. The undesired molecules were removed from the gyrase protein using Discovery Studio, and a grid box was created using AutoDock Tools. The coordinates of compound QS-3 and ciprofloxacin were center_x = 37.702, center_y = 2.534, center_z = 160.609 and center_x = 37.927, center_y = 0.119, center_z = 160.682 respectively. The binding affinities were calculated using AutoDock Vina 4.0, and the 3D visualization of results was accomplished with PyMOL. The Ligplot was used for generating structures to analyse hydrophilic and hydrophobic interaction with amino acid residues. The resulting conformations, including 3D cartoon and solid surface poses of QS-3 and ciprofloxacin were analyzed to assess their interactions with amino acid residues and evaluate their binding energies. All the software used in this study were freeware.49,50
3.4 General procedure
3.4.1.1 N-(7-Chloro-quinolin-4-yl)-benzene-1,4-diamine QS-(i). Yellow powder, yield: 94%; mp > 211 °C; MS: 270 (M + 1); 1H NMR (300 MHz, DMSO-d6): d 8.54 (d, 1H, J = 5.31 Hz, Ar–H), 8.04 (d, 1H, J = 2.04 Hz, Ar–H), 8.85 (d, 1H, J = 8.97 Hz, Ar–H), 7.54 (d, 2H, J = 8.74 Hz, Ar–H), 7.49 (dd, 1H, J = 2.04, 8.97 Hz, Ar–H), 7.10 (d, 2H, J = 8.74 Hz, Ar–H), 6.83 (d, 1H, J = 5.31, Ar–H).
3.4.1.2 N-(7-Chloro-quinolin-4-yl)-benzene-1,3-diamine QS-(ii). Yellow greenish powder, yield: 92%; mp 206–207 °C; MS: 270 (M + 1); 1H NMR (300 MHz, DMSO-d6): d 8.56 (d, 1H, J = 5.32 Hz, Ar–H), 8.06 (d, 1H, J = 1.98 Hz, Ar–H), 7.94 (s, 1H, Ar–H), 7.87 (d, 1H, J = 8.94 Hz, Ar–H), 7.44 (dd, 1H, J = 1.98, 8.94 Hz, Ar–H), 7.34 (t, 1H, J = 7.98 Hz, Ar–H), 7.18 (d, 1H, J = 7.21 Hz, Ar–H), 6.96 (d, 1H, J = 7.22 Hz, Ar–H), 6.89 (d, 1H, J = 5.32 Hz, Ar–H).
3.4.2.1 N-(4-((7-Chloroquinolin-4-yl)amino)phenyl)quinoline-5-sulfonamide (QS-1). Greenish yellow powder, mp: 248 °C, yield: 85%, Rf (5% MeOH in DCM): 0.41, FTIR-3249, 3059, 1976, 1994, 1618, 1547, 1570, 1514, 1331.1149. 1H NMR (400 MHz, DMSO d6) δ 10.12 (s, 1H), 9.19 (dd, J = 4.2, 1.8 Hz, 1H), 8.92 (s, 1H), 8.54 (dd, J = 8.4, 1.8 Hz, 1H), 8.42–8.24 (m, 4H), 7.85 (d, J = 2.2 Hz, 1H), 7.79–7.69 (m, 2H), 7.51 (dd, J = 9.0, 2.3 Hz, 1H), 7.11 (s, 4H), 6.62 (d, J = 5.4 Hz, 1H). 13C NMR (101 MHz, DMSO d6) δ 152.20, 151.96, 149.73, 148.62, 143.21, 137.50, 136.36, 135.69, 134.71, 134.49, 134.36, 132.60, 128.87, 127.93, 126.14, 125.31, 124.73, 124.18, 123.15, 121.83, 118.44, 101.70. Chemical formula: C24H17ClN4O2S, exact mass: 460.0761, MS: 461.15 [M + H]+.
