Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The role of phosphate in silk fibroin self-assembly: a Hofmeister study†
Caleb
Wigham
ab,
Vrushali
Varude
ab,
Henry
O’Donnell
a and
R. Helen
Zha
 *ab
*ab
aDepartment of Chemical and Biological Engineering, Rensselaer Polytechnic Institute, Troy, NY 12180, USA. E-mail: zhar@rpi.edu
bCenter for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, Troy, NY 12180, USA
First published on 26th February 2025
Abstract
Silk fibroin is the primary protein component of the threads of Bombyx mori silkworm cocoons. Previous work has demonstrated that silk fibroin can self-assemble at solid–liquid interfaces to form dense, nanothin coatings that grow continuously from a substrate surface when exposed to potassium phosphate, a kosmotropic salt. Herein, the role of potassium phosphate in promoting silk fibroin self-assembly in solution and on surfaces is studied and compared to other salts in the Hofmeister series. Results show that strong kosmotropes, such as ammonium sulfate and potassium phosphate, promote a bimodal distribution of assembled species in solution that is indicative of a nucleation-growth mechanism. Interestingly, silk fibroin assemblies formed by potassium phosphate contain the highest β-sheet content, suggesting that phosphate-specific interactions play a role in silk fibroin self-assembly. In the presence of kosmotropic salts, silk fibroin nanoaggregates continuously accumulate at solid–liquid interfaces with varying early- and late-stage adsorption rates. Interfacial coatings formed in the presence of potassium phosphate are smooth, dense, and completely cover the underlying substrate without evidence of large-scale aggregation, whereas other kosmotropes generate rough, heterogeneous coatings. These studies thus decouple the kosmotropic effects of phosphate (via disruption of the protein hydration shell) from ion-specific behavior in driving silk fibroin self-assembly.
Introduction
Silk fibroin, a protein naturally derived from the threads spun by Bombyx mori silkworms, is often utilized by biomedical researchers for its biocompatibility, biodegradability, and excellent mechanical properties.1–6 Under certain conditions, this protein undergoes self-assembly, transitioning from a soluble protein with predominantly random coil secondary structure to an insoluble material rich in β-sheets.7–11 Researchers have leveraged this structural transition to fabricate silk materials into biomedically relevant morphologies for surgical, drug delivery, tissue engineering, and food preservation applications1,12–17During the natural B. mori silk spinning process, the spinning dope experiences a confluence of compositional changes and mechanical stresses, such as ion exchange,18 solution acidification,19,20 dehydration,11 and shear flows.20–24 Metallic cations, including K+, Na+, Mg2+, Cu2+, and Zn2+ increase in concentration towards the anterior of the middle division of the B. mori silk gland, while Ca2+ ions decrease in concentration.18 The pH changes throughout the spinning process as well, decreasing from ∼8.2 to below ∼6.2,19 closer to the theoretical isoelectric point of silk fibroin (4.2).20 Collectively, these changes trigger silk fibroin self-assembly and the protein transition from a predominantly random coil soluble protein to β-sheet rich material.
On a benchtop setting, B. mori silk fibroin self-assembly is often triggered by treating the protein with methanol or ethanol, promoting β-sheet formation by dehydrating the protein backbone.2,7,9,25–29 A less commonly utilized but more biomimetic route towards triggering silk fibroin assembly ex vivo leverages phosphate anions,9 which have been found to play a role in promoting β-sheet formation of spidroins during spider silk spinning.30 Phosphate is considered a kosmotropic ion, positioned in the Hofmeister series ahead of acetate and citrate and behind sulfate in its ability to precipitate (salt out) proteins.31 The salting out behavior of kosmotropes is conventionally attributed to their favorable interaction with water molecules,32–35 which disrupts the hydration layer surrounding proteins and promotes intra- and inter-protein interactions. The ability of phosphate to promote self-assembly and β-sheet formation has been demonstrated using naturally derived and recombinant spidroins.27,36–39 Studies have shown that kosmotropic ions, including phosphate, acetate, and citrate, induce liquid–liquid phase separation in spidroin solutions, resulting in protein-rich droplets that create hierarchically organized silk fibers enriched with β-sheets upon acidification and mechanical drawing.36,37,40 More specific investigations into the role of phosphate, which increases in concentration along a spider's spinning duct, have suggested that phosphate pre-orders spidroins by affecting the ring packing of tyrosine residues flanking β-sheet-forming polyalanine regions.36 Other studies have suggested kosmotropic ions also encourages aggregation of C-terminal domains, potentially by driving hydrophobic interactions and tyrosine ring stacking within the linker region of the C-terminal domain.40
The effect of kosmotropic ions on silk fibroin self-assembly is far less understood, and phosphate has not been identified in the natural B. mori spinning process. Nevertheless, this topic merits study, as kosmotropic ions such as phosphate can offer a biocompatible route towards creating tunable silk fibroin materials for a variety of biomedical applications. For example, Lammel et al. utilized concentrated potassium phosphate to prepare silk fibroin particles for drug delivery, demonstrating control over size, morphology, and secondary structure of the protein particles.9 Recent work in our lab has demonstrated that a mild phosphate buffer can trigger self-assembly of silk proteins at a solid–liquid interface into adherent and defect-free nanothin coatings. Our work demonstrated that both the recombinant dragline spidroin eADF4(C16)41 and naturally derived B. mori silk fibroin42–44 can form continuously growing coatings via phosphate-induced interfacial self-assembly without requiring covalent chemistry, surface activation (e.g. high energy radiation or plasma treatment), or specific surface chemistry or geometry. We found that these coatings form by a two-stage process that leverages non-specific protein–surface adhesive interactions in combination with cohesive protein–protein interactions.44 We also demonstrated that these coatings are a robust and versatile method for modifying cell-surface interactions41,42 (i.e. increasing nerve cell adhesion and neurite extension, preventing bacteria attachment) and for creating drug-eluting bioactive surfaces.43 Structural analysis of phosphate-induced silk fibroin self-assembly at surfaces and in solution strongly suggest that phosphate promotes β-sheet formation.44 However, it remains unclear whether the nature of this behavior is simply kosmotropic (i.e. hydration of kosmotropic ions by water, leading to dehydration of the silk fibroin chains) or whether there exists a phosphate-specific effect. Here, we explore the role of phosphate in silk fibroin self-assembly in solution and on surfaces by evaluating its effects against the effects of other salts on the Hofmeister series.
