Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Microbial degradation of bioplastic (PHBV) is limited by nutrient availability at high microplastic loadings†
Michaela K.
Reay‡
 *a,
Martine
Graf‡
*a,
Martine
Graf‡
 b,
Lucy M.
Greenfield
b,
Lucy M.
Greenfield
 b,
Rafael
Bargiela
b,
Rafael
Bargiela
 c,
Charles
Onyije
c,
Charles
Onyije
 a,
Charlotte E. M.
Lloyd
a,
Charlotte E. M.
Lloyd
 ad,
Ian D.
Bull
ad,
Ian D.
Bull
 a,
Richard P.
Evershed
a,
Richard P.
Evershed
 a,
Peter N.
Golyshin
a,
Peter N.
Golyshin
 c,
David R.
Chadwick
c,
David R.
Chadwick
 a and
Davey L.
Jones
a and
Davey L.
Jones
 b
b
aOrganic Geochemistry Unit, School of Chemistry, University of Bristol, Bristol, BS8 1TS, UK. E-mail: michaela.reay@bristol.ac.uk
bSchool of Environmental and Natural Sciences, Bangor University, Bangor, Gwynedd LL57 2UW, UK
cCentre for Environmental Biotechnology, Bangor University, Bangor, Gwynedd LL57 2UW, UK
dSchool of Geography, University of Bristol, Bristol, BS8 1TS, UK
First published on 29th October 2024
Abstract
Biodegradable plastic offers an alternative to conventional plastic for use in agriculture. However, slower degradation in the environment compared to industrial composting and high production of microplastics is of growing concern and poses the question whether they represent a viable replacement. It remains unclear whether observed effects of biodegradable plastics on the soil microbial community and plant nutrient uptake are from biodegradation or from the abiotic effects of the microplastics themselves. The aim of this study was to quantify the biodegradation of the bioplastic poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV), at increasing microplastic loadings (0.06–3.2% w/w) via pyrolysis/gas chromatography-mass spectrometry (Py/GC-MS) alongside effects on soil health and plant growth (Zea mays L.). Between 1.5 and 5% of PHBV microplastic was degraded in soil after 8 weeks, with the rate declining with increasing PHBV concentrations due to microbial nitrogen (N) limitation, demonstrated by increased investment in N-cycling enzymes. Plants were also limited by both N and phosphorus (P). Greater extractable soil ammonium and nitrate contradicted N limitation, however, increases in soil hydrophobicity likely limited mobility, and thus plant and microbial utilisation. As a result, increased C from PHBV degradation did not result in a concurrent increase in microbial biomass, which was reduced under higher PHBV microplastic loading, indicating low microbial carbon use efficiency. While high PHBV microplastic loadings resulted in significant effects on the microbial community size and structure, soil properties and plant growth, there were minimal effects at low PHBV concentrations (0.06% w/w). Observations of nutrient limitation at higher plastic loadings has significant implications for the design of standard biodegradation assays, which must consider both abiotic and biotic effects of microplastic on soil nutrient cycling.
Environmental significanceBiodegradable plastics may be a significant source of microplastics in the environment due to abiotic fragmentation, and potentially slow degradation rates under soil conditions compared to standardised biodegradation tests. Studies have suggested high inputs of biodegradable microplastics significantly disrupt soil biogeochemistry and plant development, yet it remains unclear the mechanism behind this: carbon released from biodegradation, or the effect of hydrophobic microplastic themselves. To address this, we combined direct quantification of bioplastic degradation using pyrolysis-gas chromography-mass spectrometry with assessments of soil nutrient cycling and plant nutrient uptake. Our study highlights that the hydrophobic environment created by biodegradable microplastics alters soil and plant nutrient availability, however, not at realistic plastic loadings in agricultural soils. The finding of nutrient limitation of biodegradation has significant implications for the design of standard biodegradation assays. |
Introduction
Microplastic (≤5 mm) contamination in soil is of growing concern, and the terrestrial environment represents a potential sink for microplastics, with inputs into soils far exceeding that of aquatic settings.1 Microplastics formed from conventional plastics, such as polyethylene (PE) and polypropylene (PP) have long residence times, ranging from decadal to centurial-scales.2 Therefore, biodegradable plastics, particularly biobased ones such as polyhydroxyalkanoates (PHAs), are of interest as an alternative to petroleum based and persistent polymers. One promising PHA is poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV), which can be produced by bacteria from sugars and lipids.3 PHBV has comparable Young's modulus (i.e. stiffness) and tensile strength to PP and PE, with further improvement of this mechanical property via inclusion of natural fibres or other biodegradable polymers.4,5As a natural polymer, PHBV is used for carbon (C) storage in a range of bacterial taxa, and the ability to degrade PHAs has been observed in a range of bacteria and fungi.6,7 During biodegradation, biodegradable plastics, including PHBV, should be converted to carbon dioxide (CO2) and microbial biomass.8 However, conditions (e.g. moisture, temperature, pH, microbial community, nutrient availability) in the natural environment may not be optimal to facilitate degradation.9 Instead, abiotic fragmentation from physical damage and UV irradiation may yield more microplastics on a shorter timescale than conventional counterparts.10–12 Several studies have demonstrated changes in the soil microbial community and biogeochemistry in the presence of biodegradable microplastics. C released during biodegradation of PHBV microplastics has been shown to prime the soil microbial community and increase nitrogen (N) immobilisation and microbial biomass, which may limit plant available N.13 Divergent effects of PHBV on microbial community activity have been observed, with both increases13,14 and decreases,15 alongside the same trends in bacterial alpha diversity. This is despite consistent increases in the abundance of Acidobacteria and Verrucomicrobia phyla under PHBV microplastic addition.13–15 In field-based experiments of PHBV at lower loadings (0.01% w/w) under winter barley (Hordeum vulgare L.), no effects on available N or microbial community structure were observed,16 indicating that at more realistic microplastic input levels affects of PHBV microplastics on soil function may be minimal. However, it remains unclear if the effects on the soil microbial community structure and function are a result of biodegradation of the bioplastic input, or indirect effects arising from the microplastics, such as change in moisture or transport of nutrients.17 Varying rates of biodegradation of PHBV in soil have been observed, controlled by moisture, temperature, and soil type.7,18–20 However, there has not been a combined study quantifying the effects of PHBV at varying concentrations on soil function and microbial community structure, plant development and the degree of degradation of biodegradable microplastics.
