Open Access Article
Open Access ArticlePolyhydroxyalkanoate (PHA) production by thermophilic Caldimonas thermodepolymerans comb. nov. from xylan†
Wen
Zhou‡
a,
Salvador Bertrán
Llorens‡
b,
Peter J.
Deuss
 b,
Gert-Jan W.
Euverink
b,
Gert-Jan W.
Euverink
 a and
Janneke
Krooneman
a and
Janneke
Krooneman
 *ac
*ac
aProducts and Processes for Biotechnology, Engineering and Technology Institute Groningen, Faculty of Science and Engineering, University of Groningen, Groningen, The Netherlands. E-mail: j.krooneman@rug.nl; j.krooneman@pl.hanze.nl
bChemical Technology, Engineering and Technology Institute Groningen, Faculty of Science and Engineering, University of Groningen, Groningen, The Netherlands
cBioconversion and Fermentation Technology, Research Centre Biobased Economy, Hanze University of Applied Sciences, Groningen, The Netherlands
First published on 26th February 2025
Abstract
For the future circular economy, renewable carbon feedstocks manifest considerable promise for synthesizing sustainable and biodegradable polyhydroxyalkanoate (PHA). In this study, 16 wt% and 30 wt% PHA (cell dry weight) are respectively produced by thermophilic Caldimonas thermodepolymerans from beechwood xylan and wheat arabinoxylan as the sole carbon source. Moreover, an in silico study of the potential xylan-degrading proteins was conducted using proteome sequencing and CAZyme specialized bioinformatic tools. This study demonstrates the feasibility of utilizing complex polysaccharide substrates for PHA biosynthesis, thereby potentially eliminate additional processing steps and reducing overall production costs for sustainable plastic.
Sustainability spotlightMicrobial production of polyesters such as polyhydroxyalkanoates (PHA) provides a promising alternative for plastics from fossil origin. Compared to conventional production of fossil-based plastics, biobased polymers like PHA have the important advantage of being produced from renewable resources. In addition, production processes are performed under milder conditions with concomitant lower energy costs and a significant lower CO2 footprint. Moreover, PHA are fully biodegradable, thereby significantly reducing environmental contamination with microplastics. This study shows the direct microbial PHA-production from more complex, and less biorefined biobased polymers like xylans: a promising route for cost-effective industrial valorization of renewable resources into PHAs. This research corresponds to SDGs 9, 12 and 14, 15. |
1. Introduction
Plastics are ubiquitous, but most are derived from petroleum and are non-biodegradable. The dependence on oil-based plastics has led to many issues, including marine pollution and potential health risks.1 Transitioning to biodegradable plastics could address these challenges within a sustainable circular carbon economy.2 Polyhydroxyalkanoates (PHAs) are a microbiological-produced polyester from bio-based substrates. Known for their biodegradability and biocompatibility, PHAs are advantageous for mitigating plastic waste. However, despite these environmental advantages, the production costs of PHA still restrict scale-up industrialization due to the need for pure monosaccharide precursors.3To reduce the high production costs of PHA, it is essential to find a cheap and abundant carbon source for their production.4 The conversion of biomass and waste materials into biodegradable products has emerged as a promising strategy to address global challenges such as resource depletion, environmental pollution, and climate change.5–7 The valorization of these waste streams into value-added biodegradable materials, such as polylactic acid (PLA), PHA, and other bio-based polymers, represents a cornerstone of green chemistry and circular economy principles.8 Lignocellulose is abundant and globally distributed as a byproduct of industrial and agricultural activities, presents a promising option.9 Utilizing lignocellulose is considered pivotal for achieving a carbon-neutral society. Lignocelluloses comprise cellulose (25–40%), hemicellulose (25–50%), and lignin (10–30%). Hemicellulose, in particular, is a promising carbon source for PHA production due to its underutilization and lower competition for products use to glucose.10 Hemicellulose primarily consists of glucuronoarabinoxylan (GAX) in most lignocellulosic biomasses, which is composed mainly of xylose residues linked via β-D (1→4) bonds and substituted with various C5 and C6 sugars, hydroxycinnamic acid, acetic acid, and methylglucuronic acid. The abstention of monosaccharides from biomass that are able to be directly utilized without the production of inhibitor compounds such as furfural or acids it still remains a challenge.11 Therefore, to avoid the dehydratation reaction of the sugars milder conditions need to be used, creating a range of more complex xylan polymeric fractions.12–14 However, xylan, the primary component of hemicellulose, still presents challenges for direct utilization in PHA production by bacteria.15
Therefore, a bacterial strain can directly utilize xylan as its primary carbon source would significantly benefit PHA production from lignocellulose derived sugars. However, there is limited information on naturally occurring thermophilic wild-type strains that can utilize xylan exclusively. Caldimonas thermodepolymerans, formerly known as Schlegelella thermodepolymerans, is a thermophilic bacterium belonging to the Comamonadaceae family. Recently, this bacterium has attracted attention for its exceptional ability to produce PHA from xylose, achieving high yields of 80 wt% on cell dry weight.16 Besides, C. thermodepolymerans also can efficiently degrade extracellular PHA materials.17,18 Due to its robust xylose metabolism and capacity to degrade extracellular polymers, C. thermodepolymerans emerges as a promising candidate for PHA production from complex carbohydrate substrates.