3.4.2.2 N-(4-((7-Chloroquinolin-4-yl)amino)phenyl)-4-fluorobenzenesulfonamide (QS-2). Whitish crystalline powder, mp: 245 °C, yield: 91%, Rf (5% MeOH in DCM): 0.39, FTIR-3368, 2981, 1741, 1618, 1577, 1514, 1331, 1238, 1167; 1H NMR (400 MHz, DMSO d6) δ 10.30 (s, 1H), 9.03 (s, 1H), 8.43 (d, J = 5.3 Hz, 1H), 8.38 (d, J = 9.0 Hz, 1H), 7.91–7.79 (m, 3H), 7.56 (dd, J = 9.0, 2.3 Hz, 1H), 7.49–7.38 (m, 2H), 7.30–7.22 (m, 2H), 7.19–7.11 (m, 2H), 6.77 (d, J = 5.4 Hz, 1H); 13C NMR (101 MHz, DMSO d6) δ 166.03, 163.53, 152.39, 149.96, 148.49, 137.16, 136.29, 136.26, 134.36, 133.93, 130.29, 130.19, 128.10, 125.37, 124.80, 124.26, 122.64, 118.62, 117.04, 116.81, 101.96. Chemical formula: C21H15ClFN3O2S, exact mass: 427.0558, MS: 428.11 [M + H]+.
3.4.2.3 N-(4-((7-Chloroquinolin-4-yl)amino)phenyl)-4-methoxybenzenesulfonamide (QS-3). Yellowish cream crystalline powder, mp: 234 °C, yield: 92%, Rf (5% MeOH in DCM): 0.42, FTIR-3368, 2925, 1741, 1581, 1331, 1261, 1156. 1H NMR (400 MHz, DMSO d6)10.14 (s, 1H), 9.01 (s, 1H), 8.45–8.30 (m, 2H), 7.88 (d, J = 2.3 Hz, 1H), 7.75–7.67 (m, 2H), 7.55 (dd, J = 9.0, 2.3 Hz, 1H), 7.28–7.20 (m, 2H), 7.18–7.10 (m, 2H), 7.13–7.04 (m, 2H), 6.74 (d, J = 5.4 Hz, 1H), 3.81 (s, 3H); 13C NMR (101 MHz, DMSO d6) δ 162.87, 152.37, 149.94, 148.60, 136.67, 134.52, 134.35, 131.60, 129.38, 128.08, 125.34, 124.80, 124.36, 122.12, 118.57, 114.82, 101.85, 56.10. Chemical formula: C22H18ClN3O3S, exact mass: 439.0757, MS: 440.18 [M + H]+.
3.4.2.4 N-(4-((7-Chloroquinolin-4-yl)amino)phenyl)naphthalene-2-sulfonamide (QS-4). Yellowish crystalline powder, mp: 232 °C, yield: 92%, Rf (5% MeOH in DCM): 0.41, FTIR-3379, 3041, 2981, 1737, 1577, 1514, 1331, 1235, 1160. 1H NMR (400 MHz, DMSO d6) δ 10.39 (s, 1H), 8.98 (s, 1H), 8.45 (s, 1H), 8.41–8.31 (m, 2H), 8.14 (t, J = 9.2 Hz, 2H), 8.03 (d, J = 8.0 Hz, 1H), 7.90–7.79 (m, 3H), 7.75–7.62 (m, 1H), 7.53 (dd, J = 9.1, 2.3 Hz, 1H), 7.25–7.15 (m, 4H), 6.69 (d, J = 5.4 Hz, 1H); 13C NMR (101 MHz, DMSO d6) δ 170.80, 152.32, 149.91, 148.54, 136.92, 134.72, 134.33, 134.20, 132.02, 129.89, 129.70, 129.43, 128.47, 128.31, 128.16, 128.06, 125.32, 124.77, 124.35, 122.57, 122.45, 118.54, 101.85, chemical formula: C25H18ClN3O2S, exact mass: 459.0808, MS: 460.08 [M + H]+.
3.4.2.5 N-(4-((7-Chloroquinolin-4-yl)amino)phenyl)-4-isopropylbenzenesulfonamide (QS-5). Yellow powder, mp: 228 °C, yield: 92%, Rf (5% MeOH in DCM): 0.42, FTIR-3387, 3061, 2970, 1748, 1581, 1510, 1331, 1156. 1H NMR (400 MHz, DMSO d6) δ 10.26 (s, 1H), 9.01 (s, 1H), 8.44–8.34 (m, 2H), 7.88 (d, J = 2.3 Hz, 1H), 7.75–7.68 (m, 2H), 7.55 (dd, J = 9.1, 2.3 Hz, 1H), 7.48–7.40 (m, 2H), 7.28–7.20 (m, 2H), 7.20–7.12 (m, 2H), 6.74 (d, J = 5.4 Hz, 1H), 2.95 (hept, J = 7.1 Hz, 1H), 1.19 (d, J = 6.9 Hz, 6H); 13C NMR (101 MHz, DMSO d6) δ 154.09, 152.36, 149.95, 148.59, 137.60, 136.65, 134.44, 134.34, 128.10, 127.64, 127.32, 125.34, 124.79, 124.39, 121.93, 118.57, 101.84, 60.23, 40.60, 33.79, 23.87 (s), chemical formula: C24H22ClN3O2S, exact mass: 451.1121, MS: 452.26 [M + H]+.
