Open Access Article
Open Access ArticleTotal biosynthesis of cotylenin diterpene glycosides as 14-3-3 protein–protein interaction stabilizers†
Zhenhua
Guan
a,
Nanyu
Yao
a,
Wenling
Yuan
a,
Fengli
Li
a,
Yang
Xiao
a,
Mewlude
Rehmutulla
a,
Yuhan
Xie
a,
Chunmei
Chen
a,
Hucheng
Zhu
 a,
Yuan
Zhou
a,
Yuan
Zhou
 a,
Qingyi
Tong
a,
Zheng
Xiang
a,
Qingyi
Tong
a,
Zheng
Xiang
 *bc,
Ying
Ye
*a and
Yonghui
Zhang
*bc,
Ying
Ye
*a and
Yonghui
Zhang
 *a
*a
aHubei Key Laboratory of Natural Medicinal Chemistry and Resource Evaluation, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, Hubei Province, People's Republic of China. E-mail: zhangyh@mails.tjmu.edu.cn; ying_ye@hust.edu.cn
bState Key Laboratory of Chemical Oncogenomics, Shenzhen Key Laboratory of Chemical Genomics, School of Chemical Biology and Biotechnology, Peking University Shenzhen Graduate School, Shenzhen, Guangdong 518055, P. R. China
cInstitute of Chemical Biology, Shenzhen Bay Laboratory, Shenzhen, Guangdong 518132, China. E-mail: zxiang@pku.edu.cn
First published on 28th November 2024
Abstract
Cotylenins (CNs) are bioactive fungal diterpene glycosides that exhibit stabilizing activity on 14-3-3 protein–protein interactions (PPIs), which has significant therapeutic potential. Although CNs were isolated as early as 1970, their biosynthetic pathway has remained unclear, and their limited supply has hindered further research. Here, we report the identification of the biosynthetic gene cluster cty and elucidation of the biosynthetic pathway of CNs. Our investigation reveals the roles of glycosyltransferase, methyltransferase, and prenyltransferase enzymes in the assembly and modification of the saccharide moiety, as well as the multifunctional oxidation activity of the P450 enzyme CtyA. We leveraged this knowledge to achieve the total biosynthesis of not only key intermediates such as CN-C, E, F, and I, but also a novel, unnatural CN derivative using heterologous expression. This showcases the potential of pathway enzymes as catalytic tools to expand the structural diversity of diterpene glycosides. Furthermore, the stabilization effects of pathway intermediates on 14-3-3 PPIs underscore the importance of saccharide modifications in bioactivity. These findings provide a foundation for future rational synthesis of cotylenin A and other structurally diverse derivatives, broadening the scope of diterpene glycoside production.
Introduction
Cotylenins (CNs) represent a class of fungal-derived diterpene glycosides renowned for their diverse biological activities.1–3 Among them, cotylenin A (CN-A, 1) stands as the flagship molecule, derived from the fusicoccane-type diterpene cotylenol (7), featuring a distinct 5-8-5 tricyclic ring system with intricate saccharide moiety modifications (Fig. 1). Originally isolated from the plant pathogenic fungus Cladosporium sp. 501-7W in 1970, CN-A exhibits plant hormone-like activity and shows promise in inducing leukemia cell differentiation and enhancing the efficacy of various anticancer drugs.4,5 Besides 1, additional CN derivatives, such as CN-C (2) and F (3) were isolated from the same strain.6 Other structurally and biosynthetically related molecules include fusicoccin A (FC-A),1 brassicicene I (BC-I, 8)7 and their derivatives. | ||
| Fig. 1 Chemical structures of representative CN natural products and biosynthetically related FC-A and BC-I. | ||
Compound 1 exerts its biological effects by stabilizing protein–protein interactions (PPIs) involving the 14-3-3 protein, a family of regulatory proteins that bind phosphorylated targets to modulate their function. This action impacts cancer-relevant pathways such as C-RAF signaling.8 Given the integral role of PPIs in cellular processes and disease pathogenesis,9 compound 1's ability to target 14-3-3 PPIs offers a novel approach for therapeutic intervention of cancer and other diseases. Furthermore, structurally related FC glycosides also demonstrate varying activity and selectivity towards 14-3-3 PPIs.10,11 However, studies on the activity of other natural CN products on 14-3-3 PPIs have not been reported, likely due to the difficulty in isolating these molecules.
Despite its biological significance, the preparation of 1 remains challenging due to several factors. The original producing organism, Cladosporium sp. 501-7W, has lost its ability to proliferate in culture,12 rendering microbial fermentation unfeasible. Additionally, the complexity of 1's molecular structure poses a significant hurdle, with the first total synthesis reported by Masahisa Nakada's research group in 2020 requiring a 25-step process starting from geraniol.13 Many studies have attempted to shorten the total synthesis steps by employing concise synthesis or enzymatic synthesis of its aglycone 7.14,15 However, even with these efforts, the intricate saccharide modifications continue to present difficulties in chemical synthesis.
