Open Access Article
Open Access ArticleBiotin/avidin-free sandwich aptamer-based lateral flow assay (ALFA) for the diagnosis of Trichomonas vaginalis
Christine Aubrey C. Justo a,
Vasso Skouridou
a,
Vasso Skouridou a,
Piet Coolsb,
Guy Mulinganyacde,
Alexandra Ibáñez-Escribano
a,
Piet Coolsb,
Guy Mulinganyacde,
Alexandra Ibáñez-Escribano f and
Ciara K. O'Sullivan
f and
Ciara K. O'Sullivan *ag
*ag
aInterfibio Consolidated Research Group, Department of Chemical Engineering, Universitat Rovira i Virgili, Tarragona, Spain. E-mail: ciara.osullivan@urv.cat
bDepartment of Diagnostic Sciences, Faculty of Medicine and Health Sciences, Ghent University, Ghent, Belgium
cFaculty of Medicine, Catholic University of Bukavu, Bukavu, Democratic Republic of the Congo
dDepartment of Obstetrics and Gynecology, Hôpital Provincial Général de Référence de Bukavu, Bukavu, Democratic Republic of the Congo
eDepartment of Internal Medicine and Pediatrics, Faculty of Medicine and Health Sciences, Ghent University, Ghent, Belgium
fDepartment of Microbiology and Parasitology, Faculty of Pharmacy, Universidad Complutense de Madrid, Madrid, Spain
gInstitució Catalana de Recerca I Estudis Avancats (ICREA), Barcelona, Spain
First published on 22nd January 2025
Abstract
The development of low-cost, rapid point-of-care tests (POCT) for the detection of the parasite Trichomonas vaginalis is listed in the recent WHO global research priorities for sexually transmitted infections (STIs). Aiming to contribute to this call and aid in ending STIs as a public health threat, we report the development of a DNA aptamer-based POCT for T. vaginalis. Herein, we report an aptamer-based lateral flow assay (ALFA) based on dual aptamers for the detection of T. vaginalis. This is the first report of an ALFA that does not rely on biotin/streptavidin for either capture or reporter molecule. Aptamer capture and recognition relies on the use of an UV-crosslinked aminated capture aptamer on the nitrocellulose membrane and gold nanoparticles (AuNPs) functionalized with a thioctic acid-modified reporter aptamer. The developed ALFA has an estimated laboratory production cost of less than 1 € per test, with the running buffer and the ALFA strip stable for at least 1 year at room temperature (22 °C). The assay requires three simple operator steps from sample procurement to result with a 15-minute runtime. The developed ALFA can detect down to 1.6 × 105 T. vaginalis per mL and shows no cross-reactivity to common vaginal microorganisms and no matrix effect from clinical cervicovaginal lavage (CVL) samples is observed. Preliminary clinical evaluation using CVLs demonstrated that the assay has perfect concordance with wet-mount assay on matching vaginal swabs. The developed rapid test offers a simple, stable, and low-cost diagnostic test for T. vaginalis.
Introduction
The World Health Organization (WHO) estimated that since 2021, one million new cases of sexually transmitted infections (STIs) occur daily worldwide.1 To control the wide spread of STIs, the target product profile for the STI point-of-care tests (POCTs)2 was created and is continually updated to guide developers in fulfilling the REASSURED criteria (real-time connectivity, ease of sample preparation, affordable, sensitive, specific, user-friendly, robust/rapid, equipment-free and delivered to end users).3 However, only a few POCTs fully comply with the set guidelines. Accessibility, affordability, and integration within health systems of available POCTs remain a challenge in the global implementation of POCTs for STI management.1,4,5In 2020, a total of 374.3 million new cases of non-viral curable STIs were recorded among adults aged 15–46 years, with 42% or 156.3 million cases due to infection with the Trichomonas vaginalis parasite.1 This parasite is an anaerobic/microaerophilic flagellate that typically invades the human urogenital tract, although extragenital cases such as oral6 and rectal7,8 have also been reported. Several assays are available for the laboratory testing for T. vaginalis including the traditional wet-mount microscopy and commercially available tests for culture, nucleic acid amplification assays, and immunoassays.9,10 However, access to testing, even by high-risk groups is very limited, leading to the still widely practiced syndromic management of STIs, including trichomoniasis. The development of low-cost rapid diagnostic tests for T. vaginalis has been listed in the recently released WHO global research priorities for STIs.11 The target product profile for T. vaginalis POCT includes a minimal specificity of 99% and a clinical sensitivity of 85% for clinician-collected samples. Moreover, the minimal sample is vaginal swabs, urine is the optimal option, and the assay should be completed in at most three simple operator steps with minimal instructions for interpretation.2 To the best of our knowledge, there are only two commercial Clinical Laboratory Improvement Amendments (CLIA)-waived POCTs for T. vaginalis, namely the OSOM Trichomonas Rapid Test12 and the Visby Medical Sexual Health Test.13,14 The OSOM Trichomonas rapid test is an immunochromatographic capillary-flow T. vaginalis rapid antigen test that takes only 10 minutes to perform, while the Visby Medical Sexual Health Test is a single-use, fully integrated, automated PCR in vitro diagnostic test for the rapid detection and differentiation of DNA from Chlamydia trachomatis, Neisseria gonorrhoeae, and T. vaginalis. Both assays are so far only intended for use with vaginal swab samples, and there is still a need for rapid and simple T. vaginalis testing in males. A serodiagnostic POCT for T. vaginalis in women and men has been recently reported15 whilst more complex, near-patient tests that are compatible with female and male specimens include the Xpert TV assay16 and the Truenat® Trich chip-based real time PCR test.17
Our group is working on the development of new T. vaginalis diagnostic assays to fill the gaps in the T. vaginalis POCT field and contribute to the 2030 goal of eliminating STIs as a public health threat.18 We previously reported the identification and application of aptamers with dual binding to T. vaginalis in a microplate-based enzyme linked aptamer assay.19 While the microplate assay is high throughput, it is not compatible for use as a POCT. In this work, the microplate-based aptamer assay was translated to an aptamer-based lateral flow assay (ALFA), the first of its kind for T. vaginalis testing. It fully exploits the flexibility of DNA, and the final design does not rely on the use of biotin/avidin either for immobilisation or in the gold nanoparticle reporter conjugate, thus reducing costs and increasing stability. The developed assay can be used for the analysis of clinical CVLs and has great potential for further development as an alternative POCT for screening for T. vaginalis.
