Open Access Article
Open Access ArticleShearinines U–Y, indole diterpenoids from an entomogenous fungus, Penicillium sp.†
Peinan Fu a,
Feng Guoa,
Tingnan Zhoub,
Hongxia Beia,
Huiling Tanga,
Jiaxin Lia and
Zhiyang Lv*a
a,
Feng Guoa,
Tingnan Zhoub,
Hongxia Beia,
Huiling Tanga,
Jiaxin Lia and
Zhiyang Lv*a
aSchool of Pharmacy, Jiangsu Food and Pharmaceutical Science College, Huaian 223003, People's Republic of China. E-mail: lvpharm@163.com
bInstitute of Medicinal Biotechnology, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100730, People's Republic of China
First published on 24th January 2025
Abstract
Five new indole diterpenoids, shearinines U–Y (2–6), and the known compound 22,23-dehydro-shearinine A (1) were isolated from cultures of the entomogenous fungus Penicillium sp. Their structures were elucidated primarily by NMR experiments. The absolute configurations of 2–6 were assigned by electronic circular dichroism calculations, respectively. Compounds 1–3 showed moderate cytotoxicity, with IC50 values of 11.7–26.2 μM.
Introduction
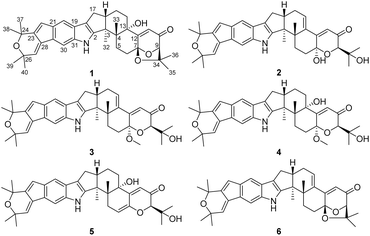
Indole diterpenoids (IDTs) represent a class of structurally diverse and biologically active natural products.1,2 Most reported IDTs are isolated from fungi within the genera Penicillium,3–5 Aspergillus,6Claviceps,7 and Epichloe.8 These intriguing molecules are characterized by a fused diterpenoid backbone and an indole ring, which can be further elaborated through cyclization, oxidation, and prenylation to yield a plethora of complex architectures.9–12 These natural products have been reported to exhibit a range of biological activities, such as antibacterial, antifungal, antitumor, and antiviral effects.13–17The species of fungal genus Penicillium are known to be prolific producers of highly diverse secondary metabolites, which contribute to their multifaceted biological profiles.18–22 During our continuous search for new cytotoxic metabolites from the entomogenous fungus fungi,23–25 a strain of Penicillium sp., isolated from a wheat cyst nematode in Xiao County, Anhui, People's Republic of China, was grown in a solid-substrate fermentation culture. An ethyl acetate (EtOAc) extract of the culture showed cytotoxic effects towards a small panel of four tumor cell lines. Fractionation of the extract afforded five new indole diterpenoids, which we named shearinines U–Y (2–6; Fig. 1), and the known compound 22,23-dehydro-shearinine A (1; Fig. 1). Details of the isolation, structure elucidation, and cytotoxicity evaluation of these compounds are reported herein.
Results and discussion
The known compound 1 was identified as 22,23-dehydro-shearinine A by comparison of its MS and NMR data (Fig. S55 and S56†) with those previously reported.26Shearinine U (2) was assigned a molecular formula of C37H43NO5 by HRESIMS, indicating 17° of unsaturation. The 1H and 13C NMR data for 2 (Tables 1 and 2) closely resembled those for 1, apart from two additional hydroxy signals resonating at δH 6.55 and 4.31. The HMBC correlations from OH-7 to C-7 and C-12, from OH-34 to C-9 and C-34 suggested cleavage of the 1,3-dioxolane ring in the structure of 1. Moreover, 2 displays an olefinic signal (a sp2 carbon at δC 140.7 and δH 6.06/δC 130.4), instead of a 13-OH group, that were assigned to the double bond at Δ13(14), which was confirmed by the HMBC correlation from H2-15 to C-13. On the basis of these data, the planar structure of 2 was elucidated.
| No. | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|
| δHa (J in Hz) | δHa (J in Hz) | δHa (J in Hz) | δHa (J in Hz) | δHa (J in Hz) | |
| a Recorded at 600 MHz. | |||||
| 5a | 2.06, m | 1.92, m | 2.39, m | 3.00, brd (16.5) | 1.98, m |
| 5b | 1.81, m | 2.41, dd (17.3, 6.2) | |||
| 6a | 2.08, m | 2.08, m | 2.49, m | 5.64, m | 1.84, m |
| 6b | 1.97, m | 1.87, m | |||
| 9 | 4.14, s | 3.92, s | 3.92, s | 4.02, s | 4.43, s |
| 11 | 5.85, s | 5.90, s | 5.78, s | 5.80, s | 6.15, s |
| 14 | 6.06, dd (5.0, 2.3) | 6.03, dd (5.0, 2.3) | 1.78, m | 1.93, m | 6.52, dd (4.9, 2.6) |
| 1.66, m | 1.82, m | ||||
| 15 | 2.27, m | 2.27, m | 1.91, m | 1.92, m | 2.38, m |
| 1.60, m | 1.65, m | ||||
| 16 | 2.80, m | 2.81, m | 2.71, m | 2.70, m | 2.87, m |
| 17 | 2.74, dd (13.0, 6.7) | 2.74, dd (13.0, 6.7) | 2.60, dd (12.9, 6.1) | 2.60, dd (12.9, 6.2) | 2.73, dd (13.2, 6.7) |