Biodegradation of microplastics can be observed via indirect approaches, such as CO2 evolution or oxygen demand, standardised for plastics in soil under ISO17556.21 However, the input of biodegradable plastic-derived C may result in mining of soil organic matter (SOM) for N and phosphorus (P), and not all CO2 evolved may be plastic-derived,22 resulting in the overestimation of biodegradation. Direct quantification of biodegradation, e.g. using stable isotope22,23 or radiolabelling,24 can enable source attribution for evolved CO2, although these methods are often limited by substrate availability and cost.25 An alternative approach is to directly quantify the remaining PHBV in soil via pyrolysis/gas chromatography-mass spectrometry (Py/GC-MS).26 This approach is widely applied to microplastics in environmental settings for conventional plastics (e.g. LDPE, PP) in soils.27–31 Furthermore, this approach can also be used to detect nanoplastics (<1 μm) in environmental matrices.32 Therefore, the application of Py/GC-MS to biodegradable plastics in soils will enable direct monitoring of biodegradation and quantification of remaining plastic contamination.
The primary aim of this study was to quantify degradation of PHBV, with increasing microplastic addition and the effects on soil microbial community diversity and function and Zea mays L. development. Microplastic loadings represented realistic plastic loadings (0.06% w/w), a hotspot (0.6% w/w), and future accumulation under low degradation scenarios (1.6%, 3.2% w/w). Degradation of PHBV was quantified via Py/GC-MS. Soil nutrient and potential extracellular enzyme activity measurements were used to reveal effects on microbial nutrient cycling, while phospholipid fatty acid (PLFA) analyses were used to indicate effects on microbial biomass, combined with 16S rRNA gene amplicon sequencing for microbial community structure and diversity. Plant development was monitored throughout the experimental period, and nutrient analyses used to reveal effects on plant nutrient acquisition. We hypothesise that: (i) PHBV degradation will be a contributor of C to the microbial community, but degradation will be limited at higher concentrations due to nutrient limitation, (ii) C inputs from degradation will prime the microbial community, with increased enzymatic activity to acquire other essential nutrients (N, P), and (iii) plant nutrient uptake will be reduced with higher plastic loadings, due to microbial immobilisation of nutrients. An alternative hypothesis is that the introduction of hydrophobic microplastics into soil will reduce soil moisture, which will feedback on available nutrients, plant nutrient acquisition and the size of the microbial community.
Materials and methods
PHBV degradation experiment
Soil was collected to a depth of 10 cm of a Lolium perenne L. dominated grassland located at Henfaes Research Station (Abergwyngregyn, Wales, 53°14′19.7′′ N, 4°00′54.4′′ W). The site is a flat lowland field with no notable surface runoff, and has no prior history of external microplastic input, except unavoidable atmospheric deposition (37 microplastic particles per 1 g soil at a depth of 0–10 cm). The most prevalent polymer types present were polyamide, polypropylene, and rubber. The soil is classified as a Eutric Cambisol,33 with a sandy clay loam texture and crumb structure. Soil was sieved to 9 mm to remove stones and roots, and air-dried. General soil properties are presented in Table S1.†The PHBV microplastics were between 1 and 80 μm in diameter (average particle size = 23.5 μm; ENMAT Y1000; Ningbo Tianan Biologic Materials Ltd, Ningbo City, China). The ratio of PHB to PHV was 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Four plastic concentrations were used: 0.06%, 0.6%, 1.6% and 3.2% (w/w), which represent low inputs e.g. single use of a biodegradable plastic (0.06%, based on the mass of plastic mulch film to cover the surface area of the mesocosm experiment), a hotspot of plastic pollution (0.6%), and future potential pollution extremes (1.6% and 3.2%).34 While we acknowledge that biodegradable plastics are, by definition, intended to decompose to microbial biomass and CO2, previous studies (between 40 and 100 days) in soils have indicated slow degradation of biodegradable plastics,7,20 hence, there is potential for a net accumulation at sites with repeated use. Microplastics were homogenised into the soil by adding the equivalent weight required to soil, resulting in a combined total weight of 1.5 kg in a 2 L terracotta pot. A sand treatment with 3.2% (w/w) of acid-washed quartz sand (125–300 μm) (Sigma-Aldrich, UK) was also included to account for potential changes in soil structure, alongside a negative soil control with no plastic or sand. No difference was observed between the sand treatment relative to the negative control, hence these results are presented together. All treatments were conducted with 5 replicates.