Results and discussion
Silk fibroin solution-phase assembly in kosmotropic salts
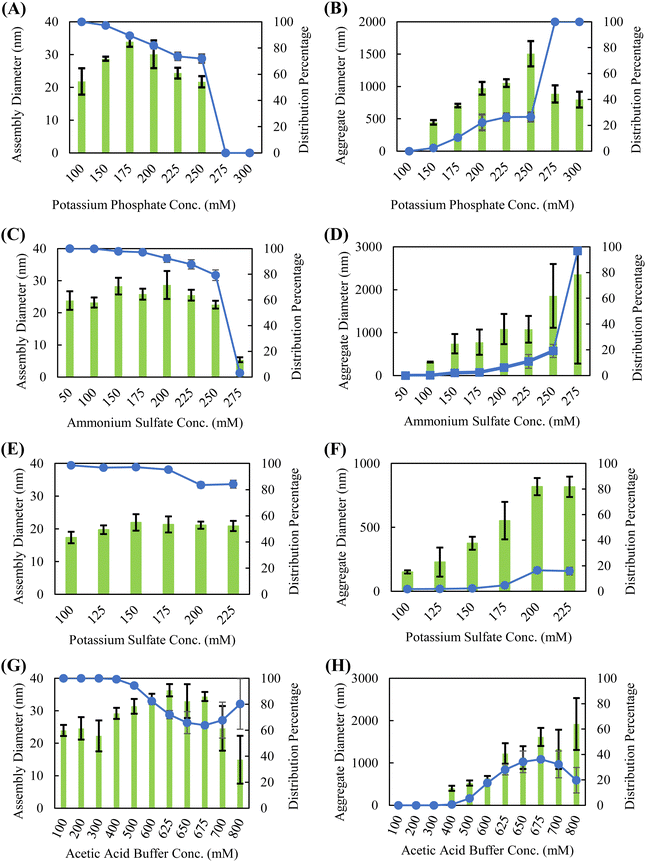
Though the position of a salt on the Hofmeister series is generally intended to indicate how readily it precipitates proteins from solution, exact positioning is often protein-dependent, as ions can have varying affinities for interacting with proteins through charge–charge interactions, direct backbone binding, or bridging.31,45,46 Dynamic light scattering (DLS) was used to characterize the solution-phase assembly and stability of silk fibroin in the presence of various Hofmeister salts. Strong chaotropic salts, such guanidine thiocyanate and lithium bromide, disrupt the intermolecular interactions required for self-assembly to occur and are commonly used to dissolve silk fibroin into aqueous solutions.2,12,14,47 Therefore, this study is confined to kosmotropic salts and “neutral” salts–salts that do not display strong kosmotropic or chaotropic behavior. The salts selected in this study were chosen to span across the kosmotropic and neutral (center) sections of the Hofmeister series. In particular, potassium phosphate and ammonium sulfate are commonly used in downstream bioprocessing for protein isolation. We also examined potassium sulfate as a comparison against potassium phosphate (anion effect) and sodium phosphate as a comparison to potassium phosphate (cation effect), though prior studies investigating salt-induced phase separation of recombinant spidroins showed that the cationic species did not affect the outcome.37 Consistent with these studies, Dicko et al. showed that Na+ and K+ cations induce similar changes in protein structure when investigating the role of cations in spidroin collected from Nephila edulis spiders.48 We also work with dilute silk fibroin solutions (0.05 wt%) compared to other published studies of silk fibroin self-assembly, as we have found that dilute concentrations lead to improved shelf stability and coating formation.Silk fibroin in the presence of kosmotropic salts, such as potassium phosphate, sodium phosphate, ammonium sulfate, potassium sulfate, and acetic acid, forms a bimodal distribution of assembled sizes indicative of a nucleation-growth aggregation mechanism (Fig. 1 and Fig. S1, ESI†). The presence of a smaller species, ranging from approximately 25–40 nm in diameter, is modulated by salt concentration. These smaller species are consumed at higher salt concentrations to form the larger, presumably inactive (not able to form coatings) protein aggregates. This bimodal distribution of assembly sizes is illustrated in Fig. S2 (ESI†); sizes below 200 nm are considered the small, active species, whereas sizes above 200 nm are considered large, inactive aggregates. To determine the position of a salt on a silk fibroin specific Hofmeister series, the salt concentration is simply increased until a precipitation event occurs. Ranking the kosmotropic salts tested, the concentrations required to precipitate silk fibroin in ascending order are potassium sulfate (∼225 mM), potassium phosphate (∼250 mM), ammonium sulfate (∼275 mM), sodium phosphate (>300 mM), and acetic acid (>800 mM). The ability of kosmotropic salts to promote solution-phase assembly is not surprising, as they promote hydrophobic intermolecular interactions by disrupting the hydration shell surrounding the protein.