| 2.37, dd (13.0, 10.1) | 2.38, dd (13.0, 10.2) | 2.30, dd (12.9, 11.4) | 2.31, dd (12.9, 11.4) | 2.37, dd (13.2, 10.6) | |
| 20 | 7.11, s | 7.11, s | 7.07, s | 7.07, s | 7.11, s |
| 22 | 6.44, d (1.8) | 6.45, d (1.8) | 6.43, d (1.7) | 6.43, d (1.2) | 6.45, d (1.6) |
| 27 | 6.74, d (1.8) | 6.74, d (1.8) | 6.71, d (1.7) | 6.71, d (1.2) | 6.74, d (1.6) |
| 30 | 7.49, s | 7.50, s | 7.46, s | 7.47, s | 7.52, s |
| 32 | 0.92, s | 0.90, s | 1.25, s | 1.29, s | 0.97, s |
| 33 | 1.09, s | 1.12, s | 0.94, s | 1.01, s | 1.17, s |
| 35 | 1.19, s | 1.23, s | 1.23, s | 1.13, s | 1.39, s |
| 36 | 1.19, s | 1.22, s | 1.20, s | 1.25, s | 1.14, s |
| 37 | 1.45, s | 1.46, s | 1.45, s | 1.45, s | 1.46, s |
| 38 | 1.45, s | 1.46, s | 1.45, s | 1.45, s | 1.46, s |
| 39 | 1.40, s | 1.40, s | 1.39, s | 1.39, s | 1.39, s |
| 40 | 1.40, s | 1.40, s | 1.39, s | 1.39, s | 1.40, s |
| 1-NH | 10.86, s | 10.87, s | 10.74, s | 10.76, s | 10.87, s |
| 7-OH | 6.55, s | ||||
| 7-OMe | 3.26, s | 3.37, s | |||
| 13-OH | 4.57, s | 4.90, s | |||
| 34-OH | 4.31, s | 4.40, s | 4.45, s | 4.54, s | |
| No. | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|
| δCa, type | δCa, type | δCa, type | δCa, type | δCa, type | |
| a Recorded at 150 MHz. | |||||
| 2 | 148.9, qC | 148.8, qC | 152.8, qC | 152.9, qC | 148.7, qC |
| 3 | 47.9, qC | 47.9, qC | 50.4, qC | 50.3, qC | 49.0, qC |
| 4 | 42.1, qC | 42.0, qC | 42.3, qC | 42.8, qC | 38.9, qC |
| 5 | 28.1, CH2 | 26.7, CH2 | 24.8, CH2 | 30.6, CH2 | 29.0, CH2 |
| 6 | 32.9, CH2 | 27.4, CH2 | 27.2, CH2 | 110.9, qC | 30.6, CH2 |
| 7 | 93.5, qC | 96.5, qC | 97.0, qC | 145.1, qC | 103.9, qC |
| 9 | 78.3, CH | 78.6, CH | 78.4, CH | 85.9, CH | 87.4, CH |
| 10 | 198.7, qC | 197.1, qC | 197.0, qC | 195.1, qC | 195.1, qC |
| 11 | 120.7, CH | 121.2, CH | 121.2, CH | 116.0, CH | 117.6, CH |
| 12 | 161.4, qC | 159.8, qC | 160.2, qC | 154.5, qC | 163.9, qC |
| 13 | 140.7, qC | 140.5, qC | 75.6, qC | 73.9, qC | 138.4, qC |
| 14 | 130.4, CH | 130.7, CH | 32.5, CH2 | 31.8, CH2 | 132.9, CH |
| 15 | 27.5, CH2 | 27.5, CH2 | 20.9, CH2 | 21.1, CH2 | 28.2, CH2 |
| 16 | 44.9, CH | 44.9, CH | 49.1, CH | 49.2, CH | 44.7, CH |
| 17 | 26.7, CH2 | 26.3, CH2 | 26.8, CH2 | 27.0, CH2 | 26.8, CH2 |
| 18 | 117.0, qC | 117.0, qC | 115.8, qC | 115.9, qC | 117.1, qC |
| 19 | 124.2, qC | 124.1, qC | 124.3, qC | 124.5, qC | 124.2, qC |
| 20 | 109.8, CH | 109.7, CH | 109.5, CH | 109.6, CH | 109.7, CH |
| 21 | 135.5, qC | 135.5, qC | 135.3, qC | 135.3, qC | 135.6, qC |
| 22 | 121.3, CH | 121.3, CH | 121.2, CH | 121.3, CH | 121.3, CH |
| 23 | 140.9, qC | 140.9, qC | 140.7, qC | 140.9, qC | 141.0, qC |
| 24 | 72.5, qC | 72.5, qC | 72.3, qC | 72.5, qC | 72.5, qC |
| 26 | 72.4, qC | 72.4, qC | 72.2, qC | 72.4, qC | 72.4, qC |
| 27 | 129.1, CH | 129.1, CH | 128.6, CH | 128.9, CH | 129.1, CH |
| 28 | 133.5, qC | 133.5, qC | 133.4, qC | 133.5, qC | 133.5, qC |
| 29 | 127.6, qC | 127.6, qC | 127.0, qC | 127.2, qC | 127.6, qC |
| 30 | 104.9, CH | 104.9, CH | 104.7, CH | 104.9, CH | 105.8, CH |
| 31 | 138.5, qC | 138.5, qC | 137.7, qC | 138.0, qC | 138.5, qC |
| 32 | 15.0, CH3 | 15.0, CH3 | 16.2, CH3 | 16.5, CH3 | 14.4, CH3 |
| 33 | 22.5, CH3 | 22.6, CH3 | 19.0, CH3 | 19.7, CH3 | 27.5, CH3 |
| 34 | 71.4, qC | 71.1, qC | 70.9, qC | 73.0, qC | 77.7, qC |
| 35 | 26.5, CH3 | 26.0, CH3 | 26.1, CH3 | 26.8, CH3 | 28.8, CH3 |
| 36 | 25.8, CH3 | 26.0, CH3 | 25.6, CH3 | 27.1, CH3 | 22.9, CH3 |
| 37 | 31.4, CH3 | 31.4, CH3 | 31.3, CH3 | 31.4, CH3 | 31.5, CH3 |
| 38 | 31.4, CH3 | 31.4, CH3 | 31.3, CH3 | 31.5, CH3 | 31.4, CH3 |
| 39 | 30.7, CH3 | 30.7, CH3 | 30.6, CH3 | 30.8, CH3 | 30.8, CH3 |
| 40 | 30.7, CH3 | 30.7, CH3 | 30.6, CH3 | 30.7, CH3 | 30.7, CH3 |
| 7-OMe | 48.5, CH3 | 48.8, CH3 | |||
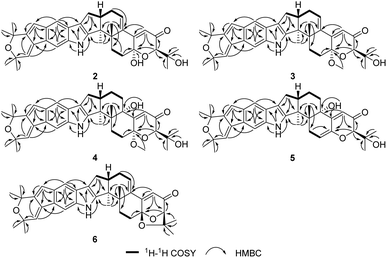
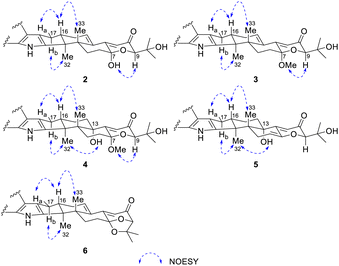
The relative configuration of 2 was proposed by analysis of NOESY correlations (Fig. 2 and 3). NOESY correlations of H-16 with H-17a and H3-33 indicated that these protons are all on the same face of the ring system, whereas those of H-17b with H3-32 placed the protons on the opposite face. The other NOESY correlations of OH-7 with H-9 placed these protons on the same face. Therefore, the relative configuration of 2 was deduced as shown.