1. Four plastic concentrations were used: 0.06%, 0.6%, 1.6% and 3.2% (w/w), which represent low inputs e.g. single use of a biodegradable plastic (0.06%, based on the mass of plastic mulch film to cover the surface area of the mesocosm experiment), a hotspot of plastic pollution (0.6%), and future potential pollution extremes (1.6% and 3.2%).34 While we acknowledge that biodegradable plastics are, by definition, intended to decompose to microbial biomass and CO2, previous studies (between 40 and 100 days) in soils have indicated slow degradation of biodegradable plastics,7,20 hence, there is potential for a net accumulation at sites with repeated use. Microplastics were homogenised into the soil by adding the equivalent weight required to soil, resulting in a combined total weight of 1.5 kg in a 2 L terracotta pot. A sand treatment with 3.2% (w/w) of acid-washed quartz sand (125–300 μm) (Sigma-Aldrich, UK) was also included to account for potential changes in soil structure, alongside a negative soil control with no plastic or sand. No difference was observed between the sand treatment relative to the negative control, hence these results are presented together. All treatments were conducted with 5 replicates.
All treatments received 50 kg N ha−1 (as NH4NO3) one week prior to sowing the seeds; no P and potassium (K) were initially applied as the soil had sufficient P and K concentration, according to RB209 guidelines (Table S1†).35 A random block design was used in watering trays in the greenhouse, and the terracotta pots were rotated weekly to account for any position effects. Watering was conducted every 2 to 3 days from the base of the pots, to keep the absorbent fabric at the base of the pots saturated and allow sufficient water uptake from plants throughout the experiment. Maize (Zea mays L. cv. Humboldt) seeds were germinated for 4 days before sowing in the mesocosms (2 per pot). The experiment was conducted in a greenhouse for 8 weeks with a 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 light–dark cycle and an average light intensity of 207 W m−2 before being destructively harvested. The average temperature during this growing period was 24.5 °C and the relative humidity was 34.6%. At 5 weeks, mesocosms were fertilised at a rate equivalent to 50 kg N ha−1 (NH4NO3), 55 kg P ha−1 (Na2H2PO4) and 150 kg K ha−1 (KCl), due to the appearance of foliar anthocyanin production. Throughout the experiment, plant height and leaf chlorophyll content were determined weekly. Chlorophyll content was determined using a Soil Plant Analysis Development (SPAD-502 PLUS) chlorophyll meter (Konica-Minolta, Japan) on the newest fully extended leaf. The experiment was ended after 8 weeks by removing the plants.
8 light–dark cycle and an average light intensity of 207 W m−2 before being destructively harvested. The average temperature during this growing period was 24.5 °C and the relative humidity was 34.6%. At 5 weeks, mesocosms were fertilised at a rate equivalent to 50 kg N ha−1 (NH4NO3), 55 kg P ha−1 (Na2H2PO4) and 150 kg K ha−1 (KCl), due to the appearance of foliar anthocyanin production. Throughout the experiment, plant height and leaf chlorophyll content were determined weekly. Chlorophyll content was determined using a Soil Plant Analysis Development (SPAD-502 PLUS) chlorophyll meter (Konica-Minolta, Japan) on the newest fully extended leaf. The experiment was ended after 8 weeks by removing the plants.