These challenges underscore the need for alternative approaches, such as identifying the biosynthetic gene cluster (BGC) from other strains or employing synthetic biology strategies to develop more efficient and scalable methods to produce 1 and other CN derivatives. One such attempt involved knocking out genes in the FC-A BGC, thereby generating analogous glucosylated precursors for semisynthetic production.12 FC-A, which is structurally similar to CN-A, benefits from an available producing strain, and its BGC and biosynthetic pathway have been extensively studied by the Dairi group.7,12,16,17
In this study, we identified a BGC and achieved the production of crucial intermediates such as CN-C (2) and F (3) through heterologous expression in Aspergills oryzae NSAR1 (AO).18,19 Additionally, we characterized the glycosyltransferase and the modifying enzymes for the saccharide moiety, enabling enzymatic synthesis of previously unidentified intermediates and unnatural CN derivatives. Our research will make significant contributions to the ultimate biosynthesis of 1 and the preparation of its derivatives.
Results
Identification of the cotylenin BGC
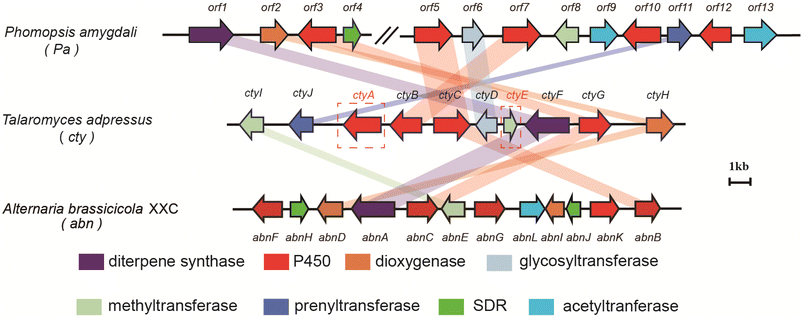
The original producing strain of CNs has lost its ability to proliferate, and there are no reports of its genome, thus making the search for its BGC challenging. Considering the structural similarity between CNs and FCs, the BGC of CNs should have a high degree of similarity with the known BGC of FCs (Pa in Fig. 2).12 The fusicoccadiene synthase (FS) and glycosyltransferase, as well as prenyltransferase, are its indispensable components. At the same time, the BGC of CNs should also have its uniqueness, possessing a saccharide methyltransferase (MTase) gene and lacking the acetyltransferase gene found in the Pa cluster.Based on these characteristics, we identified an Pa-like cluster from the genomic library of our own fungus Talaromyces adpressus (TA), which encodes the FS and glycosyltransferase. However, it lacks the prenyltransferase. Since this cluster is located at the end of the scaffold, it is suspected to be incomplete. Using cblaster,20 we found a highly similar but longer BGC in the Talaromyces verruculosus TS63-9 strain (Fig. S1†), which encodes both a prenyltransferase and a MTase. Subsequently, we performed a local BLAST search for these two enzymes in our TA strain and found two homologous enzymes located next to each other at the end of another scaffold. By connecting the ends of these two scaffolds, we obtained a more complete BGC, which we named the cty cluster (Fig. 2). We then conducted a comparative analysis of this cluster with the Pa cluster and the brassicicenes BGC abn (Fig. 2).21
BC-I (8) is a common intermediate for both CN and BC compounds, formed by the five genes abnABCDE21,22 in abn cluster (Fig. S2 and S16†), which have highly similar counterparts ctyCFGHI in the cty cluster (Fig. 2 and Table S3†). After the formation of BC-I, the subsequent steps of 9-hydroxylation, glucosylation, and prenylation are similar for FCs and CNs. Not surprisingly, we found that the three modifying enzymes in the FC pathway, Pa-orf6 (glycosyltransferase), Pa-orf7 (C-9 hydroxylase), and Pa-orf11 (prenyltransferase), have highly similar homologous genes in the cty cluster: ctyD, ctyB, and ctyJ, respectively (Fig. 2 and Table S3†). Additionally, we identified two genes unique to the cty cluster, including ctyE, which encodes a MTase annotated as a ribosomal RNA small subunit methyltransferase, and ctyA, a P450 enzyme with low homology to the P450 enzymes in the other two clusters, possibly catalyzing a novel oxidation reaction (Table S3†). Based on this information, we propose that the cty cluster is likely responsible for the biosynthesis of CNs.