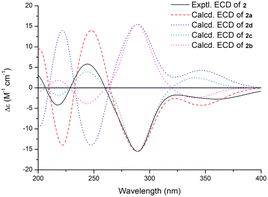
The absolute configuration of 2 was deduced by comparison of the experimental and simulated electronic circular dichroism (ECD) spectra calculated using the time-dependent density functional theory (TDDFT).27 The ECD spectra of the four possible isomers 2a–d (Fig. S11†) were calculated. A systematic conformational analysis was performed for 2a–d with the Molecular Operating Environment (MOE) software package using the MMFF94 molecular mechanics force field calculation. The selected conformers were then reoptimized using TDDFT at the B3LYP/6-311G(d,p) level to afford the lowest energy conformers. The conformers were further filtered based on the Boltzmann-population rule, resulting in one significant conformer for each configuration (Fig. S11†). The overall calculated ECD spectra of 2a–d were then generated by Gaussian broadening (Fig. 4). The experimental ECD curve of 2 was nearly identical to that calculated for 2a, suggesting the 3S,4S,7S,9R,16S absolute configuration for 2.
The molecular formula of Shearinine V (3) was determined to be C38H45NO5 (17° of unsaturation) based on HRESIMS and the NMR data (Tables 1 and 2), which is 14 mass units higher than that of 2. Analysis of the 1H and 13C NMR data for 3 revealed the presence of structural features similar to those found in 2, except that OH-7 (δH 6.55) was replaced by a methoxy unit (δH/δC 3.26/48.5), and this observation was supported by the HMBC correlations from these newly observed methoxy protons to C-7. Therefore, the gross structure of 3 was established. The relative configuration of 3 was also deduced by analysis of NOESY data, and by analogy to 2. The ECD spectrum of 3 was nearly identical to that of 2 (Fig. S10 and S21†), indicating that the absolute configuration of 2 was the same as that of 2.
Shearinine W (4) was assigned a molecular formula of C38H47NO6 (16° of unsaturation) by HRESIMS and the NMR data (Tables 1 and 2), which is 18 mass units higher than that of 3, except that the C-13 oxygenated tertiary carbons (δC 75.6) units and C-14 methylene (δH/δC 1.78, 1.66/32.5) in 4 instead of an olefin moiety in 3, which was confirmed by the HMBC correlations from H-11 to C-13 and from 13-OH to C-12 and C-13. Therefore, the gross structure of 4 was proposed.
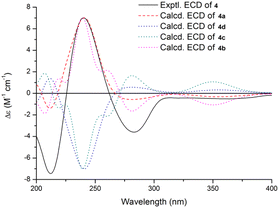
The relative configuration of 4 was also proposed by analysis of NOESY correlations (Fig. 3). The NOESY correlations of H3-32 with 13-OH revealed that these protons are all on the same face of the ring system. The relative configuration of other stereogenic centers were proposed in a similar fashion to 2 and 3 as shown in Fig. 3. The absolute configuration for 4 was also deduced by comparison of the experimental and calculated ECD spectra for the four possible enantiomers 4a–d. The lowest energy conformers for 4a–d were generated by Gaussian broadening (Fig. 5). The experimental ECD spectrum of 4 matched reasonably well with the calculated one for 4a, suggesting that 4 has the 3S,4R,7S,9R,13S,16S absolute configuration.
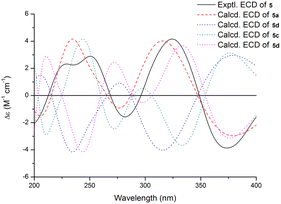
Shearinine X (5) was determined to have a molecular formula of C37H43NO5 (17° of unsaturation) based on HRESIMS and the NMR data (Tables 1 and 2). The 1H and 13C NMR spectra of 5 showed resonances similar to those of 4, revealed that 5 displays an olefinic signal (δH 5.64/δC 110.9 and a sp2 carbon at δC 145.1), instead of a 7-OMe group, that were assigned to the double bond at Δ6(7),accounting for the additional degree of unsaturation of 5, which was confirmed by the HMBC correlation from H-5 to C-7. The relative configurations of all stereogenic centers in 5 were proposed to be the same as their counterparts in 4 by analysis of the NOESY data (Fig. 3) for relevant protons, and the absolute configuration was deduced to be 3S,4R,9R,13S,16S based on the results of experimental and calculated ECD (Fig. 6).
On the IR spectra, the higher wavenumber region of the spectrum of compounds 2–5 did not contain a diagnostic strong and sharp band above 3200 cm−1 that is typical of the N–H stretching vibration of an indole N–H group. Instead, a very broad band was attributed to the O–H (H-bonded).28
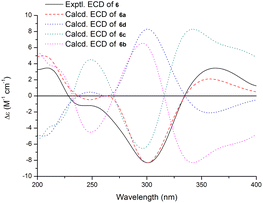
Shearinine Y (6) gave a molecular formula of C37H41NO4 (18° of unsaturation) by analysis of its HRESIMS and the NMR data (Tables 1 and 2), which is 18 mass units less than that of 2. Analysis of the 1H and 13C NMR data for 6 revealed the presence of structural features similar to those found in 2, except that 7-OH (δH 6.55) and 34-OH (δH 4.31) were absent. Considering the unsaturation requirement for 6, allowed the deduction of a dioxolane moiety fused to the pyranone at C-7 and C-9. Therefore, the gross structure of 6 was established. The relative configuration of 6 was deduced as shown by analysis of the NOESY correlations (Fig. 3) and by analogy to 2, and the absolute configuration of 6 was further determined by comparison of the experimental and calculated ECD spectra. The experimental ECD of 6 correlated well to the calculated curve of 6a (Fig. 7), suggesting the 3S,4S,7S,9R,16S absolute configuration.