3.4.2.6 N-(4-((7-Chloroquinolin-4-yl)amino)phenyl)–[1,1′-biphenyl]-4-sulfonamide (QS-6). Yellow crystalline powder, mp: 236 °C, yield: 92%, Rf (5% MeOH in DCM): 0.42, FTIR-3346, 3067, 2985, 1734, 1577, 1510, 1331, 1164. 1H NMR (400 MHz, DMSO d6) δ 10.35 (s, 1H), 9.03 (s, 1H), 8.43–8.33 (m, 2H), 7.87 (d, J = 4.1 Hz, 4H), 7.73 (d, J = 8.5 Hz, 1H), 7.58–7.48 (m, 2H), 7.50–7.41 (m, 1H), 7.26 (d, J = 9.0 Hz, 2H), 7.19 (d, J = 9.0 Hz, 2H), 6.75 (d, J = 5.4 Hz, 1H); 13C NMR (101 MHz, DMSO d6) δ 152.31 (s), 149.85 (s), 148.58 (s), 144.72 (s), 138.76 (d, J = 6.4 Hz), 136.87 (s), 134.30 (d, J = 15.2 Hz), 129.60 (s), 127.86 (s), 127.53 (s), 125.37 (s), 124.80 (s), 124.35 (s), 122.23 (s), 118.57 (s), chemical formula: C27H20ClN3O2S, exact mass: 485.0965, MS: 486.10 [M + H]+.
3.4.2.7 N-(3-((7-Chloroquinolin-4-yl)amino)phenyl)quinoline-5-sulfonamide (QS-7). Greenish yellow powder, mp: 246 °C, yield: 92%, Rf (5% MeOH in DCM): 0.42, FTIR-3246, 3067, 2970, 1618, 1577, 1510, 1328, 1223, 1149. 1H NMR (400 MHz, DMSO d6) δ 10.12 (s, 2H), 9.18 (d, J = 6.1 Hz, 2H), 8.96 (s, 1H), 8.55 (d, J = 8.4 Hz, 2H), 8.41–8.26 (m, 9H), 7.85 (d, J = 2.3 Hz, 2H), 7.79–7.69 (m, 4H), 7.52 (dd, J = 9.1, 2.3 Hz, 2H), 7.10 (s, 8H), 6.61 (d, J = 5.4 Hz, 2H); 13C NMR (101 MHz, DMSO d6) δ 152.02, 151.96, 149.51, 148.76, 143.21, 137.50, 136.27, 135.69, 134.72, 134.57, 134.46, 132.60, 128.88, 127.75, 126.14, 125.37, 124.76, 124.24, 123.15, 121.81, 118.39, 101.68. Chemical formula: C24H17ClN4O2S, exact mass: 460.076, MS: 461.06 [M + H]+.
3.4.2.8 N-(3-((7-Chloroquinolin-4-yl)amino)phenyl)-4-fluorobenzenesulfonamide (QS-8). Yellow crystalline powder, mp: 246 °C, yield: 89%, Rf (5% MeOH in DCM): 0.39, FTIR-3391, 2921, 2854, 1741, 1577, 1510, 1331, 1242, 1153. 1H NMR (400 MHz, DMSO d6) δ 10.42 (s, 1H), 9.12 (s, 1H), 8.42 (dd, J = 29.0, 7.2 Hz, 2H), 7.91 (d, J = 2.2 Hz, 1H), 7.86 (dd, J = 8.9, 5.1 Hz, 2H), 7.58 (dd, J = 9.0, 2.2 Hz, 1H), 7.46 (t, J = 8.8 Hz, 2H), 7.29 (t, J = 8.1 Hz, 1H), 7.12 (s, 1H), 7.04 (d, J = 8.0 Hz, 1H), 6.89 (d, J = 8.0 Hz, 1H), 6.73 (d, J = 5.3 Hz, 1H). 13C NMR (101 MHz, DMSO d6) δ 166.11, 163.61, 152.25, 150.03, 147.97, 141.52, 138.94, 136.22, 136.19, 134.49, 130.67, 130.29, 130.19, 128.14, 125.55, 125.01, 118.91, 118.52, 117.17, 116.95, 116.13, 114.08, 102.59, chemical formula: C21H15ClFN3O2S, exact mass: 427.0558, MS: 428.07 [M + H]+.