This paper presents one of the first PHA production using xylan as the sole carbon source by a thermophilic bacterium. We investigated the optimal concentrations of xylan and nitrogen source concentrations to maximize PHA production. We also compared the PHA production yields and process from different types of xylans, specifically beechwood xylan and wheat xylan. Additionally, two proteins of unknown function (PUF) potentially involved in xylan degradation of C. thermodepolymerans have been identified based on proteomic analysis and their sequence amino acid similarity to related xylan degrading enzymes.
2. Materials and methods
2.1 Strain and culture media
Schlegelella thermodepolymerans DSM 15344, recently renamed as Caldimonas thermodepolymerans was purchased from the DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). The inoculum medium consisted of peptone (10 g L−1), yeast extract (5 g L−1), and NaCl (10 g L−1). C. thermodepolymerans was inoculated from 1.5% (w/v) agar plates and pre-grown in Erlenmeyer flasks at 150 rpm and 50 °C, following the protocol of Obruča et al.19 The cell suspension was centrifuged at 8000×g for 10 min and washed with sterilized water before being used as a 10% (v/v) inoculum for subsequent experiments.The culture medium for polyhydroxyalkanoate (PHA) production contained (NH4)2SO4 (1.1 g L−1) (if not stated otherwise), MgSO4·7H2O (0.45 g L−1), KH2PO4 (1.31 g L−1), Na2HPO4·2H2O (1.68 g L−1), and 1.5 mL L−1 of a trace element solution as described by Vishniac and Santer.20 Beechwood xylan and wheat arabinoxylan (Megazyme, Ireland) (20 g L−1) served as the initial carbon source unless stated otherwise. The pH of all initial culture media was adjusted to 7.0.
To optimize xylan utilization for maximizing PHA production, the growth conditions of C. thermodepolymerans were studied using beechwood xylan as the sole carbon source at various concentrations (1, 5, 10, 15, and 20 g L−1). Additionally, the effects of different nitrogen source, (NH4)2SO4, urea, yeast extract, and tryptone at a concentration of 4.4 g L−1, as well as varying nitrogen concentrations of (NH4)2SO4 (0.44, 1.1, 4.4 and 44 g L−1) were evaluated. The study also compared two types of xylans (beechwood xylan and wheat arabinoxylan) in the culture medium for PHA production. Experiments were conducted in 500 mL Erlenmeyer flasks with a working volume of 150 mL, incubated at 150 rpm and 50 °C.
2.2 Analytical techniques
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 10 min using a Thermo Fisher, F15-6x 100y rotor. Afterwards, the cells were washed once with sterilized water and then frozen at −60 °C and 0.1 bar for 48 h to obtain cell dry weight through lyophilization. The lyophilized cells were then depolymerized the PHA within the cells using a methanolysis protocol as previous described by Zhou et al.17 To identify the produced PHA, commercial poly(3-hydroxybutyrate) (P3HB)/poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV)/poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) (PHBHHX) was utilized as an external standard, while benzoic acid served as an internal standard. For the methanolysis process, a mixture of 2 mL chloroform, 1.7 mL methanol, and 0.3 mL 98% sulfuric acid were used at 100 °C for 4 h to methanolize the dry cell mass and commercial PHA. Following cooling, 1 mL of water was added to the mixture to separate and obtain the bottom phase solution for further determination of PHA content. To characterize and quantitative analyze the produced PHA within the cells, the corresponding methyl ester of the monomers were analyzed using a Hewlett-Packard 6890 gas chromatography (GC) system equipped with a Rxi-5Si capillary column (30 m × 0.25 mm i.d. and 0.25 μm film thickness) and a Quadrupole Hewlett-Packard 5973 mass selective detector. Besides, the methyl esters of the monomers were compared with the commercial methyl esters (3HB, 3HV, and 3HHX) to identify the types of produced PHA. All samples were filtered by a 0.22 μm PTFE membrane before analysis.