An additional experiment was performed to exclude the possibility that 3 and 4 are artifacts arising from their corresponding analogues during the isolation process. For this purpose, EtOAc extract were soaked in acetonitrile, and then analyzed by HPLC with CH3CN–H2O, which was found that there were still 3 and 4 peaks in the fermentation products. The result indicated that the new methoxylated analogues 3 and 4 are true natural products and not artifacts formed through methylation when using MeOH (Table 3).
| Compound | IC50a (μM) | |||
|---|---|---|---|---|
| Huh7 | SKBR3 | A549 | MB49 | |
| a IC50 values were averaged from at least three independent experiments.b No activity was detected at 50 μM.c Positive control. | ||||
| 1 | 11.7 ± 2.0 | 26.2 ± 3.5 | 22.0 ± 2.1 | 18.6 ± 2.3 |
| 2 | 11.8 ± 2.2 | 21.8 ± 4.0 | 22.1 ± 2.7 | 17.1 ± 0.8 |
| 3 | 18.1 ± 4.0 | 21.0 ± 4.8 | NAb | NAb |
| Cisplatinc | 5.2 ± 0.7 | 3.7 ± 0.5 | 6.5 ± 0.5 | 1.6 ± 0.4 |
Compounds 1–6 were tested for cytotoxicity against four tumor cell lines, Huh7 (human hepatocellular carcinoma cells), SKBR3 (human breast cancer cells), A549 (human lung adenocarcinoma cells), and MB49 (mouse bladder carcinoma cells).29 Compounds 1–3 showed moderate cytotoxic effects, with IC50 values of 11.7–26.2 μM (the positive control cisplatin showed IC50 values of 1.6–6.5 μM). However, compounds 4–6 did not show detectable activity at 50 μM.
Experimental
General experimental procedures
Optical rotations were measured on a Rudolph Research Analytical automatic polarimeter, and UV data were recorded on a Shimadzu Biospec-1601 spectrophotometer. CD spectra were recorded on a JASCO J-815 spectropolarimeter. IR data were recorded using a Nicolet Magna-IR 750 spectrophotometer. 1H and 13C NMR spectra were acquired with Bruker Avance III-600 spectrometers using solvent signals (DMSO-d6: δH 2.50/δC 39.52) as references. The HSQC and HMBC experiments were optimized for 145.0 and 8.0 Hz, respectively. ESIMS and HRESIMS data were obtained on an Agilent Accurate-Mass-Q-TOF LC/MS G6550 instrument equipped with an ESI source. HPLC analysis and separation were performed using an Agilent 1260 instrument equipped with a variable-wavelength UV detector.Fungal material
The culture of Penicillium sp. was isolated from a wheat cyst nematode collected in Xiao County, Anhui, People's Republic of China, in 2009. The isolate was identified based on morphology and sequence (GenBank accession no. ON307320) analysis of the ITS region of the rDNA. The fungal strain was cultured on slants of potato dextrose agar (PDA) at 25 °C for 10 days. Agar plugs were cut into small pieces (about 0.5 × 0.5 × 0.5 cm3) under aseptic conditions, and 25 pieces were used to inoculate in five 250 mL Erlenmeyer flasks, each containing 50 mL of media (0.4% glucose, 1% malt extract, and 0.4% yeast extract), and the final pH of the media was adjusted to 6.5 and sterilized by autoclave. Five flasks of the inoculated media were incubated at 25 °C on a rotary shaker at 170 rpm for 5 days to prepare the seed culture. Fermentation was carried out in 40 Fernbach flasks (500 mL) each containing 80 g of rice. Distilled H2O (120 mL) was added to each flask, and the contents were soaked overnight before autoclaving at 15 psi for 30 min. After cooling to room temperature, each flask was inoculated with 5.0 mL of the spore inoculum and incubated at 25 °C for 25 days.Extraction and isolation
The fermentation material was extracted repeatedly with EtOAc (4 × 6.0 L), and the organic solvent was evaporated to dryness under vacuum to afford 20.6 g of crude extract. The crude extract was fractionated by silica gel vacuum liquid chromatography (VLC) using petroleum ether–CH2Cl2–EtOAc–MeOH gradient elution. The fraction (1.4 g) eluted with 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CH2Cl2–EtOAc was separated by reversed-phase silica gel column chromatography (CC) eluting with a MeOH–H2O gradient. The fraction (115 mg) eluted with 80% MeOH–H2O was further separated by Sephadex LH-20 CC eluting with MeOH, and the resulting subfractions were combined and purified by semipreparative RP HPLC (Agilent Zorbax SB-C18 column; 5 μm; 9.4 × 250 mm; 85% CH3CN in H2O for 20 min; 3 mL min−1) to afford 1 (9.5 mg, tR 17.5 min). The fraction (1.8 g) eluted with 4
1 CH2Cl2–EtOAc was separated by reversed-phase silica gel column chromatography (CC) eluting with a MeOH–H2O gradient. The fraction (115 mg) eluted with 80% MeOH–H2O was further separated by Sephadex LH-20 CC eluting with MeOH, and the resulting subfractions were combined and purified by semipreparative RP HPLC (Agilent Zorbax SB-C18 column; 5 μm; 9.4 × 250 mm; 85% CH3CN in H2O for 20 min; 3 mL min−1) to afford 1 (9.5 mg, tR 17.5 min). The fraction (1.8 g) eluted with 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CH2Cl2–EtOAc was separated by reversed-phase silica gel CC eluting with a MeOH–H2O gradient. The fraction (100 mg) eluted with 80% MeOH–H2O was further separated by Sephadex LH-20 CC eluting with MeOH, and the resulting subfractions were combined and purified by semipreparative RP HPLC (Agilent Zorbax SB-C18 column; 5 μm; 9.4 × 250 mm; 75% CH3CN in H2O for 35 min; 3 mL min−1) to afford 5 (5.5 mg, tR 29.5 min). The fraction (2.0 g) eluted with 3
1 CH2Cl2–EtOAc was separated by reversed-phase silica gel CC eluting with a MeOH–H2O gradient. The fraction (100 mg) eluted with 80% MeOH–H2O was further separated by Sephadex LH-20 CC eluting with MeOH, and the resulting subfractions were combined and purified by semipreparative RP HPLC (Agilent Zorbax SB-C18 column; 5 μm; 9.4 × 250 mm; 75% CH3CN in H2O for 35 min; 3 mL min−1) to afford 5 (5.5 mg, tR 29.5 min). The fraction (2.0 g) eluted with 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 CH2Cl2–EtOAc was separated by reversed-phase silica gel CC eluting with a MeOH–H2O gradient. The fraction (105 mg) eluted with 80% MeOH–H2O was further separated by Sephadex LH-20 CC eluting with MeOH, and the resulting subfractions were combined and purified by semipreparative RP HPLC (Agilent Zorbax SB-C18 column; 5 μm; 9.4 × 250 mm; 82% CH3OH in H2O for 30 min; 3 mL min−1) to afford 2 (10.2 mg, tR 27.0 min). The fraction (185 mg) eluted with 90% MeOH–H2O was further separated by Sephadex LH-20 CC eluting with MeOH, and the resulting subfractions were combined and purified by semipreparative RP HPLC (Agilent Zorbax SB-C18 column; 5 μm; 9.4 × 250 mm; 80% CH3CN in H2O for 35 min; 3 mL min−1) to afford 4 (4.0 mg, tR 22.5 min), 3 (4.5 mg, tR 25.0 min), and 6 (1.8 mg, tR 29.5 min).