3.4.2.9 N-(3-((7-Chloroquinolin-4-yl)amino)phenyl)-4-methoxybenzenesulfonamide (QS-9). Yellowish cream, crystalline powder, mp: 226 °C, yield: 88%, Rf (5% MeOH in DCM): 0.41, FTIR-3379, 2925, 1599, 1577, 1328, 1257, 1157. 1H NMR (400 MHz, DMSO d6) δ 10.25 (s, 1H), 9.10 (s, 1H), 8.46–8.35 (m, 2H), 7.90 (d, J = 2.3 Hz, 1H), 7.72 (d, J = 9.0 Hz, 2H), 7.57 (dd, J = 9.0, 2.3 Hz, 1H), 7.27 (t, J = 8.1 Hz, 1H), 7.10 (d, J = 2.1 Hz, 1H), 6.69 (d, J = 5.4 Hz, 1H), 3.81 (s, 3H); 13C NMR (101 MHz, DMSO d6) δ 162.98, 152.23, 150.00, 148.02, 141.36, 139.34, 134.48, 131.48, 130.57, 129.38, 128.11, 125.53, 125.01, 118.87, 118.22, 115.89, 114.94, 113.78, 102.54, 56.15, chemical formula: C22H18ClN3O3S, exact mass: 439.0757, MS: 440.25 [M + H]+.
3.4.2.10 N-(3-((7-Chloroquinolin-4-yl)amino)phenyl)-4-isopropylbenzenesulfonamide (QS-10). White crystalline powder, mp: 227 °C, yield: 91%, Rf (5% MeOH in DCM): 0.41, FTIR- 3383, 3074, 3029, 2966, 1607, 1577, 1477, 1324, 1156. 1H NMR (400 MHz, DMSO d6) δ 10.50 (s, 1H), 9.07 (s, 1H), 8.48 (s, 1H), 8.34 (d, J = 9.1 Hz, 1H), 8.15 (dd, J = 6.8, 4.3 Hz, 3H), 8.05 (d, J = 7.8 Hz, 1H), 7.92–7.78 (m, 2H), 7.78–7.63 (m, 2H), 7.55 (dd, J = 9.0, 2.2 Hz, 1H), 7.27 (t, J = 8.1 Hz, 1H), 7.12 (t, J = 1.9 Hz, 1H), 6.97 (t, J = 7.8 Hz, 2H), 6.51 (d, J = 5.3 Hz, 1H). 13C NMR (101 MHz, DMSO d6) δ 152.06, 149.94, 147.94, 141.36, 139.02, 136.83, 134.77, 134.47, 132.07, 130.64, 130.02, 129.73, 129.55, 128.48, 128.37, 128.27, 128.07, 125.52, 124.96, 122.49, 118.81, 118.54, 116.30, 114.10, 102.32, chemical formula: C25H18ClN3O2S, exact mass: 459.0808, MS: 460.25 [M + H]+.