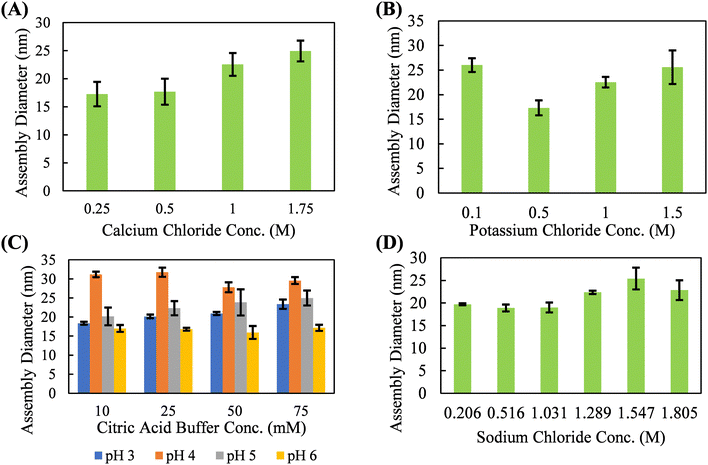
With neutral salts, such as calcium chloride, potassium chloride, and sodium chloride, silk fibroin shows solution-phase stability up to 1.5–2 M before a precipitation event occurs (Fig. 2). These three salts form a relatively monodisperse population of silk fibroin assemblies in the 15–25 nm diameter range. Neutral salts, lacking strong kosmotropic character, do not necessarily disrupt the hydration shell of the protein until multi-molar concentrations, resulting in solution phase stability of silk fibroin at much higher concentrations. Studies of silk fibroin isolated from the glands of B. mori silkworms have shown that Ca2+ may induce network formation by bridging the carboxyl groups, and that K+ may disrupt network formation by competing for carboxyl groups and by ionic shielding.49,50 However, these effects were not obvious in our studies, which utilized hydrolyzed (i.e., lower molecular weight and higher dispersity) silk fibroin and did not examine rheological behavior. The absence of large-scale aggregation indicative of a nucleation-growth mechanism suggests that there is minimal solution-phase assembly in response to potassium chloride, calcium chloride, sodium chloride, and citric acid in our system. While the diameter of the silk fibroin assemblies does slightly increase with salt concentration, this occurs over hundreds of millimolar and is likely the result of complete screening of the electric double layer and hydrophobic interactions between proteins at multi-molar salt concentrations. Citric acid deviated from the chloride salts and promoted the formation of a slightly larger silk fibroin assembly (∼25–35 nm diameter) concentrations over an order of magnitude lower than what is observed with chloride salts (∼0.05 M vs. ∼1.5 M). However, this effect is highly variable with solution pH. This solution-phase assembly into slightly larger structures may be due to the potential for citric acid to form hydrogen bonds with the protein backbone. Citric acid has previous been used to crosslink protein hydrogels through hydrogen bonding via a nucleophilic substitution reaction with the protein backbone.51,52 Due to the apparent solution-phase assembly observed for all the kosmotropic salts tested as well as the citric acid buffer, these salt solutions are evaluated for their ability to promote continuous protein accumulation at the solid–liquid interface.
Phosphate promotes the highest β-sheet content
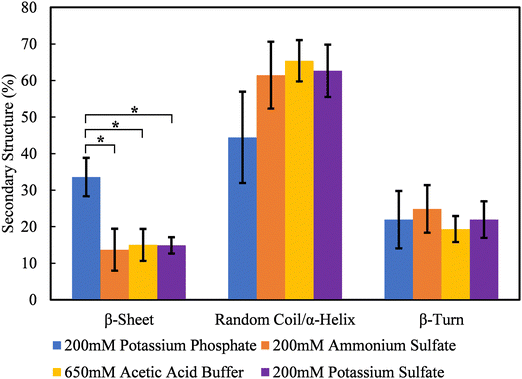
Previous published work by our group has shown that β-sheet formation drives the association of solution-phase silk assemblies and effectively crosslinks neighboring surface-bound protein assemblies.44 Therefore, the secondary structure of silk fibroin assembles formed in kosmotropic salts that promote continuous adsorption were measured using ATR-FITR after 24 hours of assembly. Deconvolution peak fitting and residuals of the raw ATR-FTIR spectra can be found in Fig. S3–S6 (ESI†). Fig. 3 shows that silk fibroin in the presence of potassium phosphate forms the highest content of β-sheets at 33.6 ± 5.3%. Sodium phosphate also promoted a similar level of β-sheet formation (Fig. S7 and S8, ESI†). The β-sheet content of silk fibroin in ammonium sulfate, potassium sulfate, and acetic acid buffer is roughly the same at 13.7 ± 5.8%, 15.9 ± 5.5%, and 15.0 ± 4.4%, respectively. This data also shows the significant increase in β-sheet content for silk in potassium phosphate is likely formed at the expense of random coil/α-helix content, in agreement with literature.7–11 Interestingly, β-sheet content does not follow the trend of kosmotropic strength of the salts tested, suggesting that phosphate-specific effects play a role in silk fibroin self-assembly.Kosmotropic salts promote continuous silk fibroin coating assembly
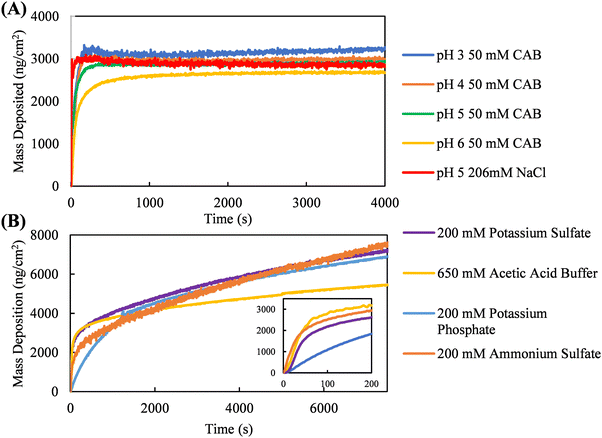
Quartz crystal microbalance with dissipation (QCM-D) was used to measure coating kinetics in kosmotropic and neutral salts selected from the Hofmeister series. To promote continuous protein accumulation at the interface, the adsorption jamming limit of silk fibroin adsorption must be surpassed. For example, Fig. 4A shows coating kinetics of silk fibroin in NaCl and citric acid solutions. These neutral salts do not promote continuous interfacial accumulation of silk fibroin, as surface saturation occurs roughly at 3000 ng cm−2. Silk fibroin in citric acid buffer was measured at one concentration but at four different solution pHs (3, 4, 5, and 6). Despite each of these solution conditions promoting different solution-phase assembly diameters (Fig. 2C), the coating kinetics are nearly identical, illustrating the size of the solution-phase assemblies formed is not necessarily predictive of continuous interfacial assembly.Fig. 4B shows silk fibroin coating kinetics in kosmotropic salts, demonstrating that potassium phosphate, potassium sulfate, ammonium sulfate, and acetic acid buffer all promote continuous protein accumulation at the solid–liquid interface, as indicated by linear mass deposition past the initial adsorption stage. Interfacial silk fibroin accumulation kinetics in the presence of kosmotropic salts can be defined by two regions: an early-stage where the surface is empty and adsorption is driven by protein–surface interactions, and a late-stage once the substrate is completely covered and further accumulation is governed by protein–protein interactions. We hypothesize that salts must promote a balance between protein–surface and protein–protein interactions for coating growth to occur. To quantitatively compare the early- and late-stage adsorption kinetics promoted by these kosmotropic salts, a two-stage interfacial assembly model developed in previous work by our group is fitted to the data.44 Briefly, the two-stage interfacial assembly model (eqn S1, ESI†) can account for the continuous, non-saturating accumulation of protein at the interface, a phenomenon not adequately described by other adsorption models in literature. From this model, we extract two parameters to characterize adsorption kinetics: the early-stage adsorption rate constant, KaC (ng cm−2 s−1), representing protein–surface interactions, and the late-stage adsorption rate constant, Kss (ng cm−2 s−1), representing protein–protein interactions. The model fits and residuals can be found in Fig. S9–S12 (ESI†) The early-stage absorption rate constants extracted from the QCM-D data in descending order are acetic acid buffer (138.2 ± 18.0 ng cm−2 s−1), ammonium sulfate (94.0 ± 16.8 ng cm−2 s−1), potassium sulfate (75.9 ± 17.2 ng cm−2 s−1), and potassium phosphate (14.5 ± 1.8 ng cm−2 s−1). Conversely, the late-stage adsorption rate constants are nearly identical for ammonium sulfate (0.7 ± 0.0 ng cm−2 s−1), potassium sulfate (0.7 ± 0.1 ng cm−2 s−1), and potassium phosphate (0.6 ± 0.1 ng cm−2 s−1); acetic acid buffer is the only outlier (0.3 ± 0.0 ng cm−2 s−1).
Since the early- and late-stage adsorption rate constants represent protein–surface and protein–protein interactions, respectively, hypotheses can be made about how these kosmotropic salts promote one interaction over the other. Acetic acid, the least kosmotropic salt tested in this set, promotes the fastest early-stage and slowest late-stage adsorption kinetics. This implies acetic acid buffer does not promote sufficiently strong protein–protein interactions, as coating formation in the late-stage is governed by these interactions. While there is little difference in the late-stage adsorption kinetics between potassium phosphate, ammonium sulfate, and potassium sulfate, the early-stage adsorption rates vary substantially. It can reasonably be postulated that the late-stage adsorption kinetics within this coating process simply has an upper limit due to solution-phase aggregation. To promote faster late-stage adsorption kinetics, stronger protein–protein interactions would be required, but this would push the system into favoring solution-phase aggregation (Fig. 1) rather than interfacial adsorption, thus disfavoring early-stage adsorption all together.