2 CH2Cl2–EtOAc was separated by reversed-phase silica gel CC eluting with a MeOH–H2O gradient. The fraction (105 mg) eluted with 80% MeOH–H2O was further separated by Sephadex LH-20 CC eluting with MeOH, and the resulting subfractions were combined and purified by semipreparative RP HPLC (Agilent Zorbax SB-C18 column; 5 μm; 9.4 × 250 mm; 82% CH3OH in H2O for 30 min; 3 mL min−1) to afford 2 (10.2 mg, tR 27.0 min). The fraction (185 mg) eluted with 90% MeOH–H2O was further separated by Sephadex LH-20 CC eluting with MeOH, and the resulting subfractions were combined and purified by semipreparative RP HPLC (Agilent Zorbax SB-C18 column; 5 μm; 9.4 × 250 mm; 80% CH3CN in H2O for 35 min; 3 mL min−1) to afford 4 (4.0 mg, tR 22.5 min), 3 (4.5 mg, tR 25.0 min), and 6 (1.8 mg, tR 29.5 min).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 225 (4.06), 282 (4.02) nm; CD (c 3.3 × 10−4 M, MeOH) λmax (Δε) 218 (−4.18), 244 (+5.80), 290 (−15.50) 360 (−2.76) nm; IR (neat) νmax 3295 (br), 2973, 2930, 1657, 1593, 1456, 1380, 1255, 1131, 1023 cm−1; 1H and 13C NMR data see Tables 1 and 2; HMBC data (DMSO-d6, 600 MHz) H2-6 → C-4, 7; H-9 → C-7, 10, 34, 35, 36; H-11 → C-7, 9, 12, 13; H-14 → C-4, 16; H-15a → C-13, 14; H-15b → C-13, 14; H-17a → C-2, 3, 16, 18; H-17b → C-2, 16, 18; H-20 → C-18, 19, 22, 29, 31; H-22 → C-21, 23, 28, 29; H-27 → C-23, 26, 29; H-30 → C-19, 21, 28; H3-32 → C-2, 3, 4, 16; H3-33 → C-3, 4, 5, 13; H3-35 → C-9, 34, 36; H3-36 → C-9, 34, 35; H3-37 → C-23, 24, 38; H3-38 → C-23, 24, 37; H3-39 → C-26, 27, 40; H3-40 → C-26, 27, 39; NH-1 → C-2, 18, 19, 31; OH-7 → C-7, 12; OH-34 → C-9, 34, 35, 36; NOESY correlations (DMSO-d6, 600 MHz) H-9 ↔ OH-7; H-16 ↔ H-17a, H3-33; H-17b ↔ H3-32; HRESIMS m/z 604.3058 [M + Na]+ (calcd for C37H43NO5Na, 604.3033).
ε) 225 (4.06), 282 (4.02) nm; CD (c 3.3 × 10−4 M, MeOH) λmax (Δε) 218 (−4.18), 244 (+5.80), 290 (−15.50) 360 (−2.76) nm; IR (neat) νmax 3295 (br), 2973, 2930, 1657, 1593, 1456, 1380, 1255, 1131, 1023 cm−1; 1H and 13C NMR data see Tables 1 and 2; HMBC data (DMSO-d6, 600 MHz) H2-6 → C-4, 7; H-9 → C-7, 10, 34, 35, 36; H-11 → C-7, 9, 12, 13; H-14 → C-4, 16; H-15a → C-13, 14; H-15b → C-13, 14; H-17a → C-2, 3, 16, 18; H-17b → C-2, 16, 18; H-20 → C-18, 19, 22, 29, 31; H-22 → C-21, 23, 28, 29; H-27 → C-23, 26, 29; H-30 → C-19, 21, 28; H3-32 → C-2, 3, 4, 16; H3-33 → C-3, 4, 5, 13; H3-35 → C-9, 34, 36; H3-36 → C-9, 34, 35; H3-37 → C-23, 24, 38; H3-38 → C-23, 24, 37; H3-39 → C-26, 27, 40; H3-40 → C-26, 27, 39; NH-1 → C-2, 18, 19, 31; OH-7 → C-7, 12; OH-34 → C-9, 34, 35, 36; NOESY correlations (DMSO-d6, 600 MHz) H-9 ↔ OH-7; H-16 ↔ H-17a, H3-33; H-17b ↔ H3-32; HRESIMS m/z 604.3058 [M + Na]+ (calcd for C37H43NO5Na, 604.3033).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 248 (3.97) nm; CD (c 2.0 × 10−4 M, MeOH) λmax (Δε) 216 (−2.35), 243 (+3.61), 290 (−10.71) 358 (−1.99) nm; IR (neat) νmax 3460 (br), 2972, 2933, 1671, 1456, 1375, 1255, 1137, 1033 cm−1; 1H and 13C NMR data see Tables 1 and 2; HMBC data (DMSO-d6, 600 MHz) H-9 → C-7, 10, 34, 35, 36; H-11 → C-7, 9, 13; H-17a → C-2, 3, 18; H-17b → C-2, 3, 18; H-20 → C-18, 22, 29, 31; H-22 → C-21, 28, 29; H-27 → C-23, 26, 29; H-30 → C-19, 21, 28; H3-32 → C-2, 3, 4; H3-33 → C-3, 4, 5, 13; H3-35 → C-9, 34, 36; H3-36 → C-9, 34, 35; H3-37 → C-23, 24, 38; H3-38 → C-23, 24, 37; H3-39 → C-26, 27, 40; H3-40 → C-26, 27, 39; NH-1 → C-2, 18, 19, 31; OCH3-7 → C-7; OH-34 → C-34, 35, 36; NOESY correlations (DMSO-d6, 600 MHz) H-9 ↔ OCH3-7; H-16 ↔ H-17a, H3-33; H-17b ↔ H3-32; HRESIMS m/z 594.3225 [M–H]− (calcd for C38H44NO5, 594.3225).