000×g for 10 min using a Thermo Fisher, F15-6x 100y rotor. Afterwards, the cells were washed once with sterilized water and then frozen at −60 °C and 0.1 bar for 48 h to obtain cell dry weight through lyophilization. The lyophilized cells were then depolymerized the PHA within the cells using a methanolysis protocol as previous described by Zhou et al.17 To identify the produced PHA, commercial poly(3-hydroxybutyrate) (P3HB)/poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV)/poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) (PHBHHX) was utilized as an external standard, while benzoic acid served as an internal standard. For the methanolysis process, a mixture of 2 mL chloroform, 1.7 mL methanol, and 0.3 mL 98% sulfuric acid were used at 100 °C for 4 h to methanolize the dry cell mass and commercial PHA. Following cooling, 1 mL of water was added to the mixture to separate and obtain the bottom phase solution for further determination of PHA content. To characterize and quantitative analyze the produced PHA within the cells, the corresponding methyl ester of the monomers were analyzed using a Hewlett-Packard 6890 gas chromatography (GC) system equipped with a Rxi-5Si capillary column (30 m × 0.25 mm i.d. and 0.25 μm film thickness) and a Quadrupole Hewlett-Packard 5973 mass selective detector. Besides, the methyl esters of the monomers were compared with the commercial methyl esters (3HB, 3HV, and 3HHX) to identify the types of produced PHA. All samples were filtered by a 0.22 μm PTFE membrane before analysis.
3. Results and discussion
3.1 Optimization of the growth conditions for PHA production
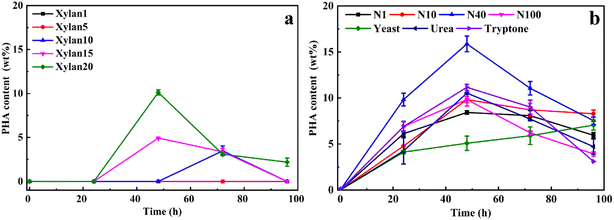
To optimize xylan utilization for maximizing PHA production, the growth conditions of Caldimonas thermodepolymerans was studied using beechwood xylan as the sole carbon source in combination with different nitrogen sources (Fig. 1). The data for xylose concentrations, cell growth, and nitrogen conditions are presented in Fig. S1 and S2,† respectively. Fig. 1a shows that no PHA was produced when the xylan concentration was 1 or 5 g L−1. PHA accumulated up to 10 wt% of the cell dry weight at a xylan concentration of 20 g L−1. No PHA accumulation was observed during the first 24 h, most likely because the nitrogen source was still available and the cells were using the carbon source for growth. Alternatively, the delay in PHA production also might be linked to the slow process of xylan degradation by C. thermodepolymerans. PHA production from xylan was shown before by Sawant et al.,21 showing a maximum PHA content of 22.7% cell dry weight (0.198 g L−1) when marine bacteria Saccharophagus degradans was grown with 20 g L−1 of beechwood xylan. However, the maximum of PHA production (34.5% of 0.267 g L−1 cell dry weight) was obtained when S. degradans was co-cultured with Bacillus cereus. Xylan was degraded into its monomers by a xylan-degrading strain (S. degradans), and a PHA-producing species (B. cereus) subsequently utilized the produced xylose as the carbon source for PHA accumulation, while no PHA was produced by B. cereus alone from xylan.3.2 PHA production process using different sources of xylans
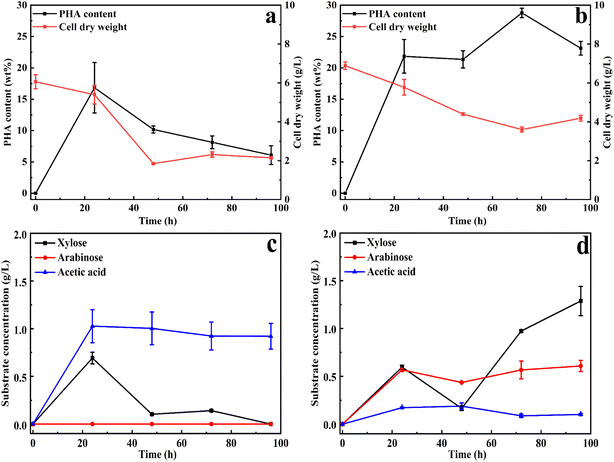
Based on the optimization of xylan utilization to maximize PHA production, a xylan concentration of 20 g L−1 and ammonium sulfate as a nitrogen source with a C/N ratio of 40 were used for both substrates. To evaluate whether xylan degradation was influenced by xylan substitutions, the study was extended from beechwood xylan to wheat arabinoxylan. The main difference between these two types of xylans is the quantity and type of substitutions. Wheat arabinoxylan is mainly substituted with arabinose (around 40%, according to the manufacturer), while beechwood xylan is mainly substituted with methyl glucuronic acid (around 19% according to the manufacturer). These