Quantification of residual PHBV in soils using Py/GC-MS
Py/GC-MS was used to directly quantify the amount of PHBV remaining in soil. The low thermal stability of PHBV enabled pyrolysis without prior extraction from the soil, as pyrolysis products from the soil at the optimum temperature (350 °C) did not interfere with the analysis. This facilitated minimal sample preparation and reduced potential for losses during extraction of remaining PHBV. Furthermore, the resulting pyrolysis products are derived from intact microplastics and any nanoplastics or free oligomers which had been produced due the degradation process. Therefore, this captures all remaining PHBV which has yet to be incorporated into microbial biomass or converted to CO2 and is not limited to a certain size range, as are many other commonly used methods.All soils and PHBV samples were weighed into quartz tubes and quartz wool, which were pre-furnaced at 1000 °C for 2 h to remove any organic contamination. Tetramethylsulfonium hydroxide (TMSH; 2.5% in methanol, 10 μL, Sigma-Aldrich, UK) was used for in situ methylation of pyrolysis products via thermochemolysis. Py/GC-MS analyses were conducted using a CDS Pyroprobe 6200 fitted with a drop autosampler and interfaced with a GC-MS (GC Trace 1310, MS ISQ 7000). The pyrolysis interface was held at 300 °C and sorbent trap at 50 °C. The pyrolysis ramp was 20 °C ms−1 and the temperature was 350 °C, held for 20 s. The pyrolysis chamber was flushed with helium (He) for 2 min to transfer pyrolysis products onto the sorbent trap. This was then desorbed at 300 °C for 4 min to transfer pyrolysis products to the GC-MS via the GC transfer line held at 310 °C. The GC inlet was split/spitless, held at 300 °C with a split flow of 40 mL min−1 and split ratio of 20. The GC was fitted with a HP-1 column (Agilent Technologies, 60 m × 0.32 mm i.d., 25 μm film thickness) with He carrier gas at 2.0 mL min−1. The temperature programme, initiated when the trap desorption started, was 40 °C (6 min) to 310 °C at 15 °C min−1 (15 min). The GC was interfaced to the MS via the MS transfer line, held at 300 °C. The scan time was 0.2 s, the scan range was m/z 50–650, the ion source was held at 310 °C and the ionisation mode was EI at 70 eV.
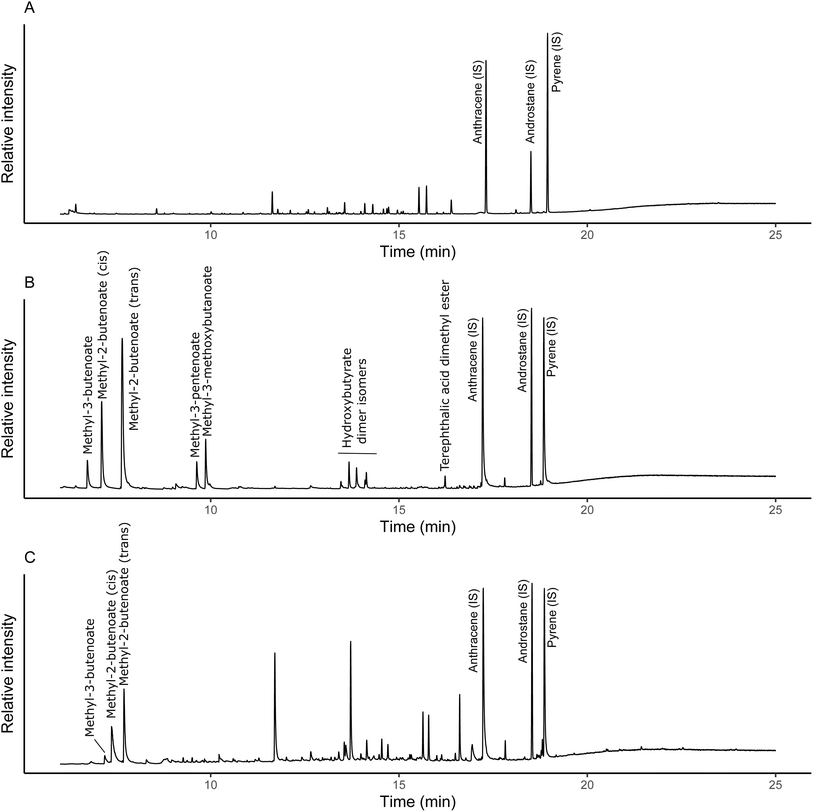
Quantification was achieved through a combination of an internal standard and external calibration. The internal standard (anthracene-d10) was used to normalise individual analyses to account for potential between-run differences in pyrolysis and transfer to the GC-MS. The external calibration of PHBV, which was also normalised to the internal standard, was then used for quantification of PHBV in soil, using the sum of methyl-3-butenoate, cis-methyl-2-butenoate and trans-methyl-2-butenoate derived from the PHB portion of the co-polymer (Fig. 1). These were the most abundant pyrolysis products and initial calibrations with PHBV indicated they were linear across the concentration range (Fig. S1†). There was low presence of PHV pyrolysis products (e.g. methyl-3-pentenoate and methyl-3-methoxybutanoate) observed in the pure PHBV standards analysed (Fig. 1b). However, due to the low ratio of PHB to PHV (99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PHB
1 PHB![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PHV), at soil-relevant concentrations, the PHV-derived pyrolysis products were not detectable (Fig. 1c). Control soils were analysed to confirm there were no natural sources of PHB pyrolysis products (Fig. 1a). Matrix effects with increasing soil mass for pyrolysis were observed (Fig. S2†), therefore, soils spiked with PHBV were used for quantification. PHBV-spiked soils were prepared by dissolution of PHBV in chloroform (0.5 mg mL−1) and spiking of soils with the PHBV solution. Chloroform was evaporated (50 °C), and soils ground to ensure PHBV was homogeneously mixed in the soil, reflecting the distribution of PHBV in the experimental soils. The dissolved PHBV standard solution was compared to a calibration of solid PHBV alone to confirm complete dissolution prior to spiking the soils to generate the soil-PHBV calibration standards. The calibration range used was 0.5 μg to 15 μg, and the respective mass of soil for each microplastic loading was 10 mg (0.06%), 1 mg (0.6%), 0.2 mg (1.6%) and 0.1 mg (3.2%). The experimental soils were lyophilised, after which any visible plant material was removed, and then the soil was ground. Individual replicates were analysed five times, to account for potential variation in the distribution of PHBV. Control soils were also analysed in the same way as the 0.06% soil, which required the highest mass for pyrolysis, to check for any natural sources of the pyrolysis products used for quantification.