Heterologous expression of cty cluster
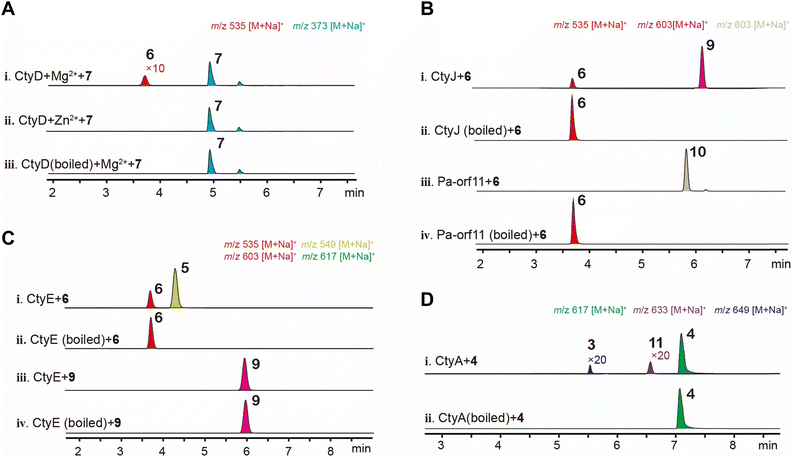
We cultured TA on various media but did not observe the production of CNs (Fig. S3†), suggesting that this cluster is likely silent. Therefore, we employed heterologous expression techniques in AO to study it.We first heterologously expressed the gene encoding terpene synthase CtyF and confirmed it produced fusicoccadiene (Fig. S15†). Given the high homology between CtyCFGHI and the BC-I (8)-producing enzymes AbnABCDE, we hypothesized that AO-ctyCFGHI also produces 8. Therefore, instead of constructing AO-ctyCFGHI, we assumed 8 as its product and used it to feed the transformants of other cty genes to quickly verify their functions. Taking the modification sequence for biosynthesizing FCs as a reference, we constructed the following transformants: AO-ctyB, AO-ctyBD, AO-ctyBDE, AO-ctyBDEJ, and AO-ctyABDEJ, and analyzed the feeding products using LC-MS. In AO-ctyB fed with 8, we detected a new peak with a molecular weight (mw) increase of 16 Da (Fig. 3A(ii)), which was isolated and identified as compound 7 through NMR spectroscopy (Fig. S17, S18 and Table S4†). Recent years have seen progress in achieving C9 beta-hydroxylation of CN glycosides through organic catalysis due to the research by Nakada, Shenvi, and Renata, who have continuously improved the stereoselectivity of the reaction.13–15 We recently reported that Orf7 in the Pa cluster can efficiently catalyze this reaction enzymatically.23 Here, we discovered that CtyB has the same function. In AO-ctyBD adding 8, we detected a new peak with a mw equals to 7 + 162 Da (Fig. 3A(iii)), which was isolated and identified as the glucosylated product 6 (Fig. S19–S21 and Table S5†). We then introduced the MTase ctyE, and after feeding 8, LC-MS detected a new peak with a mw equals to 6 + 14 Da (Fig. 3A(iv)). Scale up fermentation and isolation identified this compound as the methylated product 5 (Fig. S22–S25 and Table S6†). To verify the function of the prenyltransferase, we further introduced ctyJ into the previous transformant, and after feeding 8, LC-MS detected a new peak with a mw equals to 5 + 68 Da (Fig. 3A(v)). Scale up fermentation and isolation identified this compound as the prenylated product 4 (Fig. S26–S28 and Table S7†). Next, we introduced the last uncharacterized P450 oxidase CtyA, and expected to obtain products with oxidative modifications on the glucosyl moiety. After feeding 8, LC-MS detected two new peaks with mw equals to 4 + 32 Da and + 30 Da (Fig. 3A(vii)). Scale up fermentation and isolation identified these compounds as 3 (Fig. S29–S32 and Table S8†) and 2 (Fig. S33–S36 and Table S9†), respectively. Additionally, in the AO-ctyABDJ transformant lacking the MTase ctyE, feeding 8 resulted in small amounts of compounds 12 (mw 619 Da) and 13 (mw 635 Da) (Fig. 3A(vi)). Compound 12 was isolated, but due to its small amount, it was difficult to purify completely. However, 1D and 2D NMR allowed structural assignment (Fig. S37–S42 and Table S10†), revealing a C3′′-4′′ epoxy structure, with no oxidation at the C1′′. This indicates that CtyJ and CtyA can accept substrates without methylation at the C6′ position, forming a monooxygenated epoxy product by CtyA. Due to the low yield, 13 could not be isolated for NMR analysis., but based on its molecular formula and elution time, we infer that its structure is a further hydroxylation product of compound 12 at C1′′ position (Fig. 4).
 | ||
| Fig. 4 Reconstitution of the biosynthetic pathway of CNs in this work and the semi-synthesis of ISIR-050. | ||
The feeding results indicate that CtyA is multifunctional, capable of introducing an epoxide on the prenyl C3′′-4′′ double bond and oxidizing the prenyl C1′′ to a hydroxyl and aldehyde group. To further investigate the order of its modifications, we constructed AO-ctyA and examined the biotransformation products over a time gradient. The results showed that feeding 4 to AO-NSAR1 did not result in any detectable oxygenated products, and the substrate remained unconsumed (Fig. 3B(iv)). In AO-ctyA, small amounts of a monooxygenated compound 11 were observed at 1.5 and 2.5 days (Fig. 3B(i and ii)), but 2 and 3 were more predominant. After 3.5 days, 11 almost disappeared, with only 2 and 3 remaining (Fig. 3B(iii)). To investigate if CtyA can also transform the C1′′ hydroxyl group to an aldehyde group, 3 was fed into AO-ctyA for 3.5 days, resulting in the production of 2 (Fig. 3B(v)), while feeding 3 to AO-NSAR1 did not observe the formation of 2 (Fig. 3B(vi)). The mw of 11 corresponds to the molecular formula C33H54O10, but it could not be isolated due to its low yield. Based on the structure of 12, we speculate that 11 is the epoxidized product of 4 at the C3′′-4′′ double bond (Fig. 3B(i and ii)). These in vivo experiments demonstrate that CtyA is a multifunctional oxidase, capable of both epoxidizing the terminal double bond on the glucosylated prenyl group and performing 4-electron oxidation on the methyl group.