substitutions represent the main modifications in xylan, providing a suitable representation of natural glucuronoarabinoxylan (GAX).The PHA accumulation and the produced products (xylose, acetic acid, and arabinose) were analyzed and presented in Fig. 2. As shown in Fig. 2, compared to 16 wt% PHA from beechwood xylan (Fig. 2a), 30 wt% PHA on the cell dry weight was obtained with a wheat arabinoxylan as a carbon source (Fig. 2b). Xylans must be degraded before it could be utilized, so the cell dry weight first decreased and then increased. The molecular weight distributions of the beechwood xylan and wheat arabinoxylan are displayed in Fig. S3a and b,† respectively, indicating that both were degraded during the PHA production process. Based on GC/MS analysis, the produced PHA from xylans in cells is poly(3-hydroxybutyrate) (P3HB), as shown in Fig. S4.†Fig. 2c shows that using beechwood xylan as the sole carbon source resulted in the production of xylose and acetic acid, with the highest concentrations of xylose (0.70 g L−1) and acetic acid (1.03 g L−1) obtained after 24 h. When wheat arabinoxylan was used as the sole carbon source, arabinose was produced, as well as xylose and acetic acid (Fig. 2d).
The maximum concentrations of arabinose and acetic acid were 0.59 g L−1 and 0.17 g L−1 after 24 h, respectively, whereas the highest concentration of xylose (1.48 g L−1) was obtained after 96 h (Fig. 2d). The degradation products correspond to the monomer composition of the xylans: beechwood xylan is mainly composed of xylose residues and methyl glucuronic acid substitutions,22 while the major components of arabinoxylan are xylose and arabinose residues.23 As shown in Fig. 2d, acetic acid was produced by C. thermodepolymerans during the consumption of wheat arabinoxylan, and then was slowly consumed. Fig. S5† shows that all the obtained monomers during the xylan utilization process could be used for cell growth and PHA accumulation.
3.3 Related enzymes in xylan degradation
To better understand the xylan degradation, the enzymes involved in C. thermodepolymerans were analyzed. Several enzymes are necessary for xylan degradation based on its chemical composition. Table 1 summarizes the putative enzymes for xylan degradation in C. thermodepolymerans based on the genomic DNA sequence available in the Genbank database (accessing code NZ-CP064338.1).| CAZY family | Enzyme annotation in gene bank | GenBank accession no. | CAZY family activities |
|---|---|---|---|
| a All enzymes were annotated by at least two tolls of the dbCAN database and the CAZY genomics annotation. | |||
| GH1 | β-Glucosidase | QPC31785 | β-Xylosidase, β-glucosidase |
| GH3 | Beta-N-acetylhexosaminidase | QPC32469 | Xylan 1,4-β-xylosidase, α-l-arabinofuranosidase, β-N-acetylhexosaminidase |
| GH39 | Carbohydrate-binding domain | QPC32539 | Exo-xylanase |
| Hypothetical protein | QPC32540 | ||
| CE4 | Alpha/beta fold hydrolase | QPC31878.1 | Acetyl xylan esterase, tannase, feruloyl esterase, chitin deacetylase |
| Allantoinase PuuE | QPC32500.1 | ||
| Feruloyl esterase family | QPC31414 | ||
Even though C. thermodepolymerans can grow and degrade different xylan sources, proteins belonging to typical glycoside hydrolase families associated with endoxylanase activities like GH10, GH11, GH38, and GH5 could not be detected.24 A specific search was conducted in the dbCAN database to gain insights into carbohydrate-degrading proteins (Tables 1 and S1†). Two proteins belonging to the GH39 family (QPC32539 [ST39L] and QPC32540 [ST39S]) were identified by dbCAN and CAZY database. These proteins were labeled in NCBI as a carbohydrate-binding domain-containing protein and a hypothetical protein, respectively. Interestingly, these two proteins are localized together in the genome, forming a so-called CAZyme genetic cluster (CGC), as identified by dbCAN. An Interpro scan revealed that ST39L is