3.4.2.11 N-(3-((7-Chloroquinolin-4-yl)amino)phenyl)-4-isopropylbenzenesulfonamide (QS-11). Creamish white powder, mp: 222 °C, yield: 87%, Rf (5% MeOH in DCM): 0.41, FTIR-3372, 3031, 1607, 1577, 1328, 1265, 1156. 1H NMR (400 MHz, DMSO d6) δ 10.39 (s, 1H), 9.11 (s, 1H), 8.42 (dd, J = 24.4, 7.2 Hz, 2H), 7.91 (d, J = 2.2 Hz, 1H), 7.74 (d, J = 8.4 Hz, 2H), 7.57 (dd, J = 9.0, 2.2 Hz, 1H), 7.47 (d, J = 8.4 Hz, 2H), 7.27 (t, J = 8.1 Hz, 1H), 7.15 (s, 1H), 7.01 (d, J = 8.0 Hz, 1H), 6.91 (d, J = 8.0 Hz, 1H), 6.74 (d, J = 5.3 Hz, 1H), 2.95 (hept, J = 6.9 Hz, 1H), 1.18 (d, J = 6.9 Hz, 6H). 13C NMR (101 MHz, DMSO d6) δ 154.24, 152.25, 150.04, 147.95, 141.45, 139.30, 137.52, 134.48, 130.60, 128.13, 127.75, 127.31, 125.53, 125.01, 118.9, 118.04, 115.47, 113.34, 102.60, 33.80, 23.85, chemical formula: C24H22ClN3O2S, exact mass: 451.1121 MS: 452.26 [M + H]+.
3.4.2.12 (3-((7-Chloroquinolin-4-yl)amino)phenyl)–[1,1′-biphenyl]-4-sulfonamide (QS-12). Cream powder, mp: 225 °C, yield: 87%, Rf (5% MeOH in DCM): 0.41, FTIR-3372, 3009, 1603, 1577, 1480, 1331, 1156. 1H NMR (400 MHz, DMSO d6) δ 10.49 (s, 1H), 9.24 (s, 2H), 8.40 (d, J = 9.0 Hz, 2H), 7.95–7.85 (m, 7H), 7.73 (d, J = 8.9 Hz, 3H), 7.60 (t, J = 11.6 Hz, 1H), 7.50 (t, J = 7.4 Hz, 3H), 7.49–7.40 (m, 1H), 7.31 (t, J = 8.1 Hz, 2H), 7.17 (s, 1H), 7.04 (d, J = 5.8 Hz, 1H), 6.96 (d, J = 8.0 Hz, 1H), 6.72 (d, J = 5.5 Hz, 2H); 13C NMR (101 MHz, DMSO d6) δ 172.51, 151.69, 149.33, 148.41, 144.85, 141.31, 139.17, 138.66, 134.77, 130.72, 129.62, 129.40, 129.12, 127.95, 127.87, 127.59, 127.54, 127.19, 126.62, 126.47, 125.69, 125.09, 118.75, 118.47, 116.07, 113.89, 102.47, chemical formula: C27H20ClN3O2S, exact mass: 485.0965, MS: 486.23 [M + H]+.
4. Conclusions
In summary, a series of quinoline–sulfonamide hybrids compounds with a substituted core was synthesized in a laboratory setting and investigated for their antibacterial properties. These compounds underwent testing on both Gram-positive bacterial strains, specifically E. faecalis, and Gram-negative strains, namely E. coli, P. aeruginosa, and S. typhi. Following a sequence of experiments to evaluate the antibacterial activity of the compounds, QS3 emerged as a promising antibacterial agent. Notably, when studied in combination with Ciprofloxacin (CIP), QS3 exhibited a synergistic nature against E. faecalis, E. coli, and S. typhi. Consequently, it is evident that QS3 holds the potential for further development as a safe and effective antibacterial agent.Data availability
A data generated or analysed in this study are included in this article.Author contributions
MA, IA and AA: conceptualized and designed the study. ZS: synthesize the compounds. IH and AA: performed in vitro studies. ZS, IH and AA: wrote the original draft. MA, MK, AA and IA: reviewed and edited the manuscript. MK, AA and MA: proofread the draft. All authors have read and approved the final version of the manuscript.Conflicts of interest
The authors declared no conflict of interest.Acknowledgements
Mohammad Abid gratefully acknowledges the financial support in the form of Core Research Grant from Science & Engineering Research Board (SERB), Govt. of India (Project No. CRG/2018/003967). Zohaib Saifi acknowledges the financial support from the University Grants Commission (UGC), Govt. of India in the form of non-NET Fellowship (UGC-Ref. No. 108/(CSIR-UGC NET DEC.2017)). Asghar Ali acknowledges the Department of Health (DHR), Govt. of India Project (File No. R.12014/61/2022-HR). M. K. also acknowledges the SERB-CRG grant (Project No. CRG/2022/003895).).References
- T. Xu, X. Yan, A. Kang, L. Yang, X. Li, Y. Tian, R. Yang, S. Qin and Y. Guo, J. Med. Chem., 2024, 67, 9302–9317 CrossRef CAS PubMed.