Converting 2 to 1 requires only one more oxidation step to form an oxygen bridge between C4′ and C1′′. We proceeded by introducing genes from the extended region of the cty cluster (Fig. S4(i, ii, iii, iv) and Table S3†), particularly focusing on the FAD enzyme hp5 with potential oxidase functions. Unfortunately, none of the transformants produced CN-A (Fig. S4(v)†). This suggests two possibilities: either the final catalytic enzyme is outside the cty cluster, or the enzyme required for the formation of 1 is absent in the TA strain. To verify these possibilities, we fed the previously obtained pathway intermediates 8, 6, 4, and 2 to TA in multiple medium, but 1 was still not detected (Fig. S3†). This result suggests that while the TA strain harbors the BGC for CNs, the exact capability of the strain to produce CN-A remains unclear. Further efforts will be needed to identify new strains or novel oxidases to achieve the complete biosynthesis of CN-A.
Functional characterization of pathway enzymes in vitro
CNs and FCs undergo extensive modifications beyond glucosylation, with CNs involving prenylation, methylation, and oxidation, and FCs involving prenylation and acetylation. We proceeded to examine their in vitro activities. Biotransformation results indicate the glycosyltransferase CtyD converts 7 into 6. Alphafold2 modeling indicates that it belongs to the GT-A family of glycosyltransferases (Fig. S5†).24 We purified CtyD and performed in vitro reactions with 7. In the presence of the divalent metal ion Mg2+, CtyD introduces glucose at the C9 hydroxyl position (Fig. 5A(i)), while no activity was observed in the presence of Zn2+ (Fig. 5A(ii)). The poor in vitro activity is likely due to the low solubility of CtyD produced in E. coli (Fig. S14†).In vivo experiments showed that the prenyltransferase CtyJ catalyzes the prenylation of the C4′-hydroxyl group on the saccharide of 5, whereas the Pa-orf11 (H7CE84.1) from the Pa cluster is reported to prenylate the C6′-position on the FC's saccharide moiety. To investigate the substrate specificity of these PTs and try to prepare artificial combinatorial molecules, we reacted CtyJ and Pa-orf11 with 6 separately. Both enzymes exhibited regioselectivity by recognizing the substrate and catalyzing prenylation reactions at specific sites, producing 9 (Fig. S43–S48 and Table S11†) and an unnatural molecule 10 (Fig. S49–S54 and Table S12†), respectively (Fig. 5B). Prenylation on saccharides is rare; a recent example is the work by Zou's group, which reported that the prenyltransferase CosD can introduce a prenyl group onto linear saccharide.25 Phylogenetic analysis revealed that CtyJ, Pa-orf11, and CosD belong to typical fungal prenyltransferases and share high homology (Fig. S6†).
In vivo experiments showed that the MTase CtyE converts 6 into 5. Purified CtyE exhibited the same activity, but it cannot recognize the prenylated product 9 (Fig. 5C). This result clarifies that methylation occurs before prenylation in the pathway (Fig. 4). According to the protein structure predicted by Alphafold2, CtyE belongs to the Type I MTase family (Fig. S7†).26 MTases acting on saccharides are rare in fungi but are more common in bacteria, appearing in pathways such as the polyketide glycoside elloramycin27 and the macrolide mycinamicin.28 Phylogenetic analysis of representative MTases from fungal and bacterial secondary metabolites showed that CtyE has higher homology with bacterial MTases (Fig. S8†). This interesting phenomenon suggests that bacteria might have first evolved MTases for metabolizing 6, and this capability was later acquired by fungi through gene transfer.
The P450 enzyme CtyA exhibited consecutive oxidation activity on the prenyl group in vivo. We introduced its cDNA into S. cerevisiae RC01 which contains an integrated copy of cytochrome P450 reductase (AtCPR) from Aspergillus terreus,29 extracted the microsomes, and tested the in vitro activity (Fig. 5D). The results showed that CtyA can oxidize 4 to 11 and 3, but the efficiency is much lower than that in AO, and 2 was not detected. The in vitro results confirmed CtyA's ability to oxidize two distinct positions on the prenyl group, suggesting that after the formation of the epoxide, a change in the relative position of the intermediate brings the C1′′ position closer to the Heme iron center (Fig. S9†). We also examined CtyA's activity on 10, which has a shifted prenyl position, but no conversion was observed (Fig. S10†).
Based on the results from the in vivo and in vitro experiments, we have elucidated for the first time the biosynthetic pathways of several reported CN diterpene glycosides, including cotylenin E (5), I (4), F (3), and C (2) (Fig. 4), remaining only one oxidation step away from forming the oxygen bridge in CN-A (1). Additionally, by recombining the relevant genes in AO, we have isolated and identified previously unreported intermediates 6, 9, and 12 in the CNs pathway, as well as an artificial molecule 10 by borrowing the prenyltransferase from Pa cluster.