PHV), at soil-relevant concentrations, the PHV-derived pyrolysis products were not detectable (Fig. 1c). Control soils were analysed to confirm there were no natural sources of PHB pyrolysis products (Fig. 1a). Matrix effects with increasing soil mass for pyrolysis were observed (Fig. S2†), therefore, soils spiked with PHBV were used for quantification. PHBV-spiked soils were prepared by dissolution of PHBV in chloroform (0.5 mg mL−1) and spiking of soils with the PHBV solution. Chloroform was evaporated (50 °C), and soils ground to ensure PHBV was homogeneously mixed in the soil, reflecting the distribution of PHBV in the experimental soils. The dissolved PHBV standard solution was compared to a calibration of solid PHBV alone to confirm complete dissolution prior to spiking the soils to generate the soil-PHBV calibration standards. The calibration range used was 0.5 μg to 15 μg, and the respective mass of soil for each microplastic loading was 10 mg (0.06%), 1 mg (0.6%), 0.2 mg (1.6%) and 0.1 mg (3.2%). The experimental soils were lyophilised, after which any visible plant material was removed, and then the soil was ground. Individual replicates were analysed five times, to account for potential variation in the distribution of PHBV. Control soils were also analysed in the same way as the 0.06% soil, which required the highest mass for pyrolysis, to check for any natural sources of the pyrolysis products used for quantification.
Plant and soil properties
Plant and soil properties were determined as outlined in Graf et al. (2023).34 Roots were washed with water, then root and shoot biomass were determined by oven drying (80 °C, until constant mass). Dried shoot and root samples were ground (<63 μm). Total C and N was analysed using a TruSpec Leco C/N analyser (Leco Corp., St. Joseph, USA). Phosphorus (P), potassium (K) and sulphur (S) content was determined using total reflection X-ray fluorescent spectrometry (TXRF) (S2 Picofox, Bruker, Billerica, USA).Soil gravimetric water content was determined by oven drying (105 °C, 24 h). Bulk density was determined using 9.8 cm3 plastic cylinder rings as described in Rowell (1994).36 Soil hydrophobicity was determined using air dried soil (10 g), spread evenly across a Petri dish, and ethanol (50 μL) added at increasing v/v volumes in water (0, 3, 5, 8.5, 13, 24, 36, 50, 100% v/v), until drops penetrated in <3 s.37 Soil pH and electrical conductivity (EC) were measured in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (w/v) soil
2 (w/v) soil![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water suspension. Soils were extracted with 0.5 M K2SO4 (1
water suspension. Soils were extracted with 0.5 M K2SO4 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 w/v) and ammonium (NH4+), and nitrate (NO3−) concentrations were determined colorimetrically according to the salicylic acid procedure38 and vanadium chloride procedure of Miranda et al. (2001),39 respectively. Available phosphate (PO43−) was extracted using 0.5 M acetic acid (1
5 w/v) and ammonium (NH4+), and nitrate (NO3−) concentrations were determined colorimetrically according to the salicylic acid procedure38 and vanadium chloride procedure of Miranda et al. (2001),39 respectively. Available phosphate (PO43−) was extracted using 0.5 M acetic acid (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 w/v) then quantified using the molybdate blue method.40 Dissolved organic carbon (DOC) was measured on a Multi-N/C Series TOC/TN analyser (Analytik-Jena, Jena, Germany).
5 w/v) then quantified using the molybdate blue method.40 Dissolved organic carbon (DOC) was measured on a Multi-N/C Series TOC/TN analyser (Analytik-Jena, Jena, Germany).