- EClinicalMedicine, EClinicalMedicine, 2021, 41, 101221 CrossRef CAS PubMed.
- K. Sado, K. Keenan, A. Manataki, M. Kesby, M. F. Mushi, S. E. Mshana, J. R. Mwanga, S. Neema, B. Asiimwe, J. Bazira, J. Kiiru, D. L. Green, X. Ke, A. Maldonado-Barragán, M. Abed Al Ahad, K. J. Fredricks, S. H. Gillespie, W. Sabiiti, B. T. Mmbaga, G. Kibiki, D. Aanensen, V. A. Smith, A. Sandeman, D. J. Sloan and M. T. G. Holden, PLoS Glob. Public Health, 2024, 4, e0002709 CrossRef PubMed.
- C. Maria, A. M. de Matos and A. P. Rauter, Curr. Opin. Chem. Biol., 2024, 78, 102419 CrossRef CAS PubMed.
- F. Li, J. G. Collins and F. R. Keene, Chem. Soc. Rev., 2015, 44, 2529–2542 RSC.
- I.-A. Lungu, O.-L. Moldovan, V. Biriş and A. Rusu, Pharmaceutics, 2022, 14, 1749 CrossRef CAS PubMed.
- M. Hilf, V. L. Yu, J. Sharp, J. J. Zuravleff, J. A. Korvick and R. R. Muder, Am. J. Med., 1989, 87, 540–546 CrossRef CAS PubMed.
- G. Dhanda, Y. Acharya and J. Haldar, ACS Omega, 2023, 8, 10757–10783 CrossRef CAS PubMed.
- S. M. Drawz and R. A. Bonomo, Clin. Microbiol. Rev., 2010, 23, 160–201 CrossRef CAS PubMed.
- T. S. Ibrahim, A. J. Almalki, A. H. Moustafa, R. M. Allam, G. E.-D. A. Abuo-Rahma, H. I. El Subbagh and M. F. A. Mohamed, Bioorg. Chem., 2021, 111, 104885 CrossRef CAS PubMed.
- P. de Sena Murteira Pinheiro, L. S. Franco, T. L. Montagnoli and C. A. M. Fraga, Expet Opin. Drug Discov., 2024, 19, 451–470 CrossRef CAS PubMed.
- L. Drago, Microorganisms, 2024, 12, 649 CrossRef CAS PubMed.
- A. I. Anwar, L. Lu, C. J. Plaisance, C. P. Daniel, C. J. Flanagan, D. M. Wenger, D. McGregor, G. Varrassi, A. M. Kaye and A. D. Kaye, Cureus, 2024, 16, e54565 Search PubMed.
- M. P. Ambatkar, N. R. Rarokar and P. B. Khedekar, Clin. Complement. Med. Pharmacol., 2023, 3, 100102 CrossRef.
- S. Akhter, O. Concepcion, A. F. de la Torre, A. Ali, A. R. Raza, R. Eman, M. Khalid, M. F. ur Rehman, M. S. Akram and H. M. Ali, Arab. J. Chem., 2023, 16, 104570 CrossRef CAS.
- A. Saxena, S. Majee, D. Ray and B. Saha, Bioorg. Med. Chem., 2024, 103, 117681 CrossRef CAS PubMed.
- B. Çiftci, S. Ökten, Ü. M. Koçyiğit, V. E. Atalay, M. Ataş and O. Çakmak, Eur. J. Med. Chem. Res., 2024, 10, 100127 Search PubMed.
- K. El Gadali, M. Rafya, A. El Mansouri, M. Maatallah, A. Vanderlee, A. Mehdi, J. Neyts, D. Jochmans, S. De Jonghe, F. Benkhalti, Y. S. Sanghvi, M. Taourirte and H. B. Lazrek, Eur. J. Med. Chem., 2024, 268, 116235 CrossRef PubMed.
- H.-X. Li, X.-F. Luo, P. Deng, S.-Y. Zhang, H. Zhou, Y. Y. Ding, Y.-R. Wang, Y.-Q. Liu and Z.-J. Zhang, J. Agric. Food Chem., 2023, 71, 2301–2312 CrossRef CAS PubMed.