ε) 248 (3.97) nm; CD (c 2.0 × 10−4 M, MeOH) λmax (Δε) 216 (−2.35), 243 (+3.61), 290 (−10.71) 358 (−1.99) nm; IR (neat) νmax 3460 (br), 2972, 2933, 1671, 1456, 1375, 1255, 1137, 1033 cm−1; 1H and 13C NMR data see Tables 1 and 2; HMBC data (DMSO-d6, 600 MHz) H-9 → C-7, 10, 34, 35, 36; H-11 → C-7, 9, 13; H-17a → C-2, 3, 18; H-17b → C-2, 3, 18; H-20 → C-18, 22, 29, 31; H-22 → C-21, 28, 29; H-27 → C-23, 26, 29; H-30 → C-19, 21, 28; H3-32 → C-2, 3, 4; H3-33 → C-3, 4, 5, 13; H3-35 → C-9, 34, 36; H3-36 → C-9, 34, 35; H3-37 → C-23, 24, 38; H3-38 → C-23, 24, 37; H3-39 → C-26, 27, 40; H3-40 → C-26, 27, 39; NH-1 → C-2, 18, 19, 31; OCH3-7 → C-7; OH-34 → C-34, 35, 36; NOESY correlations (DMSO-d6, 600 MHz) H-9 ↔ OCH3-7; H-16 ↔ H-17a, H3-33; H-17b ↔ H3-32; HRESIMS m/z 594.3225 [M–H]− (calcd for C38H44NO5, 594.3225).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 207 (4.11), 234 (4.24) nm; CD (c 5.0 × 10−4 M, MeOH) λmax (Δε) 212 (−7.45), 240 (+7.02), 283 (−3.61) nm; IR (neat) νmax 3359 (br), 2930, 1669, 1456, 1374, 1255, 1133, 1020 cm−1; 1H and 13C NMR data see Tables 1 and 2; HMBC data (DMSO-d6, 600 MHz) H-9 → C-7, 10, 34, 35, 36; H-11 → C-7, 9, 13; H-17a → C-2, 3, 18; H-17b → C-2, 3, 18; H-20 → C-18, 22, 29, 31; H-22 → C-21, 23, 24, 28, 29; H-27 → C-23, 26, 29, 39, 40; H-30 → C-19, 21, 28; H3-32 → C-2, 3, 4, 16; H3-33 → C-3, 4, 5, 13; H3-35 → C-9, 34, 36; H3-36 → C-9, 34, 35; H3-37 → C-23, 24, 38; H3-38 → C-23, 24, 37; H3-39 → C-26, 27, 40; H3-40 → C-26, 27, 39; NH-1 → C-2, 18, 19, 31; OCH3-7 → C-7; OH-13 → C-4, 12, 13, 14; OH-34 → C-34, 35, 36; NOESY correlations (DMSO-d6, 600 MHz) H-9 ↔ 7-OH; H-16 ↔ H-17a, H3-33; H-17b ↔ H3-32; HRESIMS m/z 614.3478 [M + H]+ (calcd for C38H48NO6, 614.3476).
ε) 207 (4.11), 234 (4.24) nm; CD (c 5.0 × 10−4 M, MeOH) λmax (Δε) 212 (−7.45), 240 (+7.02), 283 (−3.61) nm; IR (neat) νmax 3359 (br), 2930, 1669, 1456, 1374, 1255, 1133, 1020 cm−1; 1H and 13C NMR data see Tables 1 and 2; HMBC data (DMSO-d6, 600 MHz) H-9 → C-7, 10, 34, 35, 36; H-11 → C-7, 9, 13; H-17a → C-2, 3, 18; H-17b → C-2, 3, 18; H-20 → C-18, 22, 29, 31; H-22 → C-21, 23, 24, 28, 29; H-27 → C-23, 26, 29, 39, 40; H-30 → C-19, 21, 28; H3-32 → C-2, 3, 4, 16; H3-33 → C-3, 4, 5, 13; H3-35 → C-9, 34, 36; H3-36 → C-9, 34, 35; H3-37 → C-23, 24, 38; H3-38 → C-23, 24, 37; H3-39 → C-26, 27, 40; H3-40 → C-26, 27, 39; NH-1 → C-2, 18, 19, 31; OCH3-7 → C-7; OH-13 → C-4, 12, 13, 14; OH-34 → C-34, 35, 36; NOESY correlations (DMSO-d6, 600 MHz) H-9 ↔ 7-OH; H-16 ↔ H-17a, H3-33; H-17b ↔ H3-32; HRESIMS m/z 614.3478 [M + H]+ (calcd for C38H48NO6, 614.3476).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 225 (3.66), 283 (3.77) nm; CD (c 1.5 × 10−4 M, MeOH) λmax (Δε) 229 (+2.32), 251 (+2.89), 282 (−1.57), 324 (+4.15), 373 (−3.87) nm; IR (neat) νmax 3390 (br), 2974, 2932, 1661, 1456, 1374, 1257, 1168, 1134 cm−1; 1H and 13C NMR data see Tables 1 and 2; HMBC data (DMSO-d6, 600 MHz) H-5a → C-4, 6, 7, 33; H-5b → C-4, 6, 7, 13, 33; H-9 → C-7, 10, 34, 35, 36; H-11 → C-7, 9, 13; H-14 → C-4, 16; H-15a → C-3, 13; H-15b → C-3, 13; H-17a → C-2, 3, 18; H-17b → C-2, 3, 18; H-20 → C-18, 19, 22, 29, 31; H-22 → C-21, 23, 28, 29; H-27 → C-23, 26, 29, 39, 40; H-30 → C-19, 21, 28; H3-32 → C-2, 3, 4, 16; H3-33 → C-3, 4, 5, 13; H3-35 → C-9, 34, 36; H3-36 → C-9, 34, 35; H3-37 → C-23, 24, 38; H3-38 → C-23, 24, 37; H3-39 → C-26, 27, 40; H3-40 → C-26, 27, 39; NH-1 → C-2, 18, 19, 31; OH-13 → C-4, 13, 14; OH-34 → C-9, 34, 35, 36; NOESY correlations (DMSO-d6, 600 MHz) H-16 ↔ H-17a, H3-33; H-17b ↔ H3-32; H3-32 ↔ OH-13 HRESIMS m/z 582.3224 [M + H]+ (calcd for C37H44NO5, 582.3214).