Potential enzyme activities in the rhizosphere were determined for the negative soil control, 0.6%, 1.6% and 3.2% treatments only according to Marx et al. (2001).41 The 0.06% treatment was excluded as there were minimal effects on soil properties compared to the soil control. Adhered soil from roots was collected after removal to obtain root-zone soil. A soil slurry (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 w/v) produced with sterile deionised water (250 rpm, 30 min). For the assay, 50 μL of soil suspension, 100 μL of 200 μM substrate and 50 μL of buffer (0.05 M Trizma, pH 7.8 for 7-amino-4-methyl coumarin (AMC), and 0.1 M MES, pH 6.1 for 4-methylumbelliferone (MUF) substrates) were added to a 96 well microplate and incubated at 20 °C. Fluorescence was measured at an excitation wavelength of 355 nm and an emission wavelength of 460 nm, and a slit width of 20 nm (Cary Eclipse Fluorescence Spectrophotometer, Agilent Corp., Santa Clara, CA). Potential enzyme activities were measured after incubation times of 0 min, 1 h and 2 h, and the difference between emission at 2 h and 1 h was used to determine AMC/MUF release and thus enzyme activity in nmol AMC/MUF per g dry soil per h. The selected enzymes, and their respective substrates were: leucine aminopeptidase (LAP, L-leucine-7-AMC), phosphatase (PHOS, 4-MUF–phosphate), β-xylosidase (XYL, 4-MUF–β-D-xylopyranoside), β-D-glucuronidase (BDG, 4-MUF–β-D-glucuronide), β-glucosidase (BG, 4-MUF–β-D-glucoside), and N-acetyl-β-glucosaminidase (NAG, 4-MUF–N-acetyl-β-glucosaminide).
100 w/v) produced with sterile deionised water (250 rpm, 30 min). For the assay, 50 μL of soil suspension, 100 μL of 200 μM substrate and 50 μL of buffer (0.05 M Trizma, pH 7.8 for 7-amino-4-methyl coumarin (AMC), and 0.1 M MES, pH 6.1 for 4-methylumbelliferone (MUF) substrates) were added to a 96 well microplate and incubated at 20 °C. Fluorescence was measured at an excitation wavelength of 355 nm and an emission wavelength of 460 nm, and a slit width of 20 nm (Cary Eclipse Fluorescence Spectrophotometer, Agilent Corp., Santa Clara, CA). Potential enzyme activities were measured after incubation times of 0 min, 1 h and 2 h, and the difference between emission at 2 h and 1 h was used to determine AMC/MUF release and thus enzyme activity in nmol AMC/MUF per g dry soil per h. The selected enzymes, and their respective substrates were: leucine aminopeptidase (LAP, L-leucine-7-AMC), phosphatase (PHOS, 4-MUF–phosphate), β-xylosidase (XYL, 4-MUF–β-D-xylopyranoside), β-D-glucuronidase (BDG, 4-MUF–β-D-glucuronide), β-glucosidase (BG, 4-MUF–β-D-glucoside), and N-acetyl-β-glucosaminidase (NAG, 4-MUF–N-acetyl-β-glucosaminide).
Soil microbial community analyses
Similarly to potential enzyme activities, the 16S rRNA gene amplicon sequencing was determined for the soil control, 0.6%, 1.6%, and 3.2% treatments only (n = 3). The analysis was conducted as outlined in Graf et al. (2023).34 Briefly, soil was frozen at −80 °C then lyophilised and prokaryotic DNA was extracted using the Zymo Research Quick DNA Fecal/Soil Microbe Miniprep Kit (Zymo Research, Irvine, CA) according to manufacturer's instructions. Bacterial 16S rRNA genes were PCR-amplified using modified forward primer F515 (5′-GTGBCAGCMGCCGCGGTAA-3′) and reverse R806 prokaryotic primer (5′-GGACTACHVGGGTWTCTAAT-3′).42 PCRs were performed using OneTaq DNA Polymerase (New England Biolabs, Ipswich, MA) with no-template negative controls.34 Following purification using the QIAEX II Gel Extraction Kit (Qiagen, Hilden, Germany), amplicons were sequenced using the Illumina MiSeq (Illumina Inc., San Diego, CA) with a 500 cycle v2 chemistry kit (2 × 250 bp paired-end reads). Raw sequences were read and processed,42,43 then taxonomically classified using Silva (v. 132) as the reference database. Analysis of the most abundant taxonomic groups was performed using in-house R scripts, for groups with a relative abundance ≥ 2%. Selection started at the genus level and groups were added to the immediate upper taxonomic level when none of the samples of that group