- P. Králová and M. Soural, Eur. J. Med. Chem., 2024, 269, 116287 CrossRef PubMed.
- R. C. Ochu, U. C. Okoro, J. Conradie and D. I. Ugwu, Eur. J. Med. Chem. Res., 2024, 10, 100136 CAS.
- B. D. Bax, P. F. Chan, D. S. Eggleston, A. Fosberry, D. R. Gentry, F. Gorrec, I. Giordano, M. M. Hann, A. Hennessy, M. Hibbs, J. Huang, E. Jones, J. Jones, K. K. Brown, C. J. Lewis, E. W. May, M. R. Saunders, O. Singh, C. E. Spitzfaden, C. Shen, A. Shillings, A. J. Theobald, A. Wohlkonig, N. D. Pearson and M. N. Gwynn, Nature, 2010, 466, 935–940 CrossRef PubMed.
- K. Douadi, S. Chafaa, T. Douadi, M. Al-Noaimi and I. Kaabi, J. Mol. Struct., 2020, 1217, 128305 CrossRef CAS.
- A. Ali, P. Hasan, M. Irfan, A. Uddin, A. Khan, J. Saraswat, R. Maguire, K. Kavanagh, R. Patel, M. C. Joshi, A. Azam, M. Mohsin, Q. M. R. Haque and M. Abid, ACS Omega, 2021, 6, 27798–27813 CrossRef CAS PubMed.
- J. Yadav and C. P. Kaushik, Synth. Commun., 2024, 54, 536–552 CrossRef CAS.
- S. Konda, S. Raparthi, K. Bhaskar, R. K. Munaganti, V. Guguloth, L. Nagarapu and D. M. Akkewar, Bioorg. Med. Chem. Lett., 2015, 25, 1643–1646 CrossRef CAS PubMed.
- J. Lal, S. K. Gupta, D. Thavaselvam and D. D. Agarwal, Eur. J. Med. Chem., 2013, 64, 579–588 CrossRef CAS PubMed.
- (a) S.-M. Wang, G.-F. Zha, K. P. Rakesh, N. Darshini, T. Shubhavathi, H. K. Vivek, N. Mallesha and H.-L. Qin, Medchemcomm, 2017, 8, 1173–1189 RSC; (b) S. Bano, K. Javed, S. Ahmad, I. G. Rathish, S. Singh and M. S. Alam, Eur. J. Med. Chem., 2011, 46, 5763–5768 CrossRef CAS PubMed.
- X. Ning, Y. Guo, X. Ma, R. Zhu, C. Tian, Z. Zhang, X. Wang, Z. Ma and J. Liu, Bioorg. Med. Chem., 2013, 21, 5589–5597 CrossRef CAS PubMed.
- H. B. Allen and D. A. Lee, Curr. Med. Res. Opin., 1973, 1, 547–553 CrossRef CAS PubMed.
- (a) X. Zhang, K. P. Rakesh, C. S. Shantharam, H. M. Manukumar, A. M. Asiri, H. M. Marwani and H.-L. Qin, Bioorg. Med. Chem., 2018, 26, 340–355 CrossRef CAS PubMed; (b) A. Thakur, S. Manohar, C. E. Vélez Gerena, B. Zayas, V. Kumar, S. V. Malhotra and D. S. Rawat, Med. Chem. Commun., 2014, 5, 576–586 RSC.
- H. I. Gul, M. Tugrak, H. Sakagami, P. Taslimi, I. Gulcin and C. T. Supuran, J. Enzyme Inhib. Med. Chem., 2016, 31, 1619–1624 CrossRef CAS PubMed.
- V. Akurathi, L. Dubois, S. Celen, N. G. Lieuwes, S. K. Chitneni, B. J. Cleynhens, A. Innocenti, C. T. Supuran, A. M. Verbruggen, P. Lambin and G. M. Bormans, Eur. J. Med. Chem., 2014, 71, 374–384 CrossRef CAS PubMed.
- (a) S. Bag, R. Tulsan, A. Sood, H. Cho, H. Redjeb, W. Zhou, H. LeVine, B. Török and M. Török, Bioorg. Med. Chem. Lett., 2015, 25, 626–630 CrossRef CAS PubMed; (b) M. Xu, Y. Peng, L. Zhu, S. Wang, J. Ji and K. P. Rakesh, Eur. J. Med. Chem., 2019, 180, 656–672 CrossRef CAS PubMed.