Alternatively, it is the difference in early-stage adsorption kinetics that lends deeper insight into the mechanism of silk fibroin interfacial assembly. It may be expected that a greater extent of solution-phase self-assembly, promoted by a more kosmotropic salt, would inhibit early-stage coating formation, as smaller, more hydrophobic species diffuse faster to a surface and more readily adopt protein–surface adhesive interactions. However, based on our DLS results, potassium sulfate is the most kosmotropic salt tested, yet it does not promote the slowest early-stage adsorption kinetics. Potassium phosphate, the second most kosmotropic salt in this analysis, promotes the slowest rate of adsorption in the early stage. The increased β-sheet content formed by potassium phosphate in relation to the other salts is likely responsible for the slower early-stage adsorption kinetics. If simply promoting stronger hydrophobic protein–protein interactions through a kosmotropic disruption of the protein hydration layer does not result in decreasing early-stage adsorption kinetics, it suggests a unique behavior induced by potassium phosphate. In the presence of potassium phosphate, silk fibroin may preferentially form intramolecular β-sheets, favoring hydrogen bonding interactions within the protein assemblies rather than interacting with the surface or neighboring assemblies at early timepoints. If silk fibroin formed more intermolecular β-sheets, this might result in potassium phosphate promoting faster late-stage adsorption kinetics compared to other kosmotropic salts, though this is not observed. A significant increase in intermolecular β-sheet formation would likely lead to increased solution-phase aggregation and a reduction in both early- and late-stage adsorption kinetics.
Potassium phosphate promotes formation of smooth, complete surface coverage coatings
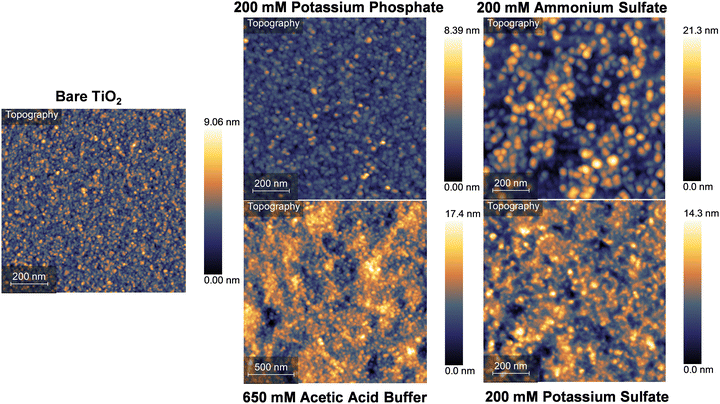
Analysis of the secondary structures promoted by various kosmotropic salts suggests that silk fibroin assemblies may interact with surfaces and neighboring surface-bound assemblies differently. It can be reasonably hypothesized that kosmotropic salts which promote less overall β-sheet content may not result in the same coating surface structure that is formed in the presence of potassium phosphate. AFM height images of silk fibroin coatings formed in potassium phosphate, ammonium sulfate, potassium sulfate, and acetic acid buffer after 24 hours of exposure to the coating solution are presented in Fig. 5. TiO2 was used as a substrate to characterize silk fibroin adsorption as this material is commonly used for biomedical devices, but it brings the challenge of distinguishing the substrate from the globular silk fibroin assemblies deposited on the surface. For all four conditions tested, it is not obvious from the AFM images how to distinguish between absorbed protein and exposed substrate. Qualitatively, it appears that there may be substrate left exposed in the coatings formed by ammonium sulfate, potassium sulfate, and acetic acid buffer. To quantitatively determine the difference between adsorbed protein and underlying substrate, masks were applied to the whole image and portions of the image where there appears to be holes in the coating to calculate surface roughness (Fig. S13, ESI†).Surface roughness of full and partial sections of the protein coatings, determined by root mean square measurements (RMS), was calculated and compared to the surface roughness of the bare TiO2 substrate. The RMS determined from the full image mask for silk fibroin coatings formed in ammonium sulfate, potassium sulfate, and acetic acid buffer were calculated to be 3.71 nm, 2.17 nm, and 2.50 nm, respectively, all resulting in rougher surfaces than the bare TiO2 substrate (1.03 nm). RMS analysis of the hypothesized exposed substrate portions of the same coatings were calculated to be 1.21 nm, 1.23 nm, and 1.47 nm for ammonium sulfate, potassium sulfate, and acetic acid buffer, respectively. While the portions of these coatings are rougher than the overall RMS of the bare TiO2, this analysis suggests coatings formed in these three kosmotropic salts do not promote uniform coating formation and likely have portions of the underlying substrate exposed to solution. It should be noted the AFM cantilever used had a 30 nm tip diameter; a sharper tip would allow for more accurate measurements within small holes of the protein coatings. The silk fibroin coating formed in potassium phosphate had a calculated RMS of between 0.71 and 0.77 nm, nearly 30% smoother than the bare substrate. Additionally, no portion of the underlying substrate was found to potentially be exposed, in agreement with prior AFM work examining the surface structure of silk fibroin coatings formed in potassium phosphate.44
Another key distinction of silk fibroin coatings formed in ammonium sulfate, potassium sulfate, and acetic acid buffer is the presence of large protein aggregates on the surface (Fig. S14, ESI†). These aggregates are anywhere from ∼500 nm to multiple micrometers in diameter, large enough to clearly see with the optical microscope used to align the AFM laser onto the cantilever, and the bare eye. These aggregates likely form in solution before landing on the surface, as large-scale aggregation in the coating solution is observed after 24 hours of assembly for these three kosmotropic salts. No large-scale aggregates have been observed on silk fibroin coatings formed with potassium phosphate, indicating the solution-phase and interfacial assembly promoted by potassium phosphate is a more controlled process. These observations align with the secondary structure analysis shown in Fig. 3; potassium phosphate promotes a higher β-sheet content, likely intramolecularly, thus internalizing the hydrophobic domains of the protein during this structural transition where they do not participate in an uncontrolled aggregation event at coating concentrations. Additionally, once the protein assemblies absorb to the surface, the higher β-sheet content may allow for neighboring assemblies to interact cohesively, allowing the assemblies to pack closer together and more fully cover the underlying substrate.