composed of two carbohydrate-binding modules (CBMs) and a C-terminal catalytic domain related to GH39 family, while ST39S only has one carbohydrate-binding module and a GH39 related catalytic domain (Fig. S6 and S7†). Interestingly, one of the CBMs in the ST39L protein (between 276 and 404 aa) has a sequence similarity with the CenC-carbohydrate binding domain family (Pfam accession number pf02018). This CBM family is usually located in the N-terminal part of thermoresistant endoxylanases,25 showcasing the correlation of this CBM domains and the mechanism of xylan degradation in thermophilic bacteria.26–28 Moreover, the blast of this protein against the SwissProt and PDB database (Tables S2 and S3†) showed that even with low similarity, the region between 400 and 650 amino acids of ST39L, where the catalytical domain is located, also had similarities with characterized thermostable endoxylanases.
The bioinformatic analysis suggests that the proteins ST39L and ST39S could play a role in xylan degradation. To confirm their involvement, we obtained proteomics data of C. thermodepolymerans growing in xylose. The extracellular enzymes related to PHA production metabolism from xylans are presented in Table S4.† Both proteins with significant xylan-degrading potential (ST39L and ST39S) were detected in the outside protein extracted from bacteria grown on wheat straw. Moreover, the proteomics analysis showcased the similarities in the metabolic routes of PHA production between xylan and the previously reported pathway with xylose. This further supports the capacity of C. thermodepolymerans to directly utilize xylans to produce PHA via xylan degradation into smaller fragments/monomers that can be utilized as carbon source.
4. Conclusions
This study shows the first example of thermophilic microbial P3HB production directly from xylans (beechwood xylan and wheat arabinoxylan) by a single wild-type thermophilic C. thermodepolymerans. The use of wheat arabinoxylan resulted in higher PHA yields compared to beechwood xylan. Most likely due to the difference in decorating molecules on beechwood xylan, making it more difficult to hydrolyze the polymeric backbone. Here, two key potentials, particularly ST39L, were identified as potentially involved in xylan backbone degradation and showed a close correlation with thermoresistant endoxylanases. Meanwhile, the related proteins to PHA production from xylose were also identified during growth on xylan. These findings suggest that C. thermodepolymerans is a promising candidate for the cost-effective industrial valorization of renewable resources into PHA.Data availability
All data that support the findings of this study are available on request. The process of proteomics sample preparation and analysis was performed according to previously described methods by Zhou in 2023, data were retrieved from mass spectrometry proteomics data previously deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD040177 and https://doi.org/10.6019/PXD040177.29 The raw data of proteomics are available on request in Excel-format.Conflicts of interest
There are no conflicts to declare.References
- S. Nanda, B. R. Patra, R. Patel, J. Bakos and A. K. Dalai, Environ. Chem. Lett., 2022, 20, 379–395 CrossRef CAS PubMed.
- K. W. Meereboer, M. Misra and A. K. Mohanty, Green Chem., 2020, 22, 5519–5558 RSC.
- Y. W. Cui, H. Y. Zhang, P. F. Lu and Y. Z. Peng, Sci. Rep., 2016, 30766 CrossRef CAS PubMed.
- K. D. Snell, V. Singh and S. M. Brumbley, Curr. Opin. Biotechnol., 2015, 32, 68–75 CrossRef CAS PubMed.
- M. I. Baig, R. Hardian, F. A. Alharthi, C. M. Fellows and G. Szekely, J. Membr. Sci., 2025, 713, 123324 CrossRef CAS.
- Y. Yoshinaka and S. A. Miller, Green Chem., 2025 10.1039/d4gc05490c.
- J. Cavalcante, D. G. Oldal, M. V. Peskov, A. K. Beke, R. Hardian, U. Schwingenschlögl and G. Szekely, ACS Nano, 2024, 18, 7433–7443 CrossRef CAS PubMed.