Experimental
Materials
The previously identified DNA aptamers pair for T. vaginalis (A1_14mer: 5′-t15-gggcgggcgggtgg-3′ and A6: 5′- agctccagaagataaattacaggggcgggggggcgggggaggcggaaggcctgctaaagtcgttgtgagcgaaccaactaggatactatgacccc-3′)19 and control oligonucleotide 33A (5′-a33) were purchased from Biomers (Ulm, Germany). The gold(III) chloride trihydrate (HAuCl4), bovine serum albumin (BSA) and radio-immunoprecipitation assay (RIPA) lysis and extraction buffer were purchased from Fisher Scientific (Barcelona, Spain). The phosphate-buffered saline (PBS; 10 mM phosphate, 137 mM NaCl, 2.7 mM KCl, pH 7.4), BSA, 3,3′,5,5′-tetramethylbenzidine (TMB), IGEPAL® CA-630, boric acid, sodium citrate, tris(2-carboxyethyl)phosphine (TCEP) and horseradish peroxidase were from Sigma (Barcelona, Spain). Sodium acetate, acetic acid, and sodium chloride were obtained from Scharlab (Barcelona, Spain). The 40 nm citrate-capped AuNPs was sourced from BBI Solutions (Crumlin, UK).The prepared buffers include binding buffer (BB; PBS with 1.5 mM MgCl2), sample solution (BB with 1% (v/v) IGEPAL® CA-630), running buffer (BB with 7.5% (v/v) BSA) and conjugate buffer (5 mM borate buffer, pH 8.8 with 10% (w/v) sucrose and 1% (w/v) BSA). All solutions were prepared using high-purity water.
The lateral flow strip material absorbent pad and nitrocellulose membrane were purchased from GE Healthcare Life Sciences (Germany), the conjugate pad (grade 8951) from Ahlström (Finland) and the backing pad from DCN DX (USA). Cassettes were purchased from TV Plastics (India).
Microbial isolates
The protozoa T. vaginalis American Type Culture Collection (ATCC) JH31A4 and PH401 were obtained from the PARADET research group of the Universidad Complutense de Madrid, Spain. Cells were inoculated in Trypticase-Yeast-Maltose medium supplemented with 10% (v/v) heat-inactivated bovine serum and incubated at 37 °C and 5% CO2.20 Parasites were subcultured every 48 hours. Cells were then collected and washed in PBS by centrifugation at 300 × g for 5 minutes. Live T. vaginalis cells were counted using trypan blue exclusion method before suspension in the sample buffer.The yeast Candida albicans IHEM 03243 and bacterial isolates Gardnerella vaginalis UGent 09.07, Klebsiella pneumoniae ATCC 700603, Lactobacillus crispatus LMG 0479T, Lactobacillus gasseri LMG 9203T, Lactobacillus iners FB 123-CNA-4, Lactobacillus jensenii LMG 6414T were provided by the Laboratory Bacteriology Research (LBR) (Ghent University, Ghent, Belgium). Prior to suspension in lysis solution, C. albicans on Sabouraud dextrose agar plate and K. pneumoniae on tryptic soy agar plate with 5% (v/v) sheep blood were incubated at 37 °C under aerobic conditions for 24 hours. Additionally, G. vaginalis on chocolate agar plate and Lactobacillus species on tryptic soy agar plates with 5% (v/v) sheep blood were incubated at 37 °C under anaerobic conditions for 48 hours and 24 hours, respectively.
Clinical samples
Four residual natural CVLs were from the AVEONS study, a prospective observational study that investigated the role of vaginal infections in pregnant women (between 16–20 weeks of gestation) in Bukavu, Democratic Republic of the Congo. Details regarding sample collection and analysis in the AVEONS study are as previously described.21,22 The AVEONS study was ethically approved by the Internal Review Board of the Catholic University of Bukavu (reference number UCB/CIE/NC/016/2016), by the Ministry of Public Health (reference number 062/CD/DPS/SK/2017) and by the Ethical Committee of Ghent University Hospital (reference number PA2014/003). An informed consent form was signed by each woman who accepted to participate in the study.A gynaecological examination, including a speculum examination with a sterile non-moistened speculum, was performed prior to sample collection. During this examination, the vaginal pH was determined by means of an indicator pH paper (Hilindicator pH paper) and vaginal swabs and CVL were taken after insertion of a sterile speculum.
Vaginal swabs (COPAN) were taken by gently rolling the tip against the midportion of the vaginal wall. One swab was used onsite in the preparation of three vaginal smears and the other one was stored for molecular testing. One of the vaginal smears was immediately analysed by wet-mount microscopy for the presence of clue cells, Candida and T. vaginalis. The other smears were fixated and shipped to LBR where they were Gram-stained for Nugent scoring to categorize the vaginal microbiota as healthy (Nugent score 0–3), intermediate (Nugent score 4–6) or bacterial vaginosis (Nugent score 7–10).23 The Gram-stained smears were also examined for the presence of Candida cells/hyphae and biofilm.
The CVL was collected by rinsing the cervical and vaginal mucosa with 10 mL of sterile physiological water and recovering as much lavage as possible into a vacutube. The CVLs were stored at −20 °C until shipment to the LBR, respecting cold chain transport.
Preparation of reporter aptamer-AuNP conjugate
Citrate-capped gold nanoparticles (AuNPs) with an approximate size of 20 nm were prepared by citrate reduction as previously described.24 The absorption spectrum of the gold suspension was measured using the Cary 100 Bio UV-visible spectrophotometer (Agilent).The AuNPs were directly functionalized with the 14mer aptamer modified at its 5′ end with thioctic acid or thiol using salt aging as previously reported,25 with minor modifications. Briefly, the thiolated 14mer aptamer (20 μL of 100 μM) was first reduced by adding 1 μL of 10 mM TCEP and 2 μL of 500 mM acetate buffer, pH 5.2 and letting it incubate for 1 hour with mild agitation before addition to 1 mL of OD 1 AuNP suspension. The 14mer aptamer with 5′ thioctic acid (20 μL of 100 μM) was added directly to 1 mL of OD 1 AuNP suspension. The AuNP and 14mer aptamer mixture was incubated overnight in dark conditions with mild agitation. Subsequently, 100 μL of 1 M NaCl and 10 μL of 500 mM Tris-acetate buffer, pH 5.2 was slowly added to the AuNP-14mer aptamer mixture at a rate of 10 μL every 20 minutes and incubated overnight in dark conditions with mild agitation. Functionalization of the 40 nm AuNPs (BBI Solutions) with thioctic acid-modified 14mer aptamer was also carried out in the same way. The AuNP-14mer aptamer conjugate was then centrifuged, washed three times and re-suspended in 50 μL conjugate buffer. The spectrum of the aptamer-functionalized AuNPs was acquired using the spectrophotometer (Cary 100 Bio UV-visible spectrophotometer, Agilent).