Stabilizing activity of CNs on 14-3-3 PPIs
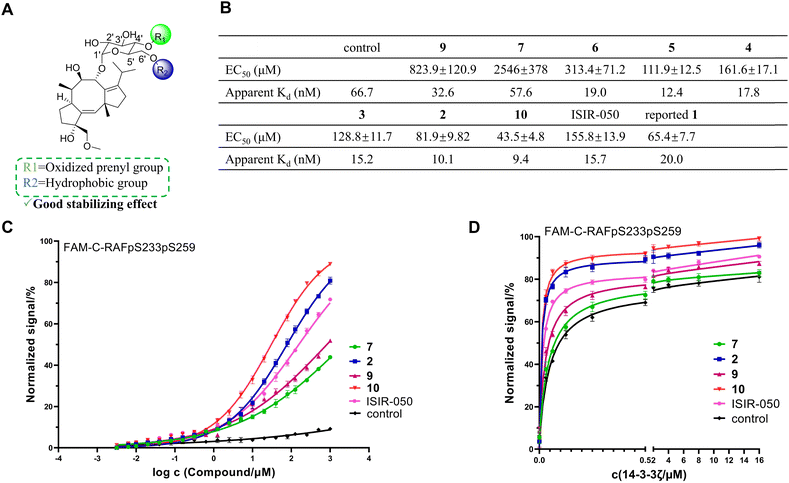
CN-A is known as a 14-3-3 protein–protein interaction (PPI) stabilizer and has been reported to stabilize the interaction between the C-RAFpS233pS259 peptide segment and 14-3-3ζ protein in vitro by Molzan et al.8 We attempted to request CN-A from various sources, but were unsuccessful. Instead, starting from compound 6, we rapidly synthesized ISIR-050 (Fig. 4, S55 and S56†), a semi-synthetic CN-A mimic. This mimic has been reported to exhibit interferon-α (IFNα)-dependent growth inhibitory activity and PPI stabilization similar to CN-A.30 Through our experiments, we confirmed its ability to stabilize the interaction between the C-RAFpS233pS259 peptide segment and the 14-3-3ζ protein (Fig. 6). Using ISIR-050 as a positive control, we tested the stabilizing effects of the CN derivatives obtained in this study on the C-RAFpS233pS259/14-3-3ζ PPI using a fluorescence polarization (FP) assay method (Fig. 6). The results showed that the aglycone 7 had the weakest stabilizing effect, while the glucosylated intermediates 2–6 and 9 exhibited significantly improved activity with 2 outperforming ISIR-050 (Fig. 6B). Notably, Molzan et al. obtained an EC50 value of 65.4 ± 7.7 μM for CN-A using the same method and conditions that we adopted in our study (Fig. 6B).8 Although a direct comparison is not possible, our results indicate that 2 exhibits a stabilizing capability close to CN-A. Interestingly, the unnatural molecule 10 had activity better than 2. We further analyzed the impact of different modifications on the glucosyl group on compound activity: compound 10, with a prenyl group at the C6′ position, had an EC50 value of 43.5 ± 4.8 μM. When the prenyl group was shifted to the C4′ position, the EC50 value of 9 sharply increased to 823.9 ± 120.9 μM. This significant difference suggests that introducing a hydrophobic group at the glycosyl C6′ position is crucial for stabilizing activity (Fig. 6A). Compound 6 had an EC50 value of 313.4 ± 71.2 μM, and after methylation at the C6′-position, the EC50 of 5 decreased to 111.9 ± 12.5 μM, supporting the previous hypothesis. On the other hand, the oxidation of the C4′ prenyl group plays a beneficial role in PPI stabilization (Fig. 6A), as reflected in the increasing EC50 values from compounds 2, 3, to 4. We further tested the impact of these compounds on the apparent affinity (Kd) of the C-RAFpS233pS259/14-3-3ζ interaction, which showed a trend consistent with the EC50 values (Fig. 6B). Taken together, these findings indicate that glycosylation and the specific modifications of the saccharide moiety are essential for stabilizing the C-RAFpS233pS259/14-3-3ζ interaction, with the unnatural derivate 10 exhibiting the most potent activity among the tested CNs.Discussion
Modulating protein–protein interactions (PPIs) is challenging due to their flat and large interfaces, making them difficult drug targets.31,32 Natural products like CNs and FCs, which stabilize PPIs, are crucial for mechanistic studies and drug development. Our discovery of the BGC for CNs and the elucidation of the biosynthetic pathways for various intermediates with PPI-stabilizing activity represent a significant advancement, particularly given the structural complexity and chemical synthesis challenges of these molecules. This is even more critical as the producing strain is no longer available.We have identified the CN BGC for the first time, enabling comparison with the known brassicicene and fusicoccin BGCs to understand the genetic basis of their structural differences. In comparing the abn and cty clusters, both share homologous genes involved in the production of BC-I (8). The abn cluster lacks glycosylation genes but contains a unique P450 enzyme for oxidative rearrangement,22 leading to the formation of the 5-9-5 scaffold as revised by our group;33 On the other hand, in the comparison between the Pa and cty clusters, both contain C9-hydroxylase and glycosyltransferase genes, facilitating similar glucosylation, but distinct modifying enzymes specific to the aglycone and sugar units drive structural variations between them. It is noteworthy that FCs primarily affect stomatal opening by stabilizing the interaction between plant 14-3-3 protein and H+-ATPase, whereas CNs promote cell elongation and division.34 The difference between FC and CN derivatives is reported to be due to the 12-hydroxyl group in FCs, which prevents complex formation with mode-1 ligands.35 Additionally, the unique enzymes CtyA (P450) and CtyE (methyltransferase) in the cty cluster for sugar moiety modifications have shown important activity-enhancing functions in our preliminary structure–activity relationship studies. These comparisons reveal critical insights into how genetic variations within BGCs influence the structure and bioactivity of these diterpenoids. Furthermore, our cblaster analysis indicates that the cty cluster is not widely distributed (Fig. S1†), which may help explain why CN-producing strains are relatively rare.