- B. Jong and V. S. Haritos, Green Chem., 2025, 1356–1364 RSC.
- Sonu, G. M. Rani, D. Pathania, Abhimanyu, R. Umapathi, S. Rustagi, Y. S. Huh, V. K. Gupta, A. Kaushik and V. Chaudhary, Sci. Total Environ., 2023, 875, 162667 CrossRef CAS PubMed.
- M. L. Gandla, C. Martín and L. J. Jönsson, Energies, 2018, 11, 2936 CrossRef CAS.
- S. Bertran-Llorens, W. Zhou, M. A. Palazzolo, D. l. Colpa, G. J. W. Euverink, J. Krooneman and P. J. Deuss, ACS Sustain. Chem. Eng., 2024, 12, 7724–7738 CrossRef CAS PubMed.
- H. Noureddini and J. Byun, Bioresour. Technol., 2010, 101, 1060–1067 CrossRef CAS PubMed.
- J. Shekiro, E. M. Kuhn, M. J. Selig, N. J. Nagle, S. R. Decker and R. T. Elander, Appl. Biochem. Biotechnol., 2012, 168, 421–433 CrossRef CAS PubMed.
- K. Rajan and D. J. Carrier, Biomass Bioenergy, 2014, 62, 222–227 CrossRef CAS.
- D. S. Naidu, S. P. Hlangothi and M. J. John, Carbohydr. Polym., 2018, 179, 28–41 CrossRef CAS PubMed.
- X. Kourilova, I. Pernicova, K. Sedlar, J. Musilova, P. Sedlacek, M. Kalina, M. Koller and S. Obruca, Bioresour. Technol., 2020, 315, 123885 CrossRef CAS PubMed.
- W. Zhou, D. I. Colpa, H. Permentier, R. A. Offringa, L. Rohrbach, G. J. W. Euverink and J. Krooneman, Resour., Conserv. Recycl., 2023, 194, 107006 CrossRef CAS.
- K. Elbanna, T. Lütke-Eversloh, S. Van Trappen, J. Mergaert, J. Swings and A. Steinbüchel, Int. J. Syst. Evol. Microbiol., 2003, 53, 1165–1168 CrossRef CAS PubMed.
- S. Obruča, P. Dvořák, P. Sedláček, M. Koller, K. Sedlář, I. Pernicová and D. Šafránek, Biotechnol. Adv., 2022, 58, 107906 CrossRef PubMed.
- W. Vishniac and M. Santer, Bacteriol. Rev., 1957, 21, 195–213 CrossRef CAS PubMed.
- S. S. Sawant, B. K. Salunke, L. E. Taylor and B. S. Kim, Appl. Sci., 2017, 7(3), 225 CrossRef.
- M. Schmoll, Fungal Biol. Biotechnol., 2018, 5, 1–20 CrossRef PubMed.
- L. Saulnier, P. E. Sado, G. Branlard, G. Charmet and F. Guillon, J. Cereal Sci., 2007, 46, 261–281 CrossRef CAS.
- E. Nordberg Karlsson, E. Schmitz, J. A. Linares-Pastén and P. Adlercreutz, Appl. Microbiol. Biotechnol., 2018, 102, 9081–9088 CrossRef CAS PubMed.
- L. Szabó, S. Jamal, H. Xie, S. J. Charnock, D. N. Bolam, H. J. Gilbert and G. J. Davies, J. Biol. Chem., 2001, 276, 49061–49065 CrossRef PubMed.
- D. Krska and J. Larsbrink, Biotechnol. Biofuels, 2020, 13, 1–13 CrossRef PubMed.
- K. Kuramochi, K. Uchimura, A. Kurata, T. Kobayashi, Y. Hirose, T. Miura, N. Kishimoto, R. Usami and K. Horikoshi, Extremophiles, 2016, 20, 471–478 CrossRef CAS PubMed.
- H. Xie, H. J. Gilbert, S. J. Charnock, G. J. Davies, M. P. Williamson, P. J. Simpson, S. Raghothama, C. M. G. A. Fontes, F. M. V. Dias, L. M. A. Ferreira and D. N. Bolam, Biochemistry, 2001, 40, 9167–9176 CrossRef CAS PubMed.
- Y. Perez-Riverol, J. Bai, C. Bandla, D. García-Seisdedos, S. Hewapathirana and S. Kamatchinathan, et al. , Nucleic Acids Res., 2022, 50, D543–D552 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5su00040h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |