Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A thermoresponsive PEG-based methacrylate triblock terpolymer as a bioink for 3D bioprinting†
Kaiwen
Zhang
a,
Anna P.
Constantinou
d,
Cathal
O’Connell
 bc,
Theoni K.
Georgiou
bc,
Theoni K.
Georgiou
 *d and
Amy
Gelmi
*d and
Amy
Gelmi
 *a
*a
aSchool of Science, RMIT University, VIC 3000, Australia. E-mail: amy.gelmi@rmit.edu.au
bSchool of Engineering, RMIT University, Melbourne, VIC 3000, Australia
cAikenhead Centre for Medical Discovery, St Vincent's Hospital Melbourne, Fitzroy, VIC 3065, Australia
dDepartment of Materials, Imperial College London, London, UK. E-mail: t.georgiou@imperial.ac.uk
First published on 11th February 2025
Abstract
Thermoresponsive polymers have been extensively reported for their use in tissue engineering and drug delivery applications. They have a wide range of thermoresponsive and rheological properties controlled by their structural characteristics, such as composition and architecture. Here, the considerable potential of a PEG based, non-ionic triblock thermoresponsive copolymer, namely OEGMA30013-b-BuMA22-b-DEGMA12 as a bioink for 3D printing with cell encapsulation is identified. The rheological tests showed that the gel transition temperature is 8 °C with 35% w/w concentration in PBS. The printability and cytotoxicity of the thermoresponsive gel were characterised and compared with those of commercial thermoresponsive polymer Pluronic®F127 in detail. Specifically, the 35% w/w triblock copolymer presented great printability with a printing speed of 450 mm min−1 at 37 °C, and was less cytotoxic than F127 at both 20% and 30% w/w concentrations. A one-layer structure of human mesenchymal stem cell (hMSC) embedded triblock copolymer was successfully printed onto a glass slide at 37 °C. This provides an option to create a scaffold for stem cell culture and programming for further tissue engineering applications via direct printing of a cell-laden thermoresponsive polymer.
1. Introduction
A thermoreponsive polymer is a smart material that responds to temperature fluctuation by changing its compatibility with a solvent and this class of smart materials has been widely studied because of their wide range of bio-applications, such as drug delivery, gene delivery, and tissue engineering.1–3 Generally, thermoresponsive polymers in aqueous media can turn from hydrophilic to hydrophobic, or from hydrophobic to hydrophilic upon heating to specific temperatures; these are known as lower critical solution temperature (LCST) or upper critical solution temperature (UCST) polymers, respectively. If the polymer design is chosen appropriately, and under suitable external conditions, a thermoresponsive polymer solution will form a gel upon temperature changes as the liquid polymer solution undergoes gelation. For most clinical applications, LCST-type gels are preferred, as they are optimised to be in the solution state at lower temperature before injection, and then form a gel at body temperature (37 °C).This behaviour has driven the development of polymer systems that can be used to effectively harness thermoresponsive properties. In the field of drug delivery, a polymer solution can be easily mixed with a drug at a lower temperature outside the body. Once injected into the body, it transforms into a gel, encapsulating the drug.4 Depending on the polymer structure, the gel would degrade or dissolve in the body to release the drug in a controlled manner, achieving local release to minimise the side effect of systemic release.5 For in vivo tissue engineering application, thermoreversible hydrogels work similarly to drug delivery agents by encapsulating cells rather than drug compounds. After injection, with body temperature higher than LCST, a gel forms and traps the cells, supporting cell proliferation and tissue formation while the gel degrades. With this, a new tissue could be generated to heal the defect site or replace the dysfunctional one.5 Thermoresponsive gels (TRGs) can also be used as scaffold materials, supporting in vitro tissue engineering applications such as 3-dimension (3D) cell culturing or forming new tissue in vitro for transplantation. Particularly, TRGs are commonly used for sacrificial printing, such as hyaluronic acid – poly(N-isopropylacrylamide) (HA-pNIPAAM),6,7 or Pluronic® F127 (F127), a commercialised thermoresponsive triblock copolymer. Upon heating, F127 changes from the liquid to gel state at physiological temperatures, which also highly depends on its concentration and solution ionic strength. As many studies reported potential inflammation or tissue irritation with F127 under certain conditions, cells are typically not encapsulated in the F127 itself for printing due to its potential cytotoxicity.8 For sacrificial printing, F127 can be printed with another more biocompatible material for encapsulating cells, such as gelatin methacrylate (GelMA) or alginate.9,10
Directly printing a cell-encapsulated scaffold with a TRG can help simplify the process significantly and collect final cell products easily via temperature control, which requires hydrogels demonstrating great printability and cytocompatibility. However, this remains a challenge due to a lack of appropriate materials.11 Only two TRGs that have reached clinical trials, Pluronic® F127 and Regel®, are not suitable, as the former presents cytotoxicity and the latter one requires UV exposure to stabilise the pattern, which could induce cell death.12 The potential of a biodegradable thermoresponsive waterborne polyurethane (PU) gel as a bioink has been investigated, reporting the stacking layer and structure maintenance was limited, resulting in difficulties for complex structure printing.13,14 Encapsulating cells within a bioink to be printed onto 3D structures allows 3D cell culture or forming new tissue with intercellular communication and bonding. Furthermore, the final products can be easily collected by removing the bioink via decreasing the temperature to lower than the gel point.
Recently, human mesenchymal stem cells (hMSCs) have been the commonly used cell type in 3D printing for tissue engineering applications, due to their extraordinary self-renewal and multipotent differentiation properties.15 At first, a 3D printed scaffold for stem cell culture could highly increase the efficiency of autologous cell expansion with no cell loss,16 which is critical when a large number of cells are required for the tissue engineering application, whereas a low number of cells are collected from patients. Secondly, it can support hMSCs to form networks and structures in 3D with appropriate extracellular matrix properties such as stiffness, mimicking in vivo conditions, which significantly affects cell differentiation and holds great promise for artificial organ creation and regenerative medicine applications.17
Here, we introduce a polyethylene glycol (PEG)-based triblock thermoresponsive polymer OEGMA30013-b-BuMA22-b-DEGMA12 (OBD)18,19 with promising thermoresponsive properties and improved non-cytotoxicity compared to F127 to be potentially used as a bioink for 3D printing. OEGMA300, BuMA, and DEGMA represent oligo(ethylene glycol) methyl ether methacrylate with 300 g mol−1 molar mass, n-butyl methacrylate, and di(ethylene glycol) methyl ether methacrylate, respectively. The printability and cytotoxicity of this triblock terpolymer were characterised using hMSCs in this study, investigating the ability of this thermoresponsive polymer as a printable ink and as a bioink carrying human stem cells. This polymer is advantageous compared to the commercially available counterparts that it is easier to be functionalised and/or tailored because it is fabricated using a living polymerisation method that enables the precise control of the comonomer position and ratio.
2. Methods and materials
2.1 Materials
Pluronic® F127 and phosphate-buffered saline (PBS) were purchased from Sigma-Aldrich, Australia.Triblock thermoresponsive polymer OEGMA30013-b-BuMA22-b-DEGMA12 was in-house synthesised via sequential group transfer polymerisation (GTP), as previously described.18,19 Briefly, 10 mg of the GTP catalyst, tetrabutylammonium bibenzoate, was added in a round bottom flask, followed by sealing with a rubber septum and purging with argon. 59 mL of freshly purified solvent, tetrahydrofuran (THF) and 0.6 mL of the GTP initiator, methyl trimethylsilyl dimethylketene acetal, (0.5 g, 3 mmol), were syringed into the flask. Each block was synthesized via sequential addition of the corresponding monomer, which was left to react for 10–15 minutes. The monomers used are: OEGMA300 (50 vol% solution in THF, 17 mL, 8.9 g, 30 mmol), BuMA (8.7 mL, 7.8 g, 55 mmol), and DEGMA (5.4 mL, 5.5 g, 29 mmol). After the exothermic reaction ceased, two aliquots of 0.1 mL were withdrawn for gel permeation chromatography (GPC) and proton nuclear magnetic resonance (1H NMR) spectroscopy analysis. OBD was purified via precipitation in cold n-hexane, followed by vacuum-drying for 10 days at room temperature. The experimental number-average molar mass, molar mass distribution, and composition were determined to be 9400 g mol−1, 1.17 and OEGMA30013-b-BuMA22-b-DEGMA12 (OBD) at 42–33–25% w/w, respectively. The GPC and 1H NMR spectroscopy results of OBD and its linear precursors are shown in Fig. S1 (ESI†). These confirm the successful sequential polymerisation, leading to an increase in molar mass values, and appearance of additional peaks corresponding to the newly added repeated units.
2.2 Polymer solution preparation
Pluronic® F127 solutions were prepared in PBS at 20% and 30% w/w concentrations. OBD solutions were prepared in PBS at 10, 15, 25, and 35% w/w. Solutions were agitated using a magnetic spinner at 300 rpm at room temperature for 24 h until fully dissolved. All polymer solutions were stored at 4 °C until use.2.3 Thermoresponsive properties – visual tests
The thermoresponsive properties of 35% w/w OBD solutions in PBS were determined by observing the phase changes of the solutions from 23 to 80 °C, at a step of 1 °C, using a basic stirrer hotplate (IKA RCT) and a continuously stirred water bath. The temperature was measured using a thermometer, and the observation was recorded 2 min after the controller reached the set temperature to ensure that the solution has reached the desired temperature.2.4 Rheological characterisation
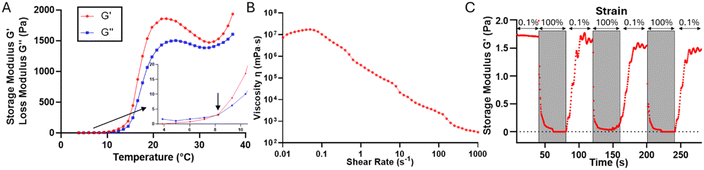
The rheological properties of the 35% w/w OBD were tested with temperature sweep, shear rate ramp and shear recovery. A Physica MCR502 rheometer (Anton Paar, Graz, Austria) with 15 mm conical plate geometry (1° angle) was used for the measurement. Rheometer calibration at room temperature was conducted before testing samples to improve the accuracy of the results. Three independent measurements were repeated for each rheological test.Temperature sweep measurements were performed at 0.1% strain and 1 rad s−1 frequency, at a temperature range from 4 to 37 °C with a temperature increase rate of 2 °C min−1. Changes of shear storage modulus (G′) and shear loss modulus (G′′) were measured during this temperature sweep. The gel point was defined as the temperature at which G′ overcomes G′′ at the first time.20–22 With the shear rate ramp test, viscosity of samples was tested at shear rate ramp from 0.01 to 1000 s−1 at 37 °C (printing temperature). The yield stress was determined at which a viscosity peak is observed. In the shear recovery test, repetitive low strain of 0.1% for 40 s, followed by a high strain of 100% for 40 s was applied to samples at 37 °C and the change of G′ over time was monitored.
2.5 Printability
The printability test followed the bio-ink characterisation protocol published by Connell et al.23 using an INKREDIBLE+ 3D printer (CELLINK, Sweden). Samples were added to a 3 mL syringe, assembled with a 250 μm nozzle. Then the syringe was loaded to the pneumatic cartridge of the 3D printer and connected with an air pressure regulator. After heating the printing head to 37 °C and wrapping it with tin foil to stabilise the temperature of the sample, the bio-ink was ready to be extruded/printed. The printing G-code is listed in S5–S7 (ESI†).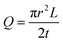 | (1) |
The printed pattern is treated as a half cylinder. Where r is the radius of the cylinder and equal to half of the width of the printed line. L is the length of the printed line within time t, which is equal to the writing speed of the nozzle head.
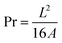 | (2) |
2.6 Stem cell culture and viability
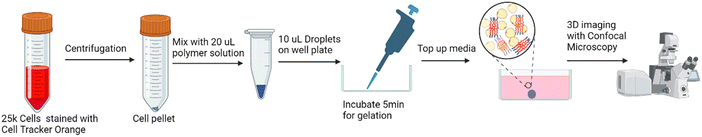
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells stained with CellTracker was collected after centrifugation. 20 μL of the cool polymer was mixed with the cell pellet. Two droplets of 10 μL solution were deposited into a well plate, and incubated at 37 °C for 5 min to solidify the polymer/cell droplets. The droplets were then immersed in complete cell media and incubated for 4 days, after which NucGreen stain was added to stain any dead cells.
000 cells stained with CellTracker was collected after centrifugation. 20 μL of the cool polymer was mixed with the cell pellet. Two droplets of 10 μL solution were deposited into a well plate, and incubated at 37 °C for 5 min to solidify the polymer/cell droplets. The droplets were then immersed in complete cell media and incubated for 4 days, after which NucGreen stain was added to stain any dead cells.
2.7 Printing with cells
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells was then collected after centrifugation and mixed with 20 μL of the cool polymer.10 μL of the cell/polymer solution was pipetted into the 12 well plate, then incubated at 37 °C for 5 min to solidify the polymer/cell droplets. The droplets were then immersed in complete cell media. Before imaging with a confocal microscope, NucGreen stain was added to stain the dead cells. Confocal imaging (20×) with Z stack allows a 3D view of how the cells are distributed inside the polymer after printing.
000 cells was then collected after centrifugation and mixed with 20 μL of the cool polymer.10 μL of the cell/polymer solution was pipetted into the 12 well plate, then incubated at 37 °C for 5 min to solidify the polymer/cell droplets. The droplets were then immersed in complete cell media. Before imaging with a confocal microscope, NucGreen stain was added to stain the dead cells. Confocal imaging (20×) with Z stack allows a 3D view of how the cells are distributed inside the polymer after printing.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 hMSCs (pre-stained with CellTracker Orange CMRA) was mixed with 1 mL cool OBD, which was still under liquid conditions. The cell-laden bioink was then transferred to the syringe and incubated at 37 °C. For printing, the syringe was assembled to the 3D printer with the method described in Section 2.5. A one-layer pattern was printed into a 6 well plate using the optimised constant writing speed of 450 mm min−1. 2 mL of warm basal medium was then added to the well plate and then returned to the incubator at 37 °C. The EVOS M5000 Imaging System was used to image the cells inside the bioink 1 h post printing.
000 hMSCs (pre-stained with CellTracker Orange CMRA) was mixed with 1 mL cool OBD, which was still under liquid conditions. The cell-laden bioink was then transferred to the syringe and incubated at 37 °C. For printing, the syringe was assembled to the 3D printer with the method described in Section 2.5. A one-layer pattern was printed into a 6 well plate using the optimised constant writing speed of 450 mm min−1. 2 mL of warm basal medium was then added to the well plate and then returned to the incubator at 37 °C. The EVOS M5000 Imaging System was used to image the cells inside the bioink 1 h post printing.
Combining the materials’ rheological properties with printing parameters, the estimated shear stress that cells experienced during printing could be calculated with eqn (3) and (4):
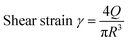 | (3) |
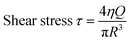 | (4) |
2.8 Statistical analysis
Data presented here are expressed as column plots with mean ± standard deviation (SD). A Welch's t test for cell viability was conducted by GraphPad Prism 9.3. Results and discussion
3.1 Material properties
The thermal properties of 35% w/w OBD were first determined by the visual test. At this concentration, polymer solutions were in the liquid state at 4 °C. At room temperature (23 °C), polymer solution already transitioned to a clear gel condition. When the temperature increased to 39 °C, the gel became cloudy, indicating increased hydrophobic interactions. Gel syneresis, the process where the solvent is expelled from the polymer network due to internal stresses, began at 45 °C and progressed to a precipitation state by 50 °C. This indicates that viscosity of polymer solution increased as the temperature increased from 4 °C to room temperature, forming a gel. The exact gel point of 35% w/w OBD was not determined by the visual test as it already reached the gel state at room temperature and was thus determined by rheology temperature sweep tests.
3.2 Printability
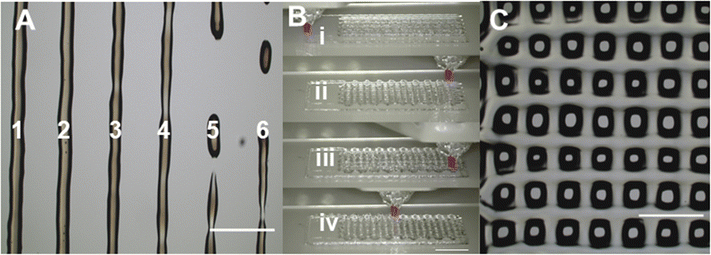
During the line test, the 35% w/w OBD was printed out on the glass slide with speed increased from 300 mm min−1 to 1050 mm min−1 (Fig. 3A, Table 1). When the printing speed was slower than 600 mm min−1, OBD performed a steady extrusion. The average width of the printed pattern at 300 mm min−1 (435 μm) was higher than that at 450 mm min−1 (401 μm), showing that slower printing speed leads to a thicker pattern at the same extrusion rate. The average width of the pattern was slightly larger than nozzle size (250 μm), which is attributed to the polymer liquefaction after exposing to a high shear rate during printing. The ideal writing speed was 450 mm min−1, being used for further printing experiments, as it was the fastest printing speed with steady extrusion. Extrusion rate Q at a 450 mm min−1 printing speed of 35% w/w OBD under 75 kPa was 0.471 μL s−1, calculated using eqn (1).
| Line | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Speed (mm min−1) | 300 | 450 | 600 | 750 | 900 | 1050 |
| Width (μm) | 435 ± 16 | 401 ± 8 | 343 ± 30 | 328 ± 60 | 328 ± 30 | 267 ± 63 |
The printing process and pattern of 4-layer stack printing are shown in Fig. 3B and C. The stacking of multiple layers can be observed clearly. It is worth mentioning that with additional layer printing, the bioink might have difficulty adhering to the bottom pattern at the turnaround point. This issue can be improved by adjusting the Z height or reducing the writing speed. Since the 35% w/w OBD was successfully printed with multiple layers, it opens up the possibility of printing more complex structures for cell culture scaffolds. However, optimising the Z height for each layer is necessary, and the gel structure is required to fully recover after printing each layer before proceeding to the next layer.
3.3 Cell viability
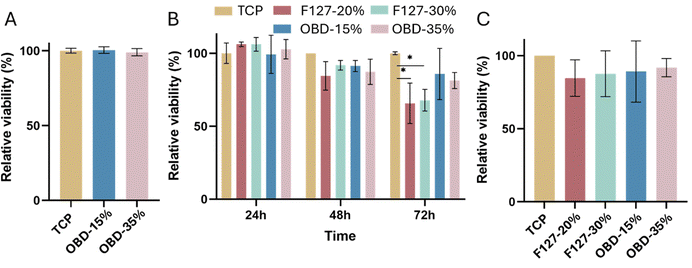
Cell viability for hMSCs seeded around droplets of OBD (15% or 35% w/w) is shown in Fig. 4A. The relative viability of cells in both concentration setting was above 99%, indicating that the OBD material did not leach any cytotoxic substances.
After culturing on the different polymers for 2 days, hMSCs were collected and reseeded, and their viability was further assessed (Fig. 4C). There was no significant difference between groups, with cell viability remaining above 84%. Culturing cells with OBD did not affect cell viability in further application, demonstrating that the OBD could be used as a 3D cell culture scaffold to allow high density cell culture.
In summary, OBD was non-cytotoxic to the hMSCs and demonstrated a higher cell viability than the commercialised product F127.
3.4 OBD printing with cells
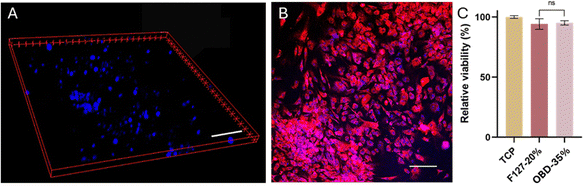
Two hours after printing, the cells were distributed throughout the polymer droplet at different heights, as evidenced by the nuclei (blue) features in the 3D image (Fig. 5A). After 24 hours, most cells were clustered on the same plane (Fig. 5B). This distribution likely occurred because the polymer gel lacked binding sites for the cells, leading them to attach to the well plate substrate. Incorporating a block of arginine–glycine–aspartate (RGD peptide) could improve cell adhesion, though it might impact the physical properties of the polymer.28
On Day 4, the polymer could no longer maintain its droplet shape, likely due to the gel dissolving into the media, which resulted in cell spreading on the substrate. The viability of cells embedded in 35% w/w OBD with droplet printing remained above 90%. In summary, 35% w/w OBD successfully passed the droplet printing test while maintaining high cell viability.
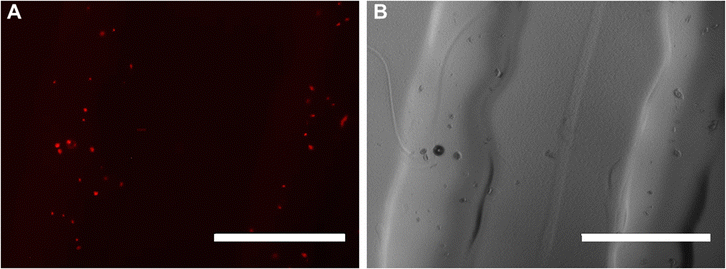 | ||
| Fig. 6 Bioink printed with cells under red excitation filter (A) and bright field (B). Cells were stained with CellTracker showing orange colour. Scale bar 1 mm. | ||
The shear stress τ that cells embedded in 35% w/w OBD experienced during printing was around 79 Pa, calculated with eqn (1)–(4). A shear stress of 79 Pa is considered a low shear stress level and is not expected to be detrimental to hMSCs. In previous research, a fluid shear stress of 89 Pa was applied to PC-3 cells while cells traveling through a needle for 6.4 ms, maintaining cell viability at 75%.30 Human dermal fibroblasts experienced an average shear stress of 190 Pa during the extrusion process with the PU gel showing 65% cell viability.31
It is worthy of note that stem cell behaviour and cell fate might be affected by the shear stress. According to previous studies on shear stress regulating hMSCs differentiation, low shear stress ranging from 0.2 to 2 Pa was commonly used.32–34 The continuous 2 Pa shear stress stimulation for 2 days induces hMSCs toward endothelial cells in the absence of chemical induction.32 Generally, during the extrusion, experiencing 75 Pa has a limited effect on stem cell fate as it is a short-term process (less than a second), with an undefined long-term effect.
With the excellent printability and non-cytotoxicity of OBD, future work should focus on optimising the cell printing process for practical applications. This includes reducing the time cells spend embedded in a high concentration polymer within the syringe without culture media and using printers that require fewer materials for printing. This could highly maintain the cell viability within the cell-laden scaffolds. Improving the binding between the polymer and cells, for example via connecting a block of arginine–glycine–aspartate (RGD peptide) to the polymer chain is another way to stabilise cells within the printed scaffold. Providing cell membrane-stabilising agents, such as hydrocortisone, glucose, and glycerol,8 might also help avoid polymers integrating into the cell membrane and induce cell membrane break through when exposing to shear stress during printing.
4. Conclusion
Here, the potential of a non-ionic triblock copolymer as a bioink has been explored. The printability of the polymer was tested across a range of concentrations, with 35% w/w OBD demonstrating great printability, characterised by steady extrusion and a stable structure. The 35% w/w OBD was not cytotoxic to hMSCs, with cell viability exceeding 80% after 3 days of seeding onto the polymer. This viability was improved compared to the commercial thermoresponsive polymer F127. Encapsulating cells within a bioprintable matrix offers exciting possibilities, such as in situ printing,35 personalised structures, and precise control over the printed design.36 The thermoresponsive properties of this material allow cells to be printed at physiologically relevant temperatures and enable gentle and chemical-free collection of encapsulated or surface-grown cells by liquefying the polymer through temperature reduction.37 This approach minimises cell stress and preserves cell function and structure, enhancing tissue engineering outcomes.38 The non-ionic triblock copolymer can also be further functionalised with cell adhesion peptides or other supportive biological motifs, showcasing the versatility of this polymer design. In summary, this thermoresponsive polymer holds significant promise for future bioprinting and tissue engineering applications.Data availability
The data supporting this article have been included in the ESI† of this manuscript.Conflicts of interest
A. P. C. and T. K. G. report that part of this work regarding the novel chemistry of the materials and their promising thermoresponsive properties have been patented.Acknowledgements
A. P. C. acknowledges the Engineering and Physical Sciences Research Council (EPSRC) for the Doctoral Prize Fellowship (EP/M506345/1) as well as the EPSRC Impact Acceleration Grant EP/R511547/1. K. Z. and A. G. acknowledge K. Z.'s PhD scholarship funded by the Australian Government through the Australian Research Council Discovery Project DP200100612.References
- M. A. Ward and T. K. Georgiou, Thermoresponsive polymers for biomedical applications, Polymers, 2011, 3(3), 1215–1242, DOI:10.3390/polym3031215.
- S. Y. Wong, J. M. Pelet and D. Putnam, Polymer systems for gene delivery-Past, present, and future, Prog. Polym. Sci., 2007, 32(8–9), 799–837, DOI:10.1016/j.progpolymsci.2007.05.007.
- N. Yamada, T. Okano, H. Sakai, F. Karikusa, Y. Sawasaki and Y. Sakurai, Thermo-responsive polymeric surfaces; control of attachment and detachment of cultured cells, Makromol. Chem., Rapid Commun., 1990, 11(11), 571–576, DOI:10.1002/marc.1990.030111109.
- E. Ruel-Gariépy and J. C. Leroux, In situ-forming hydrogels - Review of temperature-sensitive systems, Eur. J. Pharm. Biopharm., 2004, 58(2), 409–426, DOI:10.1016/j.ejpb.2004.03.019.
- A. P. Constantinou and T. K. Georgiou, Pre-clinical and clinical applications of thermoreversible hydrogels in biomedical engineering: a review, Polym. Int., 2021, 70(10), 1433–1448, DOI:10.1002/pi.6266.
- D. B. Kolesky, R. L. Truby, A. S. Gladman, T. A. Busbee, K. A. Homan and J. A. Lewis, 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs, Adv. Mater., 2014, 26(19), 3124–3130, DOI:10.1002/adma.201305506.
- M. Kesti, M. Müller and J. Becher, et al., A versatile bioink for three-dimensional printing of cellular scaffolds based on thermally and photo-triggered tandem gelation, Acta Biomater., 2015, 11(1), 162–172, DOI:10.1016/j.actbio.2014.09.033.
- S. F. Khattak, S. R. Bhatia and S. C. Roberts, Pluronic F127 as a cell encapsulation material: Utilization of membrane-stabilizing agents, Tissue Eng., 2005, 11(5–6), 974–983, DOI:10.1089/ten.2005.11.974.
- J. W. Nichol, S. T. Koshy, H. Bae, C. M. Hwang, S. Yamanlar and A. Khademhosseini, Cell-laden microengineered gelatin methacrylate hydrogels, Biomaterials, 2010, 31(21), 5536–5544, DOI:10.1016/j.biomaterials.2010.03.064.
- P. Mozetic, S. M. Giannitelli, M. Gori, M. Trombetta and A. Rainer, Engineering muscle cell alignment through 3D bioprinting, J. Biomed. Mater. Res., Part A, 2017, 105(9), 2582–2588, DOI:10.1002/jbm.a.36117.
- F. Y. Hsieh and Hsu S. Hui, 3D bioprinting: A new insight into the therapeutic strategy of neural tissue regeneration, Organogenesis, 2015, 11(4), 153–158, DOI:10.1080/15476278.2015.1123360.
- Y. Zhou, Y. Cui and L. Q. Wang, A Dual-sensitive Hydrogel Based on Poly(Lactide-co-Glycolide)-Polyethylene Glycol-Poly(Lactide-co-Glycolide) Block Copolymers for 3D Printing, Int. J. Bioprint., 2021, 7(3), 140–152, DOI:10.18063/ijb.v7i3.389.
- F. Y. Hsieh, H. H. Lin and Hsu S. Hui, 3D bioprinting of neural stem cell-laden thermoresponsive biodegradable polyurethane hydrogel and potential in central nervous system repair, Biomaterials, 2015, 71, 48–57, DOI:10.1016/j.biomaterials.2015.08.028.
- C. W. Wong, Y. T. Chen, C. L. Chien, T. Y. Yu, S. P. Rwei and Hsu S. Hui, A simple and efficient feeder-free culture system to up-scale iPSCs on polymeric material surface for use in 3D bioprinting, Mater. Sci. Eng. C, 2018, 82(1), 69–79, DOI:10.1016/j.msec.2017.08.050.
- S. V. Murphy and A. Atala, 3D bioprinting of tissues and organs, Nat. Biotechnol., 2014, 32(8), 773–785, DOI:10.1038/nbt.2958.
- K. Theodoridis, E. Aggelidou and M. E. Manthou, et al., An effective device and method for enhanced cell growth in 3D scaffolds: Investigation of cell seeding and proliferation under static and dynamic conditions, Mater. Sci. Eng., C, 2020, 114(March), 111060, DOI:10.1016/j.msec.2020.111060.
- C. S. Ong, P. Yesantharao and C. Y. Huang, et al., 3D bioprinting using stem cells, Pediatr. Res., 2017, 83(1–2), 223–231, DOI:10.1038/pr.2017.252.
- A. P. Constantinou, V. Nele and J. J. Doutch, et al., Investigation of the Thermogelation of a Promising Biocompatible ABC Triblock Terpolymer and Its Comparison with Pluronic F127, Macromolecules, 2022, 55(5), 1783–1799, DOI:10.1021/acs.macromol.1c02123.
- A. P. Constantinou, B. Zhan and T. K. Georgiou, Tuning the Gelation of Thermoresponsive Gels Based on Triblock Terpolymers, Macromolecules, 2021, 54(4), 1943–1960, DOI:10.1021/acs.macromol.0c02533.
- A. Kelarakis, V. Castelletto and C. Chaibundit, et al., Rheology and structures of aqueous gels of triblock(oxyethylene/oxybutylene/oxyethylene) copolymers with lengthy oxyethylene blocks, Langmuir, 2001, 17(14), 4232–4239, DOI:10.1021/la0101806.
- V. Cheng, B. H. Lee, C. Pauken and B. L. Vernon, Poly(N-isopropylacrylamide-co-poly(ethylene glycol))-acrylate simultaneously physically and chemically gelling polymer systems, J. Appl. Polym. Sci., 2007, 106(2), 1201–1207, DOI:10.1002/APP.26760.
- G. E. Yu, H. Li and J. P. A. Fairclough, et al., A study of lyotropic mesophases of concentrated solutions of a triblock copolymer of ethylene oxide and 1,2-Butylene oxide, E16B10E16, using rheometry, polarized light microscopy, and small-Angle X-ray scattering, Langmuir, 1998, 14(20), 5782–5788, DOI:10.1021/la9709453.
- C. O’Connell, J. Ren and L. Pope, et al., Characterizing Bioinks for Extrusion Bioprinting: Printability and Rheology, 3D Bioprinting, 2020, pp. 111–133 DOI:10.1007/978-1-0716-0520-2_7.
- J. Schindelin, I. Arganda-Carreras and E. Frise, et al., Fiji: An open-source platform for biological-image analysis, Nat. Methods, 2012, 9(7), 676–682, DOI:10.1038/nmeth.2019.
- M. Müller, J. Becher, M. Schnabelrauch and M. Zenobi-Wong, Nanostructured Pluronic hydrogels as bioinks for 3D bioprinting, Biofabrication, 2015, 7(3), 035006, DOI:10.1088/1758-5090/7/3/035006.
- K. Suman, S. Sourav and Y. M. Joshi, Rheological signatures of gel-glass transition and a revised phase diagram of an aqueous triblock copolymer solution of Pluronic F127, Phys. Fluids, 2021, 33(7), 073610, DOI:10.1063/5.0057090.
- J.-M. Brunet-Maheu, J. C. Fernandes, C. A. V. de Lacerda, Q. Shi, M. Benderdour and P. Lavigne, Pluronic F-127 as a cell carrier for bone tissue engineering, J. Biomater. Appl., 2009, 24(3), 275–287, DOI:10.1177/0885328208096534.
- S. L. Bellis, Advantages of RGD peptides for directing cell association with biomaterials, Biomaterials, 2011, 32(18), 4205–4210, DOI:10.1016/j.biomaterials.2011.02.029.
- T. Billiet, E. Gevaert, T. De Schryver, M. Cornelissen and P. Dubruel, The 3D printing of gelatin methacrylamide cell-laden tissue-engineered constructs with high cell viability, Biomaterials, 2014, 35(1), 49–62, DOI:10.1016/j.biomaterials.2013.09.078.
- J. M. Barnes, J. T. Nauseef and M. D. Henry, Resistance to Fluid Shear Stress Is a Conserved Biophysical Property of Malignant Cells, PLoS One, 2012, 7(12), e50973, DOI:10.1371/journal.pone.0050973.
- L. Ho and Hsu S. Hui, Cell reprogramming by 3D bioprinting of human fibroblasts in polyurethane hydrogel for fabrication of neural-like constructs, Acta Biomater., 2018, 70, 57–70, DOI:10.1016/j.actbio.2018.01.044.
- L. Yuan, N. Sakamoto, G. Song and M. Sato, High-level shear stress stimulates endothelial differentiation and VEGF secretion by human mesenchymal stem cells, Cell. Mol. Bioeng., 2013, 6(2), 220–229, DOI:10.1007/s12195-013-0275-x.
- Dong J. De, Y. Q. Gu and C. M. Li, et al., Response of mesenchymal stem cells to shear stress in tissue-engineered vascular grafts, Acta Pharmacol. Sin., 2009, 30(5), 530–536, DOI:10.1038/aps.2009.40.
- D. H. Kim, S. J. Heo, S. H. Kim, J. W. Shin, S. H. Park and J. W. Shin, Shear stress magnitude is critical in regulating the differentiation of mesenchymal stem cells even with endothelial growth medium, Biotechnol. Lett., 2011, 33(12), 2351–2359, DOI:10.1007/s10529-011-0706-5.
- C. Di Bella, S. Duchi and C. D. O’Connell, et al., In situ handheld three-dimensional bioprinting for cartilage regeneration, J. Tissue Eng. Regener. Med., 2018, 12(3), 611–621, DOI:10.1002/TERM.2476.
- A. V. Do, B. Khorsand, S. M. Geary and A. K. Salem, 3D Printing of Scaffolds for Tissue Regeneration Applications, Adv. Healthcare Mater., 2015, 4(12), 1742–1762, DOI:10.1002/ADHM.201500168.
- T. Takezawa, Y. Mori and K. Yoshizato, Cell Culture on a Thermo-Responsive Polymer Surface, Nat. Biotechnol., 1990, 8(9), 854–856, DOI:10.1038/nbt0990-854.
- F. Doberenz, K. Zeng, C. Willems, K. Zhang and T. Groth, Thermoresponsive polymers and their biomedical application in tissue engineering – a review, J. Mater. Chem. B, 2020, 8(4), 607–628, 10.1039/C9TB02052G.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb02572e |
| This journal is © The Royal Society of Chemistry 2025 |