reached the 2% threshold, and this process continued until the relative abundance of the examined taxonomic level was ≥2%. The NCBI BioProject accession number is PRJNA1128446.To quantify microbial biomass, PLFA analyses were conducted on lyophilized ground soil (2 g) as previously described in Graf et al. (2023).34 The total lipid extract was obtained in the organic phase following sonication extraction using a modified Bligh Dyer solution (15 mL of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.8 (v/v/v) methanol, dichloromethane (DCM), and phosphate buffer). Lipid fractionation was achieved with activated silica columns, conditioned with chloroform (5 mL). Neutral lipids (NLFAs) were eluted using 5 mL chloroform, glycolipids were eluted using 10 mL acetone, and the phospholipid fraction (PLFAs) was eluted with 5 mL methanol, with n-nonadecane added as an internal standard. The PLFA fraction underwent catalysed methylation to obtain fatty acid methyl esters (FAMEs) using 5% (v/v) HCl in methanol (5 mL, 50 °C for 2 h). The FAMEs were extracted into n-hexane (3 × 3 mL), using saturated sodium chloride solution (5 mL) to exert phase separation prior to GC and GC-MS analysis in n-hexane (50 μL). The GC was fitted with a VF23-ms column (60 m, 0.32 μm i.d., 0.15 μm film thickness), and the temperature programme was: 50 °C (1 min) to 100 °C (10 °C min−1) to 250 °C (4 °C min−1, 15 min hold), with a He carrier gas flow of 2.0 mL min−1. The injection volume was 1 μL. The MS operated in electron ionisation mode (70 eV) with a full scan range (m/z 50–650) and a scan time of 0.2 seconds. Data was acquired and analysed using Xcalibur (version 4.1). Assignment of PLFAs between C14 and C20 chain length was based on Frostegard et al. (1993) and Joergensen (2022).44,45 The sum of Firmicutes-derived PLFAs (i14:0, i15:0a, i16:0a, i17:0, i18, a15:0, a16:0, a17:0, a18:0, a19:0) and actinobacteria-derived PLFAs (10Me16:0, 10Me17:0, 10Me18:0) were used to represent Gram positive bacteria. The sum of cy17:0, cy19:0, 16:1ω7, 16:1ω9, 17:1ω8, 18:1ω7 PLFAs was used to represent Gram negative bacteria. The sum of 16:1ω5c, 18:1ω9c, 18:2ω6c, and 18:3ω6,9,12 PLFAs was used to represent fungi, and the 14:0, 15:0, 16:0, 17:0, 18:0, 20:0, 20:4ω6,9,12,15 PLFAs were classed as unspecified.
0.8 (v/v/v) methanol, dichloromethane (DCM), and phosphate buffer). Lipid fractionation was achieved with activated silica columns, conditioned with chloroform (5 mL). Neutral lipids (NLFAs) were eluted using 5 mL chloroform, glycolipids were eluted using 10 mL acetone, and the phospholipid fraction (PLFAs) was eluted with 5 mL methanol, with n-nonadecane added as an internal standard. The PLFA fraction underwent catalysed methylation to obtain fatty acid methyl esters (FAMEs) using 5% (v/v) HCl in methanol (5 mL, 50 °C for 2 h). The FAMEs were extracted into n-hexane (3 × 3 mL), using saturated sodium chloride solution (5 mL) to exert phase separation prior to GC and GC-MS analysis in n-hexane (50 μL). The GC was fitted with a VF23-ms column (60 m, 0.32 μm i.d., 0.15 μm film thickness), and the temperature programme was: 50 °C (1 min) to 100 °C (10 °C min−1) to 250 °C (4 °C min−1, 15 min hold), with a He carrier gas flow of 2.0 mL min−1. The injection volume was 1 μL. The MS operated in electron ionisation mode (70 eV) with a full scan range (m/z 50–650) and a scan time of 0.2 seconds. Data was acquired and analysed using Xcalibur (version 4.1). Assignment of PLFAs between C14 and C20 chain length was based on Frostegard et al. (1993) and Joergensen (2022).44,45 The sum of Firmicutes-derived PLFAs (i14:0, i15:0a, i16:0a, i17:0, i18, a15:0, a16:0, a17:0, a18:0, a19:0) and actinobacteria-derived PLFAs (10Me16:0, 10Me17:0, 10Me18:0) were used to represent Gram positive bacteria. The sum of cy17:0, cy19:0, 16:1ω7, 16:1ω9, 17:1ω8, 18:1ω7 PLFAs was used to represent Gram negative bacteria. The sum of 16:1ω5c, 18:1ω9c, 18:2ω6c, and 18:3ω6,9,12 PLFAs was used to represent fungi, and the 14:0, 15:0, 16:0, 17:0, 18:0, 20:0, 20:4ω6,9,12,15 PLFAs were classed as unspecified.