- H.-X. Dai, A. F. Stepan, M. S. Plummer, Y.-H. Zhang and J.-Q. Yu, J. Am. Chem. Soc., 2011, 133, 7222–7228 CrossRef CAS PubMed.
- T. Hamaguchi, T. Hirose, H. Asakawa, Y. Itoh, K. Kamado, K. Tokunaga, K. Tomita, H. Masuda, N. Watanabe and M. Namba, Diabetes Res. Clin. Pract., 2004, 66, S129–S132 CrossRef CAS PubMed.
- Y. M. Huang, N. S. Alharbi, B. Sun, C. S. Shantharam, K. P. Rakesh and H.-L. Qin, Eur. J. Med. Chem., 181, 111566 CrossRef CAS PubMed.
- R. Gawin, E. De Clercq, L. Naesens and M. Koszytkowska-Stawińska, Bioorg. Med. Chem., 2008, 16, 8379–8389 CrossRef CAS PubMed.
- (a) H.-L. Qin, Z.-W. Zhang, R. Lekkala, H. Alsulami and K. P. Rakesh, Eur. J. Med. Chem., 2020, 193, 112215 CrossRef CAS PubMed; (b) N. Boechat, L. C. S. Pinheiro, O. A. Santos-Filho and I. C. Silva, Molecules, 2011, 16, 8083–8097 CrossRef CAS PubMed.
- A. Kumar, K. Srivastava, S. Raja Kumar, M. I. Siddiqi, S. K. Puri, J. K. Sexana and P. M. S. Chauhan, Eur. J. Med. Chem., 2011, 46, 676–690 CrossRef CAS PubMed.
- L. C. S. Pinheiro, N. Boechat, M. de, L. G. Ferreira, C. C. S. Júnior, A. M. L. Jesus, M. M. M. Leite, N. B. Souza and A. U. Krettli, Bioorg. Med. Chem., 2015, 23, 5979–5984 CrossRef CAS PubMed.
- A. Daina, O. Michielin and V. Zoete, Sci. Rep., 2017, 7, 42717 CrossRef PubMed.
- L. J. Rather, Q. Zhou, A. Ali, Q. M. R. Haque and Q. Li, ACS Food Sci. Technol., 2021, 1, 427–442 CrossRef CAS.
- R. Sultana, A. Ali, C. Twala, R. Mehandi, M. Rana, D. Yameen, M. Abid and Rahisuddin, J. Biomol. Struct. Dyn., 2023, 41, 13724–13751 CrossRef CAS PubMed.
- R. A. Al-Qawasmeh, M. M. Abadleh, J. A. Zahra, M. M. El-Abadelah, R. Albashiti, F. Zani, M. Incerti and P. Vicini, J. Enzyme Inhib. Med. Chem., 2014, 29, 777–785 CrossRef CAS PubMed.
- S. Abass, S. Zahiruddin, A. Ali, M. Irfan, B. Jan, Q. M. R. Haq, S. A. Husain and S. Ahmad, Curr. Microbiol., 2022, 79, 223 CrossRef CAS PubMed.
- K. H. Rand, H. J. Houck, P. Brown and D. Bennett, Antimicrob. Agents Chemother., 1993, 37, 613–615 CrossRef CAS PubMed.
- I. Irfan, A. Ali, B. Reddi, M. A. Khan, P. Hasan, S. Ahmed, A. Uddin, M. Piatek, K. Kavanagh, Q. M. R. Haque, S. Singh, A. Addlagatta and M. Abid, Antibiotics, 2022, 11, 1126 CrossRef CAS PubMed.
- S. M. Khirallah, H. M. M. Ramadan, A. Shawky, S. H. Qahl, R. S. Baty, N. Alqadri, A. M. Alsuhaibani, M. Jaremko, A.-H. Emwas and E. M. Saied, Molecules, 2022, 27, 6271 CrossRef CAS PubMed.
- S. M. Khirallah, H. M. M. Ramadan, H. A. A. Aladl, N. O. Ayaz, L. A. F. Kurdi, M. Jaremko, S. Z. Alshawwa and E. M. Saied, Pharmaceuticals, 2022, 15, 1576 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra05069j |
| This journal is © The Royal Society of Chemistry 2025 |