Conclusion and future work
Our studies investigate the self-assembly behavior of silk fibroin in response to various ions on the Hofmeister series. Our results suggest that phosphate plays a unique role in the secondary structure transition of silk fibroin, promoting nearly double the β-sheet content formed by other kosmotropic salts. We further explore whether continuous coating self-assembly by silk fibroin is driven by a kosmotropic dehydration of the protein or a phosphate-specific effect. Kosmotropic ions, such as phosphate, sulfate, and acetate, can promote continuous accumulation of silk fibroin on a surface. Salts that do not have strong kosmotropic or chaotropic effects, such as chloride salts and citrate, lead to conventional protein adsorption phenomena, where accumulation saturates rapidly. Coatings formed using potassium phosphate are smooth and completely cover the underlying substrate, whereas coatings formed using sulfate and acetate salts are patchy and are populated with large surface aggregates. The intermolecular interactions promoted by potassium phosphate, likely the hydrogen bonding required to form β-sheet structures, is hypothesized to impart lateral cohesive interactions between surface-bound proteins, allowing the silk fibroin assemblies to pack tightly together and completely cover the surface. Overall, this work highlights the unique effect of phosphate on silk fibroin self-assembly and points to a phosphate-specific self-assembly in solution and at solid–liquid interfaces. Future work will focus on elucidating the mechanism by which phosphate ions interact with silk fibroin at the molecular scale.Materials and methods
General coating solution preparation
Commercially purchased silk fibroin (MW ∼ 100 kDa, extracted and purified from B. mori silk fibers, Advanced Biomatrix, CA, USA) is centrifuged at 8400 rcf for 30 minutes to remove insoluble protein aggregates. Protein concentration is determined using the A280 method on a NanoDrop One Spectrometer (ThermoFisher, MA, USA) assuming 1 ABS = 1 mg mL−1. This assumption is valid to the polydispersity of the protein sample caused by hydrolysis during the degumming process.53 Coating solutions are prepared by diluting silk fibroin to 0.5 mg mL−1 (unless otherwise stated) into ultrapure water and desired salt solutions. Solutions are carefully mixed to avoid shear-induced self-assembly. All solutions are buffered or titrated to pH 5 unless otherwise specified. For unbuffered salt solutions, coating solution pH is adjusted using small amounts of dilute HCl and NaOH while gently stirring with a Mettler Toledo pH probe. The following materials are used in this study: ammonium sulfate (Fisher scientific, ACS Grade ≥ 99%), potassium sulfate (Fisher scientific, ACS Grade ≥ 99%), potassium phosphate monobasic (Sigma-Aldrich, ACS Grade ≥ 99%), potassium phosphate dibasic (Fisher scientific, ACS Grade ≥ 98%), sodium phosphate monobasic monohydrate (Sigma-Aldrich, ACS Grade ≥ 98%), sodium phosphate dibasic (Sigma-Aldrich, ACS Grade ≥ 99%), glacial acetic acid (Spectrum Chemical, ACS Grade, 99.7%), sodium acetate (Sigma-Aldrich, ACS Grade, ≥ 99%), and silk fibroin (Advanced Biomatrix, 100k kDa).Attenuated total reflectance-Fourier transform infrared spectroscopy (ATR-FTIR)
Silk fibroin secondary structure content was calculated using FITR on a Vertex 70 Spectrometer (Bruker MA, USA) equipped with a Bruker Platinum attenuated total reflection (ATR) diamond crystal cell. Wavenumbers 800–3200 cm−1 were collected with 128 scans and resolution of 4 cm−1. The prepared silk fibroin solutions were allowed to self-assemble at room temperature under gentle shaking (60 rpm) for 24 hours. Approximately 30 μL of silk fibroin solution was measured on the ATR-FTIR crystal for each trial; new solution was used for each trial. To determine the secondary structure content of the silk protein, we followed a protocol by Yang et al.54 using the second derivative analysis. In this protocol, the identical Gaussian peaks were assigned to the local minima of the second derivative of the ATR-FTIR spectra. The number of initial peaks, and the location of the starting wavenumbers, were determined for each individual spectra from the number and locations of local minima of the second derivative of the spectra. Multipeak deconvolution of the amide I spectra (1600–1700 cm−1) using Igor Pro 8 (WaveMetrics Inc. OR, USA) was employed to iteratively fit the Gaussian peaks to the original spectra. Peak heights, widths, and locations were allowed to iterate without constraints until the software converged to a solution. The secondary structure content was calculated from the percent area under the deconvoluted peaks within specified secondary structure ranges. Peak assignments were based on Lammel et al.9 β-sheets were assigned to the ranges of 1610–1635 cm−1 and 1695–1700 cm−1, random coil/α-helices were assigned to 1635–1666 cm−1, and β-turns were assigned to 1667–1695 cm−1. All averaged values contain 4 replicates unless otherwise specified. Statistical analyses were performed using GraphPad Prism 9.3.1.Data availability
The data supporting this article have been included as part of the ESI.† Additionally, raw data files are available at https://osf.io/qzyax/.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Dr Rachel L. Fay for her assistance in performing statistical analysis for the ATR-FTIR data. For the use of the QCM-D, we thank Prof. Georges Belfort. We thank Dr Joel Morgan and the CBIS Analytical Biochemistry Core for use of the FTIR. We additionally thank Dr Sergey Pryshchep and the CBIS Microscopy Core for assistance with the AFM used in this work. We acknowledge funding from by the National Science Foundation (NSF DMR #2045510).References
- T. D. Fink and R. H. Zha, Macromol. Rapid Commun., 2018, 39, 1700834 CrossRef PubMed
.
- C. Fu, Z. Shao and V. Fritz, Chem. Commun., 2009, 6515–6529 RSC
.
- L. Römer and T. Scheibel, Prion, 2008, 2, 154–161 CrossRef PubMed
.
- C. Jiang, X. Wang, R. Gunawidjaja, Y.-H. Lin, M. K. Gupta, D. L. Kaplan, R. R. Naik and V. V. Tsukruk, Adv. Funct. Mater., 2007, 17, 2229–2237 CrossRef CAS
.
- D. Lin, M. Li, L. Wang, J. Cheng, Y. Yang, H. Wang, J. Ye and Y. Liu, Adv. Funct. Mater., 2024, 34, 2405255 CrossRef CAS
.
- X. Yao, S. Zou, S. Fan, Q. Niu and Y. Zhang, Mater. Today Bio, 2022, 16, 100381 CrossRef CAS PubMed
.
- X. Wang, H. J. Kim, P. Xu, A. Matsumoto and D. L. Kaplan, Langmuir, 2005, 21, 11335–11341 CrossRef CAS PubMed
.
- Y.-X. He, N.-N. Zhang, W.-F. Li, N. Jia, B.-Y. Chen, K. Zhou, J. Zhang, Y. Chen and C.-Z. Zhou, J. Mol. Biol., 2012, 418, 197–207 CrossRef CAS PubMed
.
- A. S. Lammel, X. Hu, S.-H. Park, D. L. Kaplan and T. R. Scheibel, Biomaterials, 2010, 31, 4583–4591 CrossRef CAS PubMed
.
- X. Chen, D. P. Knight and Z. Shao, Soft Matter, 2009, 5, 2777–2781 RSC
.
- C. W. P. Foo, E. Bini, J. Hensman, D. P. Knight, R. V. Lewis and D. L. Kaplan, Appl. Phys., 2006, 82, 223–233 CAS
.
- D. N. Rockwood, R. C. Preda, T. Yücel, X. Wang, M. L. Lovett and D. L. Kaplan, Nat. Protoc., 2011, 6, 1612 CrossRef CAS PubMed
.
- C. Vepari and D. L. Kaplan, Prog. Polym. Sci., 2007, 32, 991–1007 CrossRef CAS PubMed
.
- G. H. Altman, F. Diaz, C. Jakuba, T. Calabro, R. L. Horan, J. Chen, H. Lu, J. Richmond and D. L. Kaplan, Biomaterials, 2003, 24, 401–416 CrossRef CAS PubMed
.
- H. Zheng, B. Duan, Z. Xie, J. Wang and M. Yang, RSC Adv., 2020, 10, 28408–28414 RSC
.
- L. Li, S. Puhl, L. Meinel and O. Germershaus, Biomaterials, 2014, 35, 7929–7939 CrossRef CAS PubMed
.
- B. Marelli, M. A. Brenckle, D. L. Kaplan and F. G. Omenetto, Sci. Rep., 2016, 6, 25263 CrossRef CAS PubMed
.
- L. Zhou, X. Chen, Z. Shao, Y. Huang and D. P. Knight, J. Phys. Chem. B, 2005, 109, 16937–16945 CrossRef CAS PubMed
.
- L. J. Domigan, M. Andersson, K. A. Alberti, M. Chesler, Q. Xu, J. Johansson, A. Rising and D. L. Kaplan, Insect Biochem. Mol. Biol., 2015, 65, 100–106 CrossRef CAS PubMed
.
- A. Koeppel, N. Stehling, C. Rodenburg and C. Holland, Adv. Funct. Mater., 2021, 2103295 CrossRef CAS
.
- J. Sparkes and C. Holland, Nat. Commun., 2017, 8, 594 CrossRef PubMed
.
- M. Andersson, J. Johansson and A. Rising, Int. J. Mol. Sci., 2016, 17, 1290 CrossRef PubMed
.
- L. Lu, S. Fan, L. Geng, J. Lin, X. Yao and Y. Zhang, Adv. Mater. Technol., 2021, 6, 2100124 CrossRef CAS
.
- R. Wu, J. Bae, H. Jeon and T. Kim, Chem. Eng. J., 2022, 444, 136556 CrossRef CAS
.
- X. Hu, D. Kaplan and P. Cebe, Macromolecules, 2006, 39, 6161–6170 CrossRef CAS
.
- D. Huemmerich, U. Slotta and T. Scheibel, Appl. Phys., 2006, 82, 219–222 Search PubMed
.
- U. Slotta, M. Tammer, F. Kremer, P. Koelsch and T. Scheibel, Supramol. Chem., 2006, 18, 465–471 Search PubMed
.
- S. Wohlrab, K. Spieß and T. Scheibel, J. Mater. Chem., 2012, 22, 22050–22054 Search PubMed
.
- E. Metwalli, U. Slotta, C. Darko, S. V. Roth, T. Scheibel and C. M. Papadakis, Appl. Phys., 2007, 89, 655–661 CAS
.
- D. P. Knight and F. Vollrath, Naturwissenschaften, 2001, 88, 179–182 CrossRef CAS PubMed
.
- H. I. Okur, J. Hladílková, K. B. Rembert, Y. Cho, J. Heyda, J. Dzubiella, P. S. Cremer and P. Jungwirth, J. Phys. Chem. B, 2017, 121, 1997–2014 CrossRef CAS PubMed
.
- V. Yeh, J. M. Broering, A. Romanyuk, B. Chen, Y. O. Chernoff and A. S. Bommarius, Protein Sci., 2010, 19, 47–56 CrossRef CAS PubMed
.
- M. G. Cacace, E. M. Landau and J. J. Ramsden, Q. Rev. Biophys., 1997, 30, 241–277 CrossRef CAS PubMed
.
- A. C. Dumetz, A. M. Snellinger-O’Brien, E. W. Kaler and A. M. Lenhoff, Protein Sci., 2007, 16, 1867–1877 CrossRef CAS PubMed
.
- A. Barrett, J. Imbrogno, G. Belfort and P. B. Petersen, Langmuir, 2016, 32, 9074–9082 CrossRef CAS PubMed
.
- D. Stengel, M. Saric, H. R. Johnson, T. Schiller, J. Diehl, K. Chalek, D. Onofrei, T. Scheibel and G. P. Holland, Biomacromolecules, 2023, 3, 1463–1474 CrossRef PubMed
.
- A. D. Malay, T. Suzuki, T. Katashima, N. Kono, K. Arakawa and K. Numata, Sci. Adv., 2020, 6, eabb6030 CrossRef CAS PubMed
.
- M. Humenik, A. M. Smith, S. Arndt and T. Scheibel, J. Struct. Biol., 2015, 191, 130–138 CrossRef CAS PubMed
.
- N. A. Oktaviani, A. Matsugami, F. Hayashi and K. Numata, Chem. Commun., 2019, 55, 9761–9764 RSC
.
- A. Leppert, G. Chen, D. Lama, C. Sahin, V. Railaite, O. Shilkova, T. Arndt, E. G. Marklund, D. P. Lane, A. Rising and M. Landreh, Nano Lett., 2023, 23, 5836–5841 CrossRef CAS PubMed
.
- R. H. Zha, P. Delparastan, T. D. Fink, J. Bauer, T. Scheibel and P. B. Messersmith, Biomater. Sci., 2019, 7, 683–695 RSC
.
- A. M. Ziemba, T. D. Fink, M. C. Crochiere, D. L. Puhl, S. Sapkota, R. J. Gilbert and R. H. Zha, ACS Biomater. Sci. Eng., 2020, 6, 1321–1332 CrossRef CAS PubMed
.
- T. D. Fink, J. L. Funnell, R. J. Gilbert and R. H. Zha, ACS Biomater. Sci. Eng., 2024, 10, 482–496 CrossRef CAS PubMed
.
- C. Wigham, T. D. Fink, M. Sorci, P. O’Reilly, S. Park, J. Kim and R. H. Zha DOI:10.21203/rs.3.rs-4360925/v1.
- H. R. Bosshard, D. N. Marti and I. Jelesarov, J. Mol. Recognit., 2004, 17, 1–16 CrossRef CAS PubMed
.
- A. K. H. Hirsch, F. R. Fischer and F. Diederich, Angew. Chem., Int. Ed., 2007, 46, 338–352 CrossRef CAS PubMed
.
- L. S. Wray, X. Hu, J. Gallego, I. Georgakoudi, F. G. Omenetto, D. Schmidt and D. L. Kaplan, J. Biomed. Mater. Res., Part B, 2011, 99, 89–101 CrossRef PubMed
.
- C. Dicko, J. M. Kenney, D. Knight and F. Vollrath, Biochemistry, 2004, 43, 14080–14087 CrossRef CAS PubMed
.
- A. Koeppel, P. R. Laity and C. Holland, Acta Biomater., 2020, 117, 204–212 CrossRef CAS PubMed
.
- A. Ochi, K. S. Hossain, J. Magoshi and N. Nemoto, Biomacromolecules, 2002, 3, 1187–1196 CrossRef CAS PubMed
.
- T. Li, C. Wang, T. Li, L. Ma, D. Sun, J. Hou and Z. Jiang, Molecules, 2018, 23, 2383 CrossRef PubMed
.
- Z. Wang, Y. Deng, Y. Zhang, Z. Wei, Z. Wan, C. Li, X. Tang, Z. Zhao, P. Zhou, P. Li, G. Liu and M. Zhang, LWT, 2023, 183, 114872 CrossRef CAS
.
- M. Ho, H. Wang and K. Lau, Appl. Surf. Sci., 2012, 258, 3948–3955 CrossRef CAS
.
- H. Yang, S. Yang, J. Kong, A. Dong and S. Yu, Nat. Protoc., 2015, 10, 382–396 CrossRef CAS PubMed
.
- M. V. Voinova, M. Rodahl, M. Jonson and B. Kasemo, Phys. Scr., 1999, 59, 391–396 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sm01198h |
| This journal is © The Royal Society of Chemistry 2025 |