Assembly of the aptamer-based lateral flow assay (ALFA)
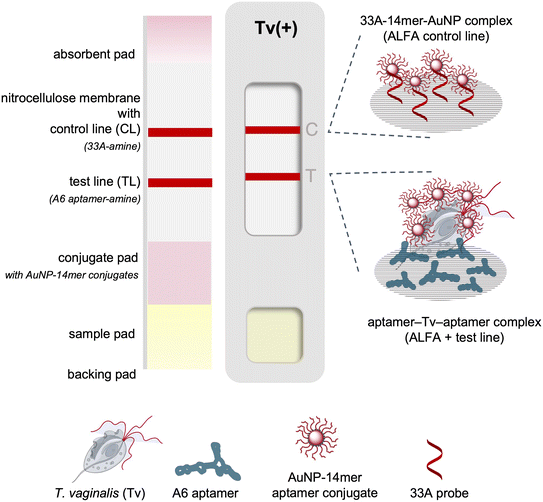
The test strip consisted of 4 overlapping pads namely the absorbent pad, detection pad, conjugate pad, and sample pad assembled on an adhesive backing pad. The CF7 cotton linter and FF170HP nitrocellulose membrane from Whatman (Germany) served as the absorbent and detection pads, respectively.The detection pad consisted of FF170HP nitrocellulose membrane with immobilized capture probes. Aminated capture A6 aptamer was dispensed at the test line and an aminated polyA (33A) oligonucleotide at the control line. The probes were either manually drawn or spotted using a pipette tip or, whenever available, automatically line dispensed using the ALFRD automated lateral flow reagent dispenser (Claremont Biosolutions). The optimum amount of capture probes was determined by testing a range of concentrations. Addition of salt in the capture probe solutions was also investigated to increase the capture zone intensities whilst maintaining the specificity of the test. The aminated oligonucleotides were then exposed to 254 nm ultraviolet (UV) light at 90 mJ cm−2 for 5 minutes (CL-1000 UV crosslinker, Analytik Jena).26 The optimum conditions for UV crosslinking were determined by testing a range of UV energy and exposure times.
The conjugate pad consisted of glass fiber (Ahlström) pre-treated with 5 mM borate buffer, pH 8.8 and 1% (w/v) BSA and dried at 37 °C for at least 2 hours before deposition of the AuNP-14mer conjugate. The addition of Tween 20 in the pre-treatment solution was also investigated. The pre-treated conjugate pad was saturated and dried with AuNP-14mer conjugate initially at OD 10 and was then optimized to achieve low to no background signal.
Bound glass fiber (Whatman, Germany) served as the sample pad. Use of cotton and Fusion 5 (proprietary composition) materials were also tested. The assembled ALFA pad was cut into strips (Advanced Sensor Systems P. Ltd.) with respect to the dimensions of the housing cassette fitting a 4 mm by 8 mm strip.
Optimization of the ALFA protocol
In the assay optimization using T. vaginalis cells, cells were cultured at 37 °C for 48 hours then harvested and resuspended in the ALFA sample buffer. The aptamer binding buffer (BB; 10 mM PBS with 1.5 mM MgCl2) with or without 1% (v/v) IGEPAL® CA-630 and RIPA buffer were evaluated as the ALFA sample buffer. For the ALFA running buffer, different ionic concentrations of BB supplemented with BSA were tested. Incorporation of BSA in different test components (addition in the membrane blocking or test running buffers or drying on the sample pad) and the use of different types of sample pad or nitrocellulose membrane were investigated. Regarding sample processing or resuspension of the cells, separate or combined preparations of the sample/cells and running buffers, adding IGEPAL and BSA together in BB, were investigated. In running the assay, sample buffer with or without cells were mixed with 3 volumes of running buffer. In the ALFA sample window, the sample mixture at 200 μL was dispensed. Different ratios of the sample buffer and running buffer were also evaluated. The sample mixture was allowed to flow through the membrane for 30 minutes, but a clear result could be observed in less than 15 minutes.Stability study
The ALFA strip and the BSA-based running buffer were stored at 4 °C and 37 °C for 90 days and their stability was evaluated at different time intervals. The ALFA pad was assembled using the CF7 cotton linter (Whatman, Germany) as absorbent pad, FF170HP nitrocellulose membrane (Whatman, Germany) as the detection pad, glass fiber pad (Ahlström) with OD 5 of AuNP-14mer conjugate, and bound glass fiber (Whatman, Germany) as the sample pad. The aminated capture probe solutions in nuclease-free water, 5 μM of polyA oligonucleotide (33A) at the control line and 200 μM of A6 aptamer at the test line, were automatically dispensed on the nitrocellulose membrane and then exposed to UV 254 nm at 90 mJ cm−2 for 10 minutes for crosslinking. The ALFA pad was cut into strips (4 mm by 8 mm), placed inside the cassette, sealed in a bag with a desiccant, and finally stored at 4 °C and 37 °C. The running buffer, consisting of 1× BB supplemented with 7.5% (w/v) BSA, was also stored at 4 °C and 37 °C and used over a 90-day period.In running the ALFA, 4.0 × 106 cells per mL of T. vaginalis was spiked in the sample buffer. A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio of sample buffer
3 ratio of sample buffer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) running buffer was then prepared and 200 μL was dispensed on the sample window. The ALFA result was recorded and photographed after 30 minutes.
running buffer was then prepared and 200 μL was dispensed on the sample window. The ALFA result was recorded and photographed after 30 minutes.
Sensitivity of the ALFA
Serial dilutions of in vitro cultured T. vaginalis in sample buffer were prepared and used to determine the lowest number of cells that could be detected with the ALFA. The cells were suspended in sample buffer to reach 2.0 × 107 cells per mL and then diluted 1/5 in sample buffer. For each cell concentration, 200 μL of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio of prepared sample
3 ratio of prepared sample![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) running buffer mixture were dispensed to the sample window. The ALFA result was recorded, photographed, and line intensities were measured using the Cube LFA reader (Biosynex Technologies, Germany) after 30 minutes. Duplicate samples were analysed.
running buffer mixture were dispensed to the sample window. The ALFA result was recorded, photographed, and line intensities were measured using the Cube LFA reader (Biosynex Technologies, Germany) after 30 minutes. Duplicate samples were analysed.
Analytical specificity
The ALFA tests were prepared as above and used to analyse T. vaginalis and other vaginal microbial isolates. The cells were suspended in the sample buffer at the following final cell concentrations: 2.0 × 107 cells per mL of T. vaginalis, 6.7 × 106 cells per mL (OD550 = 0.75) of C. albicans, and 1.5 × 108 cells per mL (OD550 = 0.125) each of Gardnerella vaginalis, Lactobacillus iners, L. crispatus, L. gasseri, L. jensenii, and Klebsiella pneumoniae. Subsequently, 200 μL of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio of cell suspension
3 ratio of cell suspension![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) running buffer mixture were dispensed to the sample window of the ALFA. The ALFA result was recorded and photographed after 30 minutes. Duplicate samples were analysed.
running buffer mixture were dispensed to the sample window of the ALFA. The ALFA result was recorded and photographed after 30 minutes. Duplicate samples were analysed.
Evaluation of the ALFA with biobanked clinical CVLs
Four residual clinical CVLs from the AVEONS study were used in the preliminary application of ALFA on clinical samples. These CVLs with at least 100 μL volume include one T. vaginalis negative sample and three T. vaginalis positive samples evaluated by wet-mount microscopy. One of the T. vaginalis positive samples was also diagnosed positive for bacterial vaginosis while the others had healthy vaginal microbiota based on the Nugent scoring.Briefly for analysis of the CVLs with the ALFA, 40 μL of each CVL was mixed with 10 μL of 5× sample buffer. This mixture was then diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 with the running buffer to reach 200 μL volume. The sample mixture was dispensed to the sample window of the ALFA. The ALFA result was recorded, photographed, and line intensities were measured using the Cube LFA reader after 30 minutes.
3 with the running buffer to reach 200 μL volume. The sample mixture was dispensed to the sample window of the ALFA. The ALFA result was recorded, photographed, and line intensities were measured using the Cube LFA reader after 30 minutes.
Results and discussion
Test design
An aptamer dual pair was exploited in a sandwich format in a paper-based chromatographic assay as a point-of-need test for the detection of T. vaginalis. The full length 95mer A6 aptamer served as the capture aptamer immobilized on the nitrocellulose membrane while the A1_14mer aptamer was used as the reporter aptamer in this aptamer lateral flow assay (ALFA). This system follows the order of aptamer-analyte binding in the microplate assay19 where the A1_14mer aptamer binds to the analyte first and followed by the A6 aptamer for detection. The schematic for the biotin/avidin-free ALFA is presented in Fig. 1.Preparation of the aptamer-AuNP conjugate
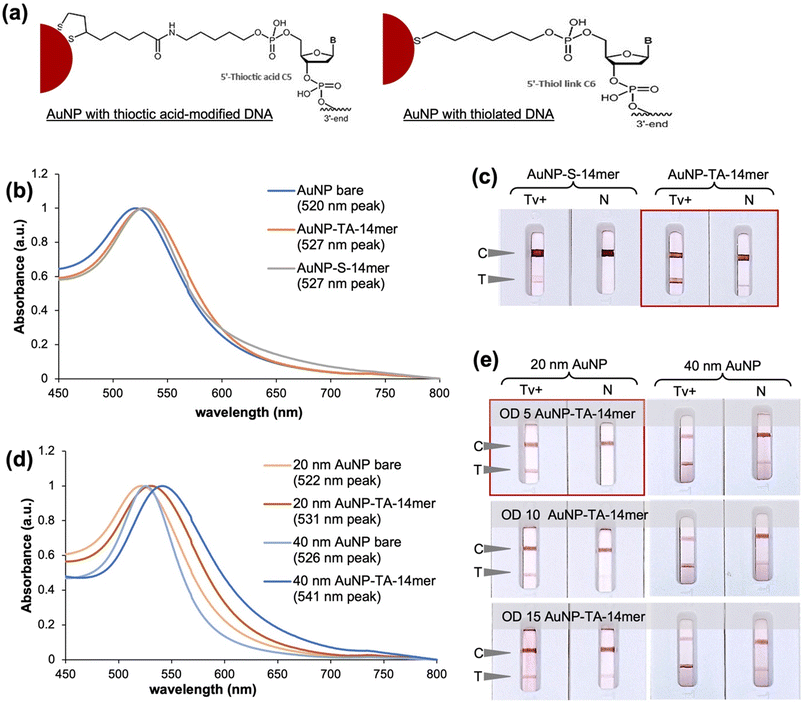
In the preparation of AuNP-aptamer conjugate, the reporter 14mer aptamer was modified at the 5′ end with thiol or thioctic acid. The thiol provides a monothiol linkage to the AuNP while thioctic acid allows dithiol linkages (Fig. 2a). The similar absorption maximum of the peaks (527 nm) of the AuNP-14mer conjugates suggest that similarly sized conjugates were achieved using the aptamer with thiol (S) or thioctic acid (TA) modifications (Fig. 2b). Modification of the 14mer with thioctic acid instead of thiol proved more suitable for the assay as it provided a significantly more intense signal (Fig. 2c). This may be attributable to less steric hindrance on the aptamer-analyte binding provided by the dithiol anchors of the thioctic acid-modified aptamer on the AuNP. A higher stability of the AuNP-14mer conjugate can also be expected with the thioctic acid compared to the thiol linker due to its bipodal nature.27 In parallel, the preparation of the AuNP-TA-14mer conjugate was optimised using 20 nm and 40 nm AuNPs. A shift in the peak absorbance of the AuNPs from 522 nm to 531 nm for the 20 nm AuNP and from 526 nm to 541 nm for the 40 nm AuNP was measured (Fig. 2d), indicating functionalization of the AuNPs.28 And as can be seen in Fig. 2e, the use of 20 nm AuNPs at OD 5 of the AuNP-TA-14mer conjugate was observed to be optimal to achieve specific test and control lines. Nonspecific signals were observed with the use of larger AuNPs (40 nm) and higher ODs of the AuNP-TA-14mer conjugates. Finally for the preparation of the aptamer-AuNP using the salt-aging method,25 the 14mer aptamer with 5′ thioctic acid (20 μL of 100 μM) was added directly to 1 mL of 20 nm AuNP suspension (OD 1). The AuNP and 14mer aptamer mixture was incubated overnight in dark conditions with mild agitation followed by slow addition of the salt solution consisting of 1 M NaCl and 500 mM Tris-acetate buffer, pH 5.2. The mixture was again incubated overnight in dark conditions with mild agitation before washing the suspension of the aptamer-functionalized AuNPs with the conjugate buffer.Optimization of ALFA buffers and material components
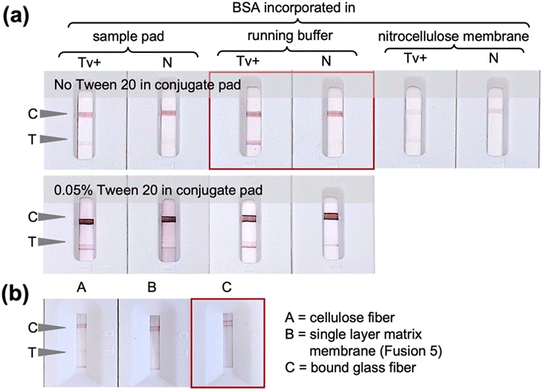
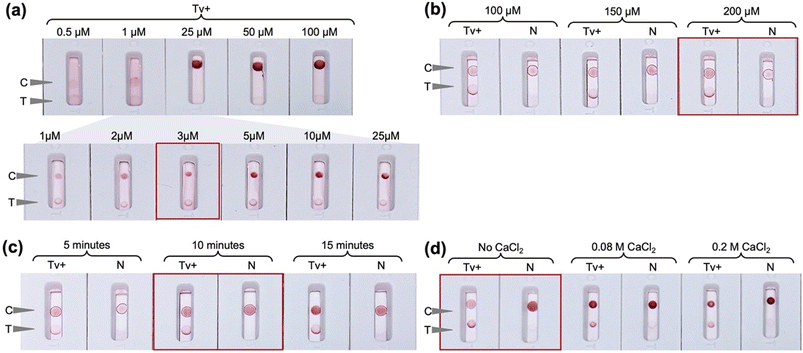
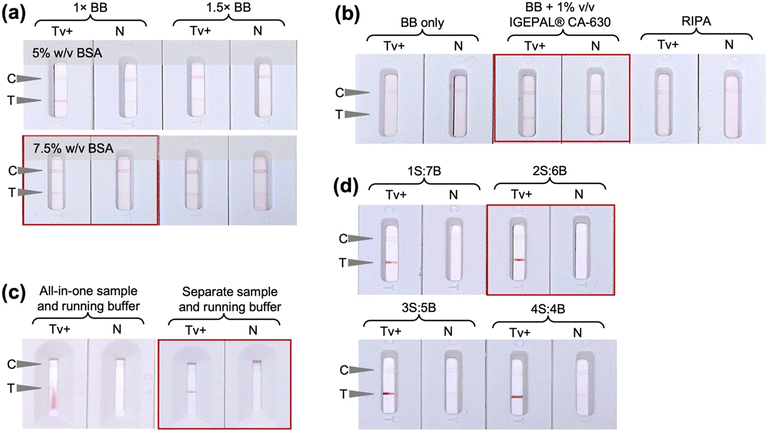
The initial ALFA experiments shown in Fig. 2 were conducted using Tween 20 surfactant in the conjugate pad and faint nonspecific test lines were observed in some cases. Whilst it is desirable to minimise the use of proteins in the ALFA so as to reduce costs, the use of BSA was explored as an alternative additive to Tween 20 to improve test specificity. Indeed, as can be seen in Fig. 3a, removal of the Tween 20 from the conjugate pad and incorporation of 5% (w/v) BSA in the running buffer led to reduction of nonspecific signal. In this case, the conjugate pad in the ALFA consisted of glass fiber pretreated with Tween 20-free solution, i.e., 5 mM borate buffer with 1% (w/v) BSA, and saturated with OD 5 conjugate prepared with 20 nm AuNPs functionalized with thioctic acid-modified 14mer aptamer via chemisorption. Alternative strategies of incorporating BSA in the assay by deposition in the oligo-functionalized nitrocellulose membrane and sample pad led to very faint positive test lines. Comparison of different sample pad materials (bound glass fiber, cellulose fibre and Fusion 5) using only assay buffer without cells (Fig. 3b) showed that bound glass fiber is the optimum sample pad with it giving the only true negative signal.The concentration and immobilization procedure of the capture probes employed for the preparation of the detection pad was then optimized. The FF170HP nitrocellulose membrane was used due to the smooth fluid flow and capture line formation as the use of more porous membranes would lead to faster fluid flow and thus less intense capture lines. A range of concentrations of both the control capture probe 33A and the test capture probe A6 aptamer were evaluated. In the case of the control line, 3 μM of 33A-amine was observed to give an adequately intense but not too intense signal (Fig. 4a), whilst for the test line a concentration of 200 μM A6-amine aptamer was required to obtain signals of strong enough intensities to be observed. The difference in the signal intensities at the control and test lines can be explained by the fact that at the control line simple hybridization is occurring, solely relying on the A–T base complementarity with polyA on the nitrocellulose and polyT spacer on the AUNP-TA-14mer conjugate. At the test line, aptamer–analyte–aptamer–AuNP complexation driven by electrostatic interactions, hydrogen bonding and hydrophobic interactions takes place and the formation of three-dimensional structures are involved. UV-mediated immobilization of DNA on the nitrocellulose membrane29,30 was also evaluated and UV exposure at 90 mJ cm−2 for 10 minutes was observed to be optimal and was thus used in all further experiments (Fig. 4c). UV-mediated covalent immobilization of aptamers on nitrocellulose membranes has been previously reported as an attractive alternative to physical or biotin/streptavidin-mediated immobilization, eliminating the use of additional proteins and potentially improving LFA consistency by promoting a more homogenous aptamer immobilization on the membrane.31,32 Finally, salt-enhanced immobilization of the aptamer was also explored in an effort to further improve its immobilization efficiency on the membrane. It was previously shown that the immobilization of DNA in the presence of high concentration of metal salts like calcium chloride improved the colorimetric signal in LFA more than ten-fold, due to partial neutralization of the negative charge of the phosphate backbone of DNA after complexing with the metal ion and consequent reduced repulsion between DNA and the nitrocellulose membrane.26 In this work it was observed that the addition of calcium chloride in the capture probe solution hinders the aptamer-analyte interaction (Fig. 4d) which may be attributable to a change in the structure and therefore aptamer-target interaction in the presence of cations.33 Consequently, the detection pad was prepared by automatic line dispensing of 3 μM control 33A oligonucleotide and 200 μM test A6 aptamer on the FF170HP nitrocellulose membrane followed by UV exposure at 90 mJ cm−2 for 10 minutes.
Another parameter studied was the ionic strength of the aptamer binding buffer (BB) which was used as the test running buffer. The incorporation of BSA in the running buffer was found to be essential during the earlier experiments and further evaluation showed that increasing the amount of BSA instead of the ionic strength of BB led to more intense test lines and a 1× BB containing 7.5% (w/v) BSA was used as running buffer in all further experiments (Fig. 5a). The sample buffer that enables cell lysis and aptamer binding was then optimized. As can be seen in Fig. 5b, the use of BB alone or the use of RIPA buffer led to false negative result, whilst the use of BB supplemented with 1% (v/v) IGEPAL® CA-630 provided a clean, specific signal. In an attempt to simplify the system and reduce the number of assay operational steps, an all-in-one sample and running buffer was tested, combining IGEPAL® CA-630 and BSA in BB but poor liquid flow on the membrane and a faint positive test line were observed (Fig. 5c). The faint positive test line may be due to inefficient cell lysis by IGEPAL® CA-630 due to the presence of BSA in the solution. It is thus essential to have a 2-step sample processing step, by first resuspending and lysing the cells in the sample buffer followed by dilution with the running buffer, prior to dispensing on the ALFA sample window. The ratio of lysed sample to running buffer were then assessed and a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio of lysis
3 ratio of lysis![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) running buffer was found to be optimal for a true positive test line (Fig. 5d).
running buffer was found to be optimal for a true positive test line (Fig. 5d).
ALFA test stability
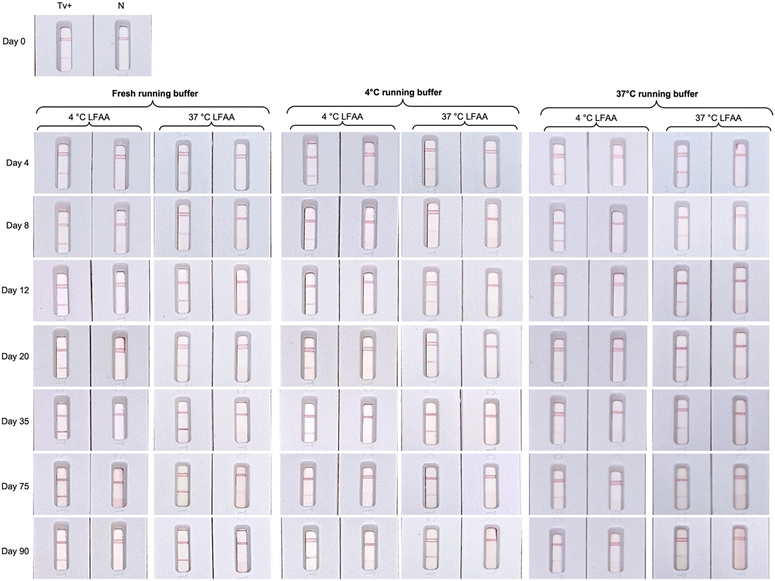
A study on the long-term stability of the developed ALFA and the running buffer at 4 °C and 37 °C storage temperature was conducted over a 90-day period using a single concentration of in vitro cultured T. vaginalis (Fig. 6). Using Arrhenius Accelerated Thermal Stability Studies34 it can be predicted that the ALFA has a predicted shelf-life of at least 1 year at room temperature (22 °C). Using freshly prepared running buffer, the ALFA remained functional when stored at 4 °C and 37 °C conditions over the 90 days studied. Regarding the stability of the running buffer containing 7.5% (w/v) BSA, the signal intensity was relatively stable and almost equal to that observed on day 0 with the freshly prepared buffer and the buffer stored at 4 °C. A general decline in line intensity was observed with the buffer stored at 37 °C for 20 days. The developed ALFA, with an estimated laboratory-level production cost of less than 1 € per test as detailed in Table 1, should be stored in dry conditions and while freshly prepared running buffer is recommended, buffer stored at 4 °C is also acceptable.| Part | Cost per unit, € | Cost per test, € |
|---|---|---|
| Cassette (excluding shipping fee) | 0.0220 | 0.0220 |
| Backing pad, pack of 50 80 mm × 300 mm pads (DCN, USA) | 88 | 0.0235 |
| Absorbent pad, roll of 22 mm × 50 m (Whatman, Germany) | 201 | 0.1005 |
| Sample pad, roll of 22 mm × 50 m (Whatman, Germany) | 218 | 0.1090 |
| Detection pad | ||
|---|---|---|
| Nitrocellulose membrane, roll of 25 mm × 50 m (Whatman, Germany) | 287 | 0.1435 |
Control capture probe, 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 pmol (Biomers, Germany) 700 pmol (Biomers, Germany) |
48.15 | 0.0010 (1 pmol) |
| Test capture zone, 6800 pmol (Biomers, Germany) | 45.95 | 0.2703 (40 pmol) |
| Conjugate pad | ||
|---|---|---|
| Glass fiber, roll of 8 mm × 100 m | 89.19 | 0.0036 |
| AuNP-aptamer conjugate, 200 μL of OD5 | 1.73 | 0.1298 |
| Buffers (sample and running) | ||
|---|---|---|
| BSA, 100 g | 387.42 | 0.0436 (0.01125 g) |
| PBS with 1.5 mM MgCl2, 10 L | 174 | 0.0035 (200 μL) |
| IGEPAL, 10 L | 130 | 0.0007 (50 μL) |
| Total | 0.8388 | |
ALFA test sensitivity and specificity
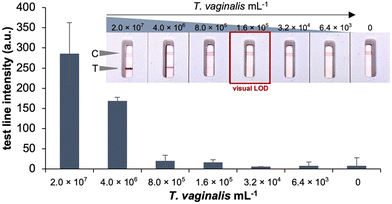
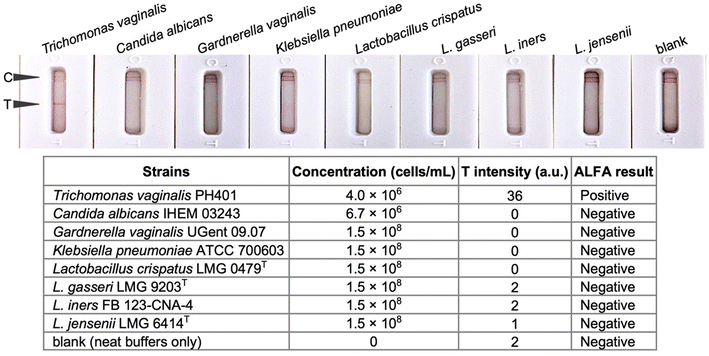
The analytical sensitivity (Fig. 7) and specificity (Fig. 8) of the developed ALFA were determined. Test of serially diluted in vitro cultured T. vaginalis cells showed that the assay can detect down to 1.6 × 105 cells per mL of T. vaginalis by visual inspection. The developed ALFA requires more T. vaginalis cells than its microplate counterpart which has a LOD of 3.02 × 104 cells per mL,19 while the commercially available immunochromatographic OSOM Trichomonas test has a LOD of 2.5 × 103 cells per mL.12 The direct detection of cells using aptamer-based colorimetric strip test have also been reported for Vibrio fischeri requiring at least 40 cells per mL (ref. 35) and for Ramos cells at a minimum of 4.0 × 103 cells by visual inspection to 800 cells with the aid of a portable strip reader.36 With antibody-based LFAs, a detection limit of 106 cells per mL was reported for the direct detection of N. gonorrhoeae cells.37The specificity of the developed ALFA was then tested using in vitro cultured microbial isolates typically found in the vaginal environment. As can be seen in Fig. 8, the test shows no cross-reactivity to C. albicans and common vaginal bacterial species such as G. vaginalis, K. pneumoniae, L. crispatus, L. gasseri, L. iners, and L. jensenii.
ALFA using clinical samples
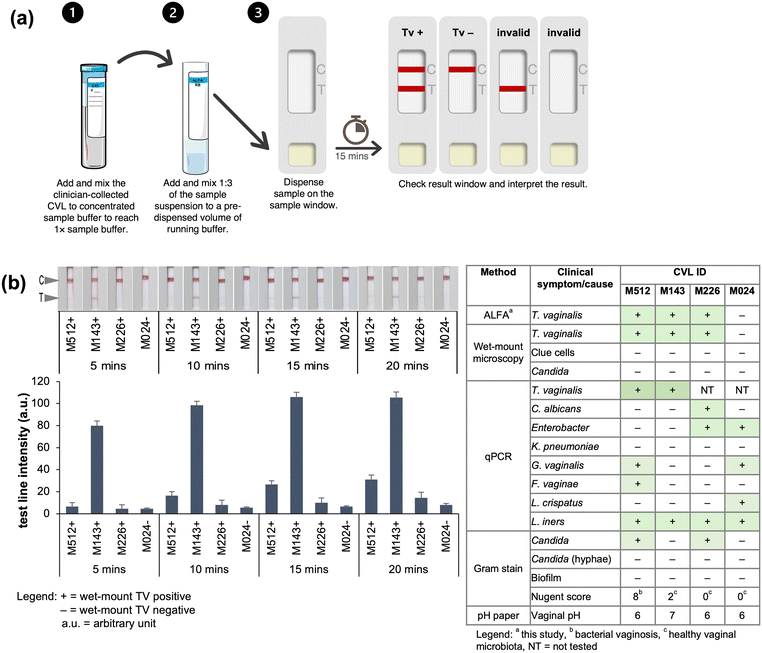
Biobanked clinical CVLs were used to demonstrate the compatibility of the ALFA with T. vaginalis testing in clinical specimens. The use of ALFA in rapid testing for the presence of T. vaginalis in the biobanked clinical CVLs require three operator steps as illustrated in Fig. 9a: (i) add and mix by finger tapping the clinician-collected CVL to concentrated sample buffer to reach 1× sample buffer, (ii) add and mix by finger tapping the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 of the sample suspension to a pre-dispensed volume of running buffer, (iii) dispense sample (200 μL) on the sample window and let run for 15 minutes wherein the sample window showing two red lines (C and T) means positive, one C line means negative, and no C line means invalid test.
3 of the sample suspension to a pre-dispensed volume of running buffer, (iii) dispense sample (200 μL) on the sample window and let run for 15 minutes wherein the sample window showing two red lines (C and T) means positive, one C line means negative, and no C line means invalid test.
Recording of the test result in 5-minute intervals after application of the sample showed that the assay selectively captured and detected T. vaginalis with a clear response observed in just 15 minutes (Fig. 9). The results obtained with the developed ALFA corroborated perfectly with those obtained with wet-mount microscopy for T. vaginalis. Moreover, the presence of other microorganisms in the CVLs revealed by previous analysis of the same set of collected samples evaluating the vaginal health21,22 did not affect the detection of T. vaginalis using the ALFA. On the other hand, the T. vaginalis negative sample according to wet-mount microscopy was found to harbour vaginal bacteria such as G. vaginalis, Lactobacillus, and Enterobacter and it was also corroborated by the ALFA as T. vaginalis negative. The test line in ALFA had different intensities possibly reflecting the different severity of T. vaginalis infection on the wet-mount T. vaginalis positive samples with abnormal vaginal pH of 6 and 7, however this cannot be confirmed considering that cell counting is not done in wet-mount microscopy and the final amount of lavage is also affected by situational conditions during collection. Interestingly, a very intense test line in ALFA was observed in the specimen with vaginal pH of 7 (CVL ID M143) and not on the specimen with bacterial vaginosis based on the Nugent score (CVL ID M512). The Nugent score and Amsel criteria are the two common diagnostic tests for bacterial vaginosis.38 An elevated vaginal pH, greater than the normal vaginal pH 4.5, is also used as an indicator of infection however it can also be due to vaginal douching, intercourse, and the presence of blood and mucus.39,40
Conclusions
The developed assay is the first aptamer-based paper chromatographic assay for T. vaginalis and the first sandwich aptamer-based paper chromatographic assay that is free of the biotin/avidin system. The developed aptamer-based LFA is specific to T. vaginalis and compatible for use with clinical CVL specimens. The assay serves as a simple alternative test for the diagnosis of trichomoniasis requiring three simple operator steps from sample-to-result. The assembled ALFA with an estimated laboratory-based production cost of less than 1 € per test should be stored in dry conditions and has a predicted shelf-life of at least 1 year at room temperature (22 °C). Freshly prepared running buffer is recommended, but buffer stored at 4 °C is also acceptable. The test can detect down to 1.6 × 105 T. vaginalis per mL without cross-reactivity to common vaginal microorganisms. Preliminary evaluation using clinical CVLs showed that the assay has perfect concordance with wet-mount assay on matching vaginal swabs. The developed rapid test offers a simple, stable, and low-cost diagnostic test for T. vaginalis.Work evaluating the assay's use on a larger number of CVL, as well as evaluation for use with other clinical specimens such as genital swabs and urine, is planned. The simple operation of the assay and protein-free design of the ALFA strip, offers a simple, stable, and low-cost diagnostic test for T. vaginalis. It has great potential to serve as near-patient test for the detection of trichomoniasis. Work is ongoing to lower the number of cells that can be detected by the developed ALFA, by incorporating a nucleic acid amplification step exploiting the nucleic acid nature of the aptamer and via the use alternative nanomaterials and test configurations.
Data availability
Data for this article are available at the URV Repository at https://repositori.urv.cat/en/.Author contributions
CACJ – investigation, formal analysis, writing –original manuscript; VS – supervision, writing – review and editing; PC – resources; GM – resources; AIE – resources; CKO – resources, funding acquisition, supervision, writing – review and editing.Conflicts of interest
The authors declare no competing interest.Acknowledgements
The pre-doctoral scholarship by Agencia Gestio d'Ajuts Universitaris i de Recerca (FI-SDU 2020 00467) to CACJ is acknowledged. The authors also thank Lisa Himschoot for the technical assistance.References
- World Health Organization, Global progress report on HIV, viral hepatitis and sexually transmitted infections, 2021. Accountability for the global health sector strategies 2016–2021: actions for impact, World Health Organization, Geneva, 2023 Search PubMed.
- World Health Organization, Point-of-care tests for sexually transmitted infections: target product profiles, World Health Organization, Geneva, 2023 Search PubMed.
- K. J. Land, D. I. Boeras, X. S. Chen, A. R. Ramsay and R. W. Peeling, Nat. Microbiol., 2019, 4, 46–54 Search PubMed.
- I. Toskin, V. Govender, K. Blondeel, M. Murtagh, M. Unemo, C. Zemouri, R. W. Peeling and J. Kiarie, Sex. Transm. Infect., 2020, 96, 342–347 CrossRef PubMed.
- T. E. Wi, F. J. Ndowa, C. Ferreyra, C. Kelly-Cirino, M. M. Taylor, I. Toskin, J. Kiarie, N. Santesso and M. Unemo, J. Int. AIDS Soc., 2019, 22, 8–18 Search PubMed.
- K. Carter-Wicker, O. Utuama and F. Omole, SAGE Open Med. Case Rep., 2016, 4, 2050313X1668213 Search PubMed.
- S. C. Francis, C. K. Kent, J. D. Klausner, L. Rauch, R. Kohn, A. Hardick and C. A. Gaydos, Sex. Transm. Dis., 2008, 35, 797–800 CrossRef PubMed.
- C. M. Hoffman, L. Fritz, O. Radebe, J. H. Dubbink, J. A. McIntyre, M. M. Kock and R. P. H. Peters, Int. J. STD AIDS, 2018, 29, 1444–1447 CrossRef PubMed.
- Association of Public Health Laboratories, Advances in Laboratory Detection of Trichomonas vaginalis (Updated), Association of Public Health Laboratories, Maryland, 2016 Search PubMed.
- Unitaid, Multi-disease diagnostic landscape for integrated management of HIV, HCV, TB and other coinfections, World Health Organization, Geneva, 2018 Search PubMed.
- S. L. Gottlieb, E. Spielman, L. Abu-Raddad, A. K. Aderoba, L. H. Bachmann, K. Blondeel, X.-S. Chen, T. Crucitti, G. G. Camacho, S. Godbole, R. G. P. de Leon, S. Gupta, J. Hermez, N. Ishikawa, J. D. Klausner, F. Kurbonov, I. Maatouk, A. Mandil, M. B. Mello, A. E. Miranda, F. S. Mosha, J. C. Okeibunor, J. J. Ong, R. P. H. Peters, F. Pérez, N. Seguy, K. L. Seib, M. Sharma, T. Sladden, B. Van Der Pol, P. J. White, T. Wi and N. Broutet, Lancet Global Health, 2024, 12, e1544–e1551 CrossRef CAS PubMed.
- Sekisui Diagnostics LLC, OSOM Trichomonas Test Package Insert, Sekisui Diagnostics, LLC, California, 2021 Search PubMed.
- S. R. Morris, C. C. Bristow, M. R. Wierzbicki, M. Sarno, L. Asbel, A. French, C. A. Gaydos, L. Hazan, L. Mena, P. Madhivanan, S. Philip, S. Schwartz, C. Brown, D. Styers, T. Waymer and J. D. Klausner, Lancet Infect. Dis., 2021, 21, 668–676 Search PubMed.
- Visby Medical Inc., Sexual Health Instructions for Use, Visby Medical, Inc., California, 2023 Search PubMed.
- J. F. Alderete and H. Chan, Pathogens, 2023, 12, 77 CrossRef CAS PubMed.
- Cepheid, Xpert® TV Instructions for use, Cepheid, California, 2022 Search PubMed.
- Molbio Diagnostics Private Limited, Truenat® Trich Package insert. Molbio Diagnostics Private Limited, Goa, 2021 Search PubMed.
- World Health Organization, Global health sector strategies on, respectively, HIV, viral hepatitis and sexually transmitted infections for the period 2022–2030, World Health Organization, Geneva, 2022 Search PubMed.
- C. A. C. Justo, M. Jauset-Rubio, M. Svobodova, V. Skouridou, P. Cools, G. Mulinganya, A. Ibáñez-Escribano, W. L. Rivera and C. K. O'Sullivan, Anal. Biochem., 2024, 695, 115656 CrossRef CAS PubMed.
- C. G. Clark and L. S. Diamond, Clin. Microbiol. Rev., 2002, 15, 329–341 CrossRef PubMed.
- G. Mulinganya, A. De Vulder, G. Bisimwa, J. Boelens, G. Claeys, K. De Keyser, D. De Vos, E. Hendwa, F. Kampara, Y. Kujirakwinja, J. Mongane, I. Mubalama, M. Vaneechoutte, S. Callens and P. Cools, PLoS One, 2021, 16, e0257939 Search PubMed.
- M. G. Mulinganya, K. De Keyser, I. J. Mongane, M. F. Kampara, A. De Vulder, J. Boelens, H. Duyvejonck, E. Hendwa, B. Y. Kujirakwinja, B. G. Bisimwa, A. Rodriguez, M. Vaneechoutte, S. Callens and P. Cools, Front. Glob. Womens Health, 2024, 5, 1–12 CrossRef PubMed.
- R. P. Nugent, M. A. Krohn and S. L. Hillier, J. Clin. Microbiol., 1991, 29, 297–301 CrossRef CAS PubMed.
- L. Liu, H. Pan, M. Du, W. Xie and J. Wang, Electrochim. Acta, 2010, 55, 7240–7245 CrossRef CAS.
- M. Jauset-Rubio, M. Svobodová, T. Mairal, C. McNeil, N. Keegan, M. S. El-Shahawi, A. S. Bashammakh, A. O. Alyoubi and C. K. O'Sullivan, Anal. Chem., 2016, 88, 10701–10709 CrossRef CAS PubMed.
- J. S. Park, S. Kim, J. Han, J. H. Kim and K. S. Park, Sens. Actuators, B, 2022, 351, 130975 CrossRef CAS PubMed.
- J. A. Dougan, C. Karlsson, W. E. Smith and D. Graham, Nucleic Acids Res., 2007, 35, 3668–3675 Search PubMed.
- L. Kaiser, J. Weisser, M. Kohl and H.-P. Deigner, Sci. Rep., 2018, 8, 5628 CrossRef PubMed.
- M. Pongsuchart, A. Sereemaspun and K. Ruxrungtham, Int. J. Life Sci. Technol., 2013, 1, 172–175 Search PubMed.
- M. Pongsuchart, A. Sereemaspun and K. Ruxrungtham, Asian Biomed., 2012, 6, 459–463 Search PubMed.
- J. Bruno, Pathogens, 2014, 3, 341–355 Search PubMed.
- T. Wang, L. Chen, A. Chikkanna, S. Chen, I. Brusius, N. Sbuh and R. N. Veedu, Theranostics, 2021, 11, 5174–5196 Search PubMed.
- E. Baldrich, A. Restrepo and C. K. O'Sullivan, Anal. Chem., 2004, 76, 7053–7063 Search PubMed.
- N. Laboria, A. Fragoso and C. K. O'Sullivan, Anal. Lett., 2011, 44, 2019–2030 CrossRef CAS.
- W. R. Shin, S. S. Sekhon, S. K. Rhee, J. H. Ko, J. Y. Ahn, J. Min and Y. H. Kim, ACS Comb. Sci., 2018, 20, 261–268 CrossRef CAS PubMed.
- G. Liu, X. Mao, J. A. Phillips, H. Xu, W. Tan and L. Zeng, Anal. Chem., 2009, 81, 10013–10018 CrossRef CAS PubMed.
- T. Oeschger, L. Kret and D. Erickson, Curr. Res. Biotechnol., 2022, 4, 359–364 CrossRef CAS.
- D. B. Nelson, Epidemiol. Rev., 2002, 24, 102–108 CrossRef PubMed.
- J. Zodzika, D. Rezeberga, I. Jermakova, O. Vasina, N. Vedmedovska and G. Donders, Acta Obstet. Gynecol. Scand., 2011, 90, 41–46 CrossRef CAS PubMed.
- Y. Lin, W. Chen, C. Cheng and C.-J. Shen, Diagnostics, 2021, 11, 1996 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2025 |