The discovery of the cty cluster provides a rich set of catalytic elements, facilitating the biosynthesis and chemoenzymatic synthesis of CNs. Additionally, combinatorial biosynthesis with the Pa cluster can generate more derivative molecules (such as compound 10 in this study). These derivatives can be employed to investigate whether their structural differences enhance the selectivity of 14-3-3 proteins for various target proteins, which is crucial for developing highly selective, druggable PPI stabilizers.
Unfortunately, the final step leading to CN-A remains unresolved in this study. Despite our attempts using various fermentation conditions and intermediate feeding in our TA strain, CN-A was not observed. This raises the possibility that the entire BGC may be silent. The cluster lacks obvious regulatory elements such as BGC-specific transcription factors, suggesting the need to explore various stimuli to activate it. Understanding the regulatory mechanisms of CN biosynthesis will provide further insight into their physiological and ecological functions.
Conclusions
In this study, through heterologous expression and functional characterization of enzymes within the cty cluster, we elucidated the biosynthetic pathways of several natural diterpene glycosides, including cotylenin C (2), F (3), I (4) and E (5). We identified and confirmed the roles of key enzymes responsible for assembling and modifying the saccharide moiety. Additionally, we demonstrated the multifunctional oxidation activity of the P450 enzyme CtyA on the prenyl group, highlighting its ability to perform sequential oxidation steps.Beyond simply elucidating the biosynthesis, our work revealed the potential of pathway enzymes as tools for rational design and the total biosynthesis of new compounds. This is exemplified by our successful creation of the unnatural cotylenin derivative 10, achieved through leveraging enzyme substrate promiscuity and regioselectivity across homologous BGCs. Furthermore, we revealed for the first time the stabilizing activity of CN biosynthetic intermediates on 14-3-3 PPIs. Future efforts will focus on identifying new strains or novel oxidases to achieve the complete biosynthesis of CN-A.
Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Y. Z., Y. Y., and Z. X. designed the research. Z. G., N. Y., Y. X., Y. X., Mewlude Rehmutulla, W. Y., and F. L. performed the experiments. C. C., H. Z., Y. Z., and Q. T. analyzed the data. Z. G., and Y. Y. wrote the manuscript. All the authors have approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are very grateful to professor Hideaki Oikawa at Division of Chemistry, Graduate School of Science, Hokkaido University for providing Aspergillus oryzae NSAR1 heterologous expression system. We thank the Medical sub-center of analytical and testing center, Huazhong University of Science and Technology for measuring NMR spectroscopic data. This work was financially supported by the National Key R&D Program of China (2021YFA0910500), the National Natural Science Foundation of China (22277035, 32000045, and 81973205), the Program for the National Natural Science Foundation for Distinguished Young Scholars (81725021), the Innovative Research Groups of the National Natural Science Foundation of China (81721005), the Fundamental Research Funds for the Central Universities (2020kfyXJJS083), the Sino-German Center for Research Promotion (M-0477), Key-Area Research and Development Program of Guangdong Province (2020B0303070002) and Shenzhen Science and Technology Program (KQTD20170330155106581).Notes and references
- A. H. de Boer and I. J. de Vries van Leeuwen, Fusicoccanes: diterpenes with surprising biological functions, Trends Plant Sci., 2012, 17, 360–368 CrossRef CAS.
- Y. Honma, Cotylenin A – a plant growth regulator as a differentiation-inducing agent against myeloid leukemia, Leuk. Lymphoma, 2002, 43, 1169–1178 CrossRef CAS.
- Y. Honma, T. Kasukabe, T. Yamori, N. Kato and T. Sassa, Antitumor effect of cotylenin A plus interferon-α: Possible therapeutic agents against ovary carcinoma, Gynecol. Oncol., 2005, 99, 680–688 CrossRef CAS.
- K.-i. Asahi, Y. Honma, K. Hazeki, T. Sassa, Y. Kubohara, A. Sakurai and N. Takahashi, Cotylenin A, a Plant-Growth Regulator, Induces the Differentiation in Murine and Human Myeloid Leukemia Cells, Biochem. Biophys. Res. Commun., 1997, 238, 758–763 CrossRef CAS PubMed.
- Y. Honma, Y. Ishii, Y. Yamamoto-Yamaguchi, T. Sassa and K. Asahi, Cotylenin A, a differentiation-inducing agent, and IFN-α cooperatively induce apoptosis and have an antitumor effect on human non-small cell lung carcinoma cells in nude mice, Cancer Res., 2003, 63, 3659–3666 CAS.
- T. Sassa and A. Takahama, Carbon-13 NMR Spectra of Cotylenins, Agric. Biol. Chem., 1979, 43, 385–387 CAS.
- Y. Ono, A. Minami, M. Noike, Y. Higuchi, T. Toyomasu, T. Sassa, N. Kato and T. Dairi, Dioxygenases, Key Enzymes to Determine the Aglycon Structures of Fusicoccin and Brassicicene, Diterpene Compounds Produced by Fungi, J. Am. Chem. Soc., 2011, 133, 2548–2555 CrossRef CAS.
- M. Molzan, S. Kasper, L. Röglin, M. Skwarczynska, T. Sassa, T. Inoue, F. Breitenbuecher, J. Ohkanda, N. Kato, M. Schuler and C. Ottmann, Stabilization of Physical RAF/14-3-3 Interaction by Cotylenin A as Treatment Strategy for RAS Mutant Cancers, ACS Chem. Biol., 2013, 8, 1869–1875 CrossRef CAS.
- Y. Aghazadeh and V. Papadopoulos, The role of the 14-3-3 protein family in health, disease, and drug development, Drug Discovery Today, 2016, 21, 278–287 CrossRef CAS.
- S. A. Andrei, P. de Vink, E. Sijbesma, L. Han, L. Brunsveld, N. Kato, C. Ottmann and Y. Higuchi, Rationally Designed Semisynthetic Natural Product Analogues for Stabilization of 14-3-3 Protein-Protein Interactions, Angew. Chem., Int. Ed., 2018, 57, 13470–13474 CrossRef CAS PubMed.
- M. Wolter, P. de Vink, J. F. Neves, S. Srdanović, Y. Higuchi, N. Kato, A. Wilson, I. Landrieu, L. Brunsveld and C. Ottmann, Selectivity via Cooperativity: Preferential Stabilization of the p65/14-3-3 Interaction with Semisynthetic Natural Products, J. Am. Chem. Soc., 2020, 142, 11772–11783 CrossRef CAS.
- M. Noike, Y. Ono, Y. Araki, R. Tanio, Y. Higuchi, H. Nitta, Y. Hamano, T. Toyomasu, T. Sassa, N. Kato and T. Dairi, Molecular Breeding of a Fungus Producing a Precursor Diterpene Suitable for Semi-Synthesis by Dissection of the Biosynthetic Machinery, PLoS One, 2012, 7, e42090 CrossRef CAS PubMed.
- M. Uwamori, R. Osada, R. Sugiyama, K. Nagatani and M. Nakada, Enantioselective Total Synthesis of Cotylenin A, J. Am. Chem. Soc., 2020, 142, 5556–5561 CrossRef CAS PubMed.
- Y. Jiang and H. Renata, Modular chemoenzymatic synthesis of ten fusicoccane diterpenoids, Nat. Chem., 2024, 16, 1531–1538 CrossRef CAS PubMed.
- S. I. Ting, D. W. Snelson, T. R. Huffman, A. Kuroo, R. Sato and R. A. Shenvi, Synthesis of (-)-Cotylenol, a 14-3-3 Molecular Glue Component, J. Am. Chem. Soc., 2023, 145, 20634–20645 CrossRef CAS PubMed.
- M. Noike, C. Liu, Y. Ono, Y. Hamano, T. Toyomasu, T. Sassa, N. Kato and T. Dairi, An enzyme catalyzing O-prenylation of the glucose moiety of fusicoccin A, a diterpene glucoside produced by the fungus Phomopsis amygdali, ChemBioChem, 2012, 13, 566–573 CrossRef CAS PubMed.
- T. Toyomasu, M. Tsukahara, A. Kaneko, R. Niida, W. Mitsuhashi, T. Dairi, N. Kato and T. Sassa, Fusicoccins are biosynthesized by an unusual chimera diterpene synthase in fungi, Proc. Natl. Acad. Sci. U.S.A., 2007, 104, 3084–3088 CrossRef CAS.
- R. Fujii, A. Minami, T. Tsukagoshi, N. Sato, T. Sahara, S. Ohgiya, K. Gomi and H. Oikawa, Total biosynthesis of diterpene aphidicolin, a specific inhibitor of DNA polymerase α: heterologous expression of four biosynthetic genes in Aspergillus oryzae, Biosci., Biotechnol., Biochem., 2011, 75, 1813–1817 CrossRef CAS PubMed.
- K. Tagami, C. Liu, A. Minami, M. Noike, T. Isaka, S. Fueki, Y. Shichijo, H. Toshima, K. Gomi, T. Dairi and H. Oikawa, Reconstitution of Biosynthetic Machinery for Indole-Diterpene Paxilline in Aspergillus oryzae, J. Am. Chem. Soc., 2013, 135, 1260–1263 CrossRef CAS.
- C. L. M. Gilchrist, T. J. Booth, B. van Wersch, L. van Grieken, M. H. Medema and Y. H. Chooi, cblaster: a remote search tool for rapid identification and visualization of homologous gene clusters, Bioinform. adv., 2021, 1, vbab016 CrossRef PubMed.
- W. Yuan, F. Li, Z. Chen, Q. Xu, Z. Guan, N. Yao, Z. Hu, J. Liu, Y. Zhou, Y. Ye and Y. Zhang, AbnI: An α-ketoglutarate-dependent dioxygenase involved in brassicicene C H functionalization and ring system rearrangement, Chin. Chem. Lett., 2024, 35, 108788 CrossRef CAS.
- A. Tazawa, Y. Ye, T. Ozaki, C. Liu, Y. Ogasawara, T. Dairi, Y. Higuchi, N. Kato, K. Gomi, A. Minami and H. Oikawa, Total Biosynthesis of Brassicicenes: Identification of a Key Enzyme for Skeletal Diversification, Org. Lett., 2018, 20, 6178–6182 CrossRef CAS.
- Z. Guan, Y. Yuan, X. Zhang, N. Yao, W. Yuan, Y. Zhang, Y. Ye and Z. Xiang, Heterologous Biosynthesis of Cotylenol and Concise Synthesis of Fusicoccane Diterpenoids, ChemRxiv, 2024, preprint, DOI:10.26434/chemrxiv-2024-d4lpr.
- D. M. Liang, J. H. Liu, H. Wu, B. B. Wang, H. J. Zhu and J. J. Qiao, Glycosyltransferases: mechanisms and applications in natural product development, Chem. Soc. Rev., 2015, 44, 8350–8374 RSC.
- G. Y. Yuan, J. M. Zhang, Q. D. Xu, H. R. Zhang, C. Hu and Y. Zou, Biosynthesis of Cosmosporasides Reveals the Assembly Line for Fungal Hybrid Terpenoid Saccharides, Angew. Chem., Int. Ed., 2023, 62, e202308887 CrossRef CAS PubMed.
- E. Abdelraheem, B. Thair, R. F. Varela, E. Jockmann, D. Popadić, H. C. Hailes, J. M. Ward, A. M. Iribarren, E. S. Lewkowicz, J. N. Andexer, P. L. Hagedoorn and U. Hanefeld, Methyltransferases: Functions and Applications, ChemBioChem, 2022, 23, e202200212 CrossRef CAS PubMed.
- E. P. Patallo, G. Blanco, C. Fischer, A. F. Braña, J. Rohr, C. Méndez and J. A. Salas, Deoxysugar Methylation during Biosynthesis of the Antitumor Polyketide Elloramycin by Streptomyces olivaceus, J. Biol. Chem., 2001, 276, 18765–18774 CrossRef CAS.
- S. Li, Y. Anzai, K. Kinoshita, F. Kato and D. H. Sherman, Functional Analysis of MycE and MycF, Two O-Methyltransferases Involved in the Biosynthesis of Mycinamicin Macrolide Antibiotics, ChemBioChem, 2009, 10, 1297–1301 CrossRef CAS PubMed.
- M. C. Tang, H. C. Lin, D. Li, Y. Zou, J. Li, W. Xu, R. A. Cacho, M. E. Hillenmeyer, N. K. Garg and Y. Tang, Discovery of Unclustered Fungal Indole Diterpene Biosynthetic Pathways through Combinatorial Pathway Reassembly in Engineered Yeast, J. Am. Chem. Soc., 2015, 137, 13724–13727 CrossRef CAS.
- T. Inoue, Y. Higuchi, T. Yoneyama, B. Lin, K. Nunomura, Y. Honma and N. Kato, Semisynthesis and biological evaluation of a cotylenin A mimic derived from fusicoccin A, Bioorg. Med. Chem. Lett., 2018, 28, 646–650 CrossRef CAS.
- L. M. Stevers, E. Sijbesma, M. Botta, C. MacKintosh, T. Obsil, I. Landrieu, Y. Cau, A. J. Wilson, A. Karawajczyk, J. Eickhoff, J. Davis, M. Hann, G. O'Mahony, R. G. Doveston, L. Brunsveld and C. Ottmann, Modulators of 14-3-3 Protein–Protein Interactions, J. Med. Chem., 2018, 61, 3755–3778 CrossRef CAS.
- H. Lu, Q. Zhou, J. He, Z. Jiang, C. Peng, R. Tong and J. Shi, Recent advances in the development of protein–protein interactions modulators: mechanisms and clinical trials, Signal Transduction Targeted Ther., 2020, 5, 213 CrossRef.
- Y. Tang, Y. Xue, G. Du, J. Wang, J. Liu, B. Sun, X.-N. Li, G. Yao, Z. Luo and Y. Zhang, Structural Revisions of a Class of Natural Products: Scaffolds of Aglycon Analogues of Fusicoccins and Cotylenins Isolated from Fungi, Angew. Chem., Int. Ed., 2016, 55, 4069–4073 CrossRef CAS PubMed.
- M. Würtele, C. Jelich-Ottmann, A. Wittinghofer and C. Oecking, Structural view of a fungal toxin acting on a 14-3-3 regulatory complex, EMBO J., 2003, 22, 987–994 CrossRef.
- J. Ohkanda, Fusicoccin: A Chemical Modulator for 14-3-3 Proteins, Chem. Lett., 2020, 50, 57–67 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Strains and methods, additional tables and figures, and NMR data. See DOI: https://doi.org/10.1039/d4sc05963h |
| This journal is © The Royal Society of Chemistry 2025 |