ε) 225 (3.66), 283 (3.77) nm; CD (c 1.5 × 10−4 M, MeOH) λmax (Δε) 229 (+2.32), 251 (+2.89), 282 (−1.57), 324 (+4.15), 373 (−3.87) nm; IR (neat) νmax 3390 (br), 2974, 2932, 1661, 1456, 1374, 1257, 1168, 1134 cm−1; 1H and 13C NMR data see Tables 1 and 2; HMBC data (DMSO-d6, 600 MHz) H-5a → C-4, 6, 7, 33; H-5b → C-4, 6, 7, 13, 33; H-9 → C-7, 10, 34, 35, 36; H-11 → C-7, 9, 13; H-14 → C-4, 16; H-15a → C-3, 13; H-15b → C-3, 13; H-17a → C-2, 3, 18; H-17b → C-2, 3, 18; H-20 → C-18, 19, 22, 29, 31; H-22 → C-21, 23, 28, 29; H-27 → C-23, 26, 29, 39, 40; H-30 → C-19, 21, 28; H3-32 → C-2, 3, 4, 16; H3-33 → C-3, 4, 5, 13; H3-35 → C-9, 34, 36; H3-36 → C-9, 34, 35; H3-37 → C-23, 24, 38; H3-38 → C-23, 24, 37; H3-39 → C-26, 27, 40; H3-40 → C-26, 27, 39; NH-1 → C-2, 18, 19, 31; OH-13 → C-4, 13, 14; OH-34 → C-9, 34, 35, 36; NOESY correlations (DMSO-d6, 600 MHz) H-16 ↔ H-17a, H3-33; H-17b ↔ H3-32; H3-32 ↔ OH-13 HRESIMS m/z 582.3224 [M + H]+ (calcd for C37H44NO5, 582.3214).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 290 (4.10), 365 (3.23) nm; CD (c 5.0 × 10−4 M, MeOH) λmax (Δε) 209 (+3.47), 301 (−8.29), 363 (+3.45) nm; IR (neat) νmax 3285, 2974, 2916, 1663, 1456, 1379, 1252, 1130, 1060 cm−1; 1H and 13C NMR data see Tables 1 and 2; HMBC data (DMSO-d6, 600 MHz) H-9 → C-7, 10; H-11 → C-7, 9, 13; H-17a → C-2, 3, 18; H-17b → C-2, 3, 18; H-20 → C-18, 22, 29, 31; H-22 → C-21, 23, 28, 29; H-27 → C-23, 26, 29; H-30 → C-19, 21, 28; H3-32 → C-2, 3, 4, 16; H3-33 → C-3, 4, 5, 13; H3-35 → C-9, 34, 36; H3-36 → C-9, 34, 35; H3-37 → C-23, 24, 38; H3-38 → C-23, 24, 37; H3-39 → C-26, 27, 40; H3-40 → C-26, 27, 39; NH-1 → C-2, 18, 19, 31; NOESY correlations (DMSO-d6, 600 MHz) H-16 ↔ H-17a, H3-33; H-17b ↔ H3-32; HRESIMS m/z 564.3106 [M + H]+ (calcd for C37H42NO4, 564.3108).
ε) 290 (4.10), 365 (3.23) nm; CD (c 5.0 × 10−4 M, MeOH) λmax (Δε) 209 (+3.47), 301 (−8.29), 363 (+3.45) nm; IR (neat) νmax 3285, 2974, 2916, 1663, 1456, 1379, 1252, 1130, 1060 cm−1; 1H and 13C NMR data see Tables 1 and 2; HMBC data (DMSO-d6, 600 MHz) H-9 → C-7, 10; H-11 → C-7, 9, 13; H-17a → C-2, 3, 18; H-17b → C-2, 3, 18; H-20 → C-18, 22, 29, 31; H-22 → C-21, 23, 28, 29; H-27 → C-23, 26, 29; H-30 → C-19, 21, 28; H3-32 → C-2, 3, 4, 16; H3-33 → C-3, 4, 5, 13; H3-35 → C-9, 34, 36; H3-36 → C-9, 34, 35; H3-37 → C-23, 24, 38; H3-38 → C-23, 24, 37; H3-39 → C-26, 27, 40; H3-40 → C-26, 27, 39; NH-1 → C-2, 18, 19, 31; NOESY correlations (DMSO-d6, 600 MHz) H-16 ↔ H-17a, H3-33; H-17b ↔ H3-32; HRESIMS m/z 564.3106 [M + H]+ (calcd for C37H42NO4, 564.3108).Computational details
Conformational analyses for 2 and 4–6 within an energy window of 3.0 kcal mol−1 were performed by using the OPLS3 molecular mechanics force field. The conformers were then further optimized with the software package Gaussian 09 at the B3LYP/6-311G(2d,2p) level. Then the 60 lowest electronic transitions for the obtained conformers were calculated using time-dependent density functional theory (TD-DFT) methods at the CAM-B3LYP/6-311G(2d,2p) level. ECD spectra of the conformers were simulated using a Gaussian function. The overall theoretical ECD spectra were obtained according to the Boltzmann weighting of each conformer.30Conclusions
In summary, five new indole diterpenoids, shearinines U–Y (2–6), and a structurally related known compound 22,23-dehydro-shearinine A (1) were isolated from cultures of the entomogenous fungus Penicillium sp. Their structures were elucidated based on NMR spectroscopic data and electronic circular dichroism (ECD) calculations. Compounds 2–5 differ from the known precedent by lacking the 1,3-dioxolane ring.26 In addition, compound 3 is a C-7 methoxy product of 2, 4 is a C-13 hydroxy product and C-14 alkylation of 3, while 5 is a C-6–C-7 alkenyl product of 4. Compound 6 is a C-13–C-14 alkenyl product of 1. Compounds 1–3 showed moderate cytotoxicity, with IC50 values of 11.7–26.2 μM. The shearinines exhibit certain chemical structures or properties that are shared among Penicillium strains and others. These shared chemical characteristics could be an indication of a common origin or a close evolutionary relationship.8,31 Biogenetically, 1–6 could be originated from the copalyl diphosphate (CPP) synthases via the precursor (E,E,E)-geranylgeranyl diphosphate (GGDP) and tryptophan,10,32,33 and the hypothetical biosynthetic pathways leading to the generation of these metabolites are illustrated in Scheme S1.†Data availability
The data supporting the findings of this study are available within the article and/or its ESI.†Conflicts of interest
The authors declare no conflict interest.Acknowledgements
We gratefully acknowledge financial support from the Jiangsu Food & Pharmaceutical Science College (302320240033 and 302320230555) and the Natural Sciences Basic Research of Huaian (HAB2024065).Notes and references
- M. A. Corsello, J. Kim and N. K. Garg, Chem. Sci., 2017, 8, 5836–5844 RSC.
- J. Niu, J. Qi, P. Wang, C. Liu and J. Gao, Nat. Prod. Bioprospect., 2023, 13, 3 CrossRef CAS PubMed.
- X. Hu, L. Meng, X. Li, S. Yang, X. Li and B. Wang, Mar. Drugs, 2017, 15, 137 CrossRef PubMed.
- O. F. Smetanina, A. I. Kalinovsky, Y. V. Khudyakova, M. V. Pivkin, P. S. Dmitrenok, S. N. Fedorov, H. Ji, J. Kwak and T. A. Kuznetsova, J. Nat. Prod., 2007, 70, 2054 CrossRef CAS.
- T. Yamaguchi, K. Nozawa, T. Hosoe, S. Nakajima and K. Kawai, Phytochemistry, 1993, 32, 1177–1181 CrossRef CAS.
- P. Zhang, X. Li, X. Li and B. Wang, Phytochem. Lett., 2015, 12, 182–185 CrossRef CAS.
- S. Uhlig, W. Egge-Jacobsen, T. Vrålstad and C. O. Miles, Rapid Commun. Mass Spectrom., 2014, 28, 1621–1634 CrossRef CAS PubMed.
- C. A. Young, B. A. Tapper, K. May, C. D. Moon, C. L. Schardl and B. Scott, Appl. Environ. Microbiol., 2009, 75, 2200–2211 CrossRef CAS PubMed.
- T. Kawahara, A. Nagai, M. Takagi and K. Shin-ya, J. Antibiot., 2012, 65, 535–538 CrossRef CAS PubMed.
- Y. Fan, Y. Wang, P. Liu, P. Fu, T. Zhu, W. Wang and W. Zhu, J. Nat. Prod., 2013, 76, 1328–1336 CrossRef CAS PubMed.
- G. N. Belofsky, J. B. Gloer, D. T. Wicklow and P. F. Dowd, Tetrahedron, 1995, 51, 3959–3968 CrossRef CAS.
- N. P. Ariantari, E. Ancheeva, C. Wang, A. Mándi, T. Knedel, T. Kurtán, C. Chaidir, W. E. G. Müller, M. U. Kassack, C. Janiak, G. Daletos and P. Proksch, J. Nat. Prod., 2019, 82, 1412–1423 CrossRef CAS PubMed.
- L. Dai, L. Yang, F. Kong, Q. Ma, Q. Xie, H. Dai, Z. Yu and Y. Zhao, Mar. Drugs, 2021, 19, 613 CrossRef CAS PubMed.
- S. Pang, Z. Guo, L. Wang, Q. Guo and F. Cao, Nat. Prod. Res., 2022, 37, 586–591 CrossRef PubMed.
- B. Chaiyosang, K. Kanokmedhakul, N. Yodsing, S. Boonlue, J. Yang, Y. A. Wang, R. J. Andersen, J. Yahuafai and S. Kanokmedhakul, Nat. Prod. Res., 2021, 36, 1–9 Search PubMed.
- G. Zhou, C. Sun, X. Hou, Q. Che, G. Zhang, Q. Gu, C. Liu, T. Zhu and D. Li, J. Org. Chem., 2021, 86, 2431–2436 CrossRef CAS PubMed.
- Z. Liang, N. Shen, X. Zhou, Y. Zheng, M. Chen and C. Wang, Chem. Nat. Compd., 2020, 56, 379–382 CrossRef CAS.
- A. C. D. Carvalho, C. Y. O. Junior, L. D. C. Rodrigues, L. S. D. Medeiros and T. A. M. Veiga, Leuk. Lymphoma, 2021, 62, 2079–2093 CrossRef PubMed.
- R. Nicoletti, A. Andolfi, A. Becchimanzi and M. M. Salvatore, Microorganisms, 2023, 11, 1302 CrossRef CAS PubMed.
- R. M. K. Toghueo and F. F. Boyom, 3 Biotech., 2020, 10, 107 CrossRef PubMed.
- S. Liu, M. Su, S. Song and J. H. Jung, Mar. Drugs, 2017, 15, 329 CrossRef PubMed.
- M. Koul and S. Singh, Anti-Cancer Drugs, 2017, 28, 11–30 CrossRef CAS PubMed.
- E. Li, F. Zhang, S. Niu, X. Liu, G. Liu and Y. Che, Org. Lett., 2012, 14, 3320–3323 CrossRef CAS PubMed.
- Y. Feng, L. Wang, S. Niu, L. Li, Y. Si, X. Liu and Y. Che, J. Nat. Prod., 2012, 75, 1339–1345 CrossRef CAS PubMed.
- F. Ren, S. Chen, Y. Zhang, S. Zhu, J. Xiao, X. Liu, R. Su and Y. Che, J. Nat. Prod., 2018, 81, 1752–1759 CrossRef CAS PubMed.
- J. You, L. Du, J. B. King, B. E. Hall and R. H. Cichewicz, ACS Chem. Biol., 2013, 8, 840–848 CrossRef CAS PubMed.
- S. Zhu, F. Ren, Z. Guo, J. Liu, X. Liu, G. Liu and Y. Che, J. Nat. Prod., 2019, 82, 462–468 CrossRef CAS PubMed.
- A. Saidykhan, W. H. C. Martin, R. T. Gallagher, J. Kendrick and R. D. Bowen, J. Mol. Struct., 2023, 1294, 136311 CrossRef CAS.
- N. Zhang, Y. Chen, R. Jiang, E. Li, X. Chen, Z. Xi, Y. Guo, X. Liu, Y. Zhou, Y. Che and X. Jiang, Autophagy, 2011, 7, 598–612 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, O. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09, Rev D.01, Gaussian, Inc., Wallingford, CT, 2009 Search PubMed.
- X. Wang, Z. Chen, J. Zhang, L. Hai, R. Ma, X. Liu, M. Li, D. Ge, J. Bao and H. Zhang, Mar. Drugs, 2023, 21, 593 CrossRef CAS PubMed.
- C. Liu, A. Minami, T. Dairi, K. Gomi, B. Scott and H. Oikawa, Org. Lett., 2016, 18, 5026–5029 CrossRef CAS PubMed.
- R. M. McLellan, R. C. Cameron, M. J. Nicholson and E. J. Parker, Org. Lett., 2022, 24, 2332–2337 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: UV, IR, HRESIMS, NMR spectra of compounds 2–6; ECD calculations of compounds 2–6. See DOI: https://doi.org/10.1039/d4ra08249d |
| This journal is © The Royal Society of Chemistry 2025 |