Statistical analysis
All data analysis was performed in R (v4.4.0)46 unless otherwise stated. Normality of the data was determined by the Shapiro–Wilk test (p ≤ 0.05) and homogeneity of variance of the data was visually checked using residuals vs. fitted plots. One-way ANOVAs were used to test the effect of microplastic loading on end soil and plant properties (p ≤ 0.05). Mixed effect models, using the lmer function in the lmerTest package,47 with plastic loading as a fixed effect and time as a random effect, were used for plant growth measurements. Models were checked for normality, via QQ plots, and heteroscedasticity, by plotting residuals vs. fitted values. Data was transformed by natural logarithmic transformation if there was heteroscedasticity observed in the untransformed data. Microbial graphs for relative abundance were created using MS Excel and Affinity Designer (v1.10.5.1342). Dendrograms (using bray distance and ward criterion) and rarefaction curves were created using normalised data in the SHAMAN web-based application.48 The difference in species richness, Shannon diversity and Simpson diversity between treatments was determined with a Kruskal–Wallis test (p ≤ 0.05). For richness, a follow-up Dunn test was performed with Bonferroni correction. One-way ANOVAs were used to test the effect of microplastic concentration on the relative contribution of taxa, with a Tukey HSD post hoc test with a 95% confidence interval (p ≤ 0.05).Results
PHBV degradation
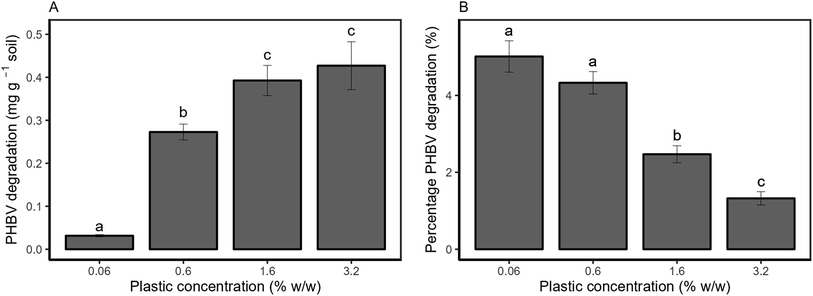
Py/GC-MS was used to directly quantify the amount of PHBV remaining in soil after 2 months (Fig. 2). Absolute degradation (Fig. 2a) increased with higher microplastic loadings, from 0.031 ± 0.003 mg g−1 of soil at 0.06% w/w microplastic loading to 0.43 ± 0.06 mg g−1 of soil at 3.2% w/w microplastic loading over the experimental period. The degradation of PHBV increased in proportion to the amount of microplastic added from 0.06% to 0.6% w/w (8.8 times higher). However, degradation in the 1.6% w/w treatment (0.39 ± 0.04 mg g−1) was only 1.4 times higher than the 0.6% w/w treatment (0.27 ± 0.02 mg g−1), and was also comparable to the 3.2% w/w treatment (0.43 ± 0.6 mg g−1). A similar trend was observed for the relative degradation of PHBV, with comparable percentage degradation for the 0.06 and 0.6% w/w treatments, and decreasing at higher plastic loadings (Fig. 2b).PHBV microplastic effects on soil properties
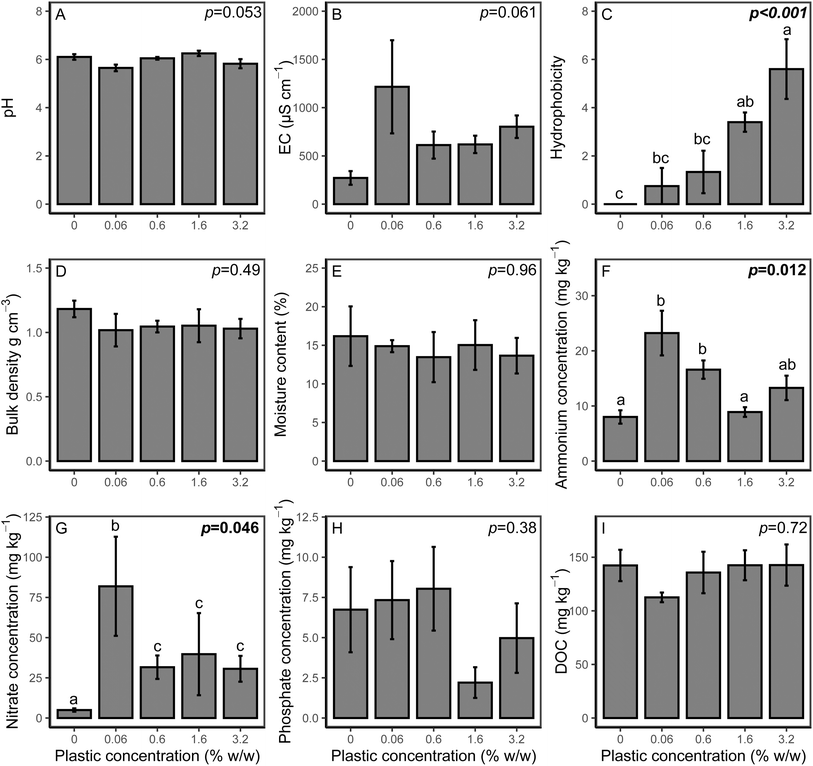
The effect of microplastics on soil properties are shown in Fig. 3. There was no effect of PHBV microplastics on pH (p = 0.813), EC (p = 0.658) or bulk density (p = 0.492). Hydrophobicity significantly varied between treatments (p < 0.001), with the 1.6% and 3.2% treatments significantly higher than the control (p = 0.006 and p < 0.001, respectively). However, there was no effect on soil moisture content (p = 0.692). Available NH4+ was significantly higher at 0.06% and 0.6% loading of PHBV (p = 0.38 and p = 0.027, respectively). Available NO3− was significantly affected by microplastics (p = 0.046) and was higher than the control at all concentrations. Extractable phosphate was lower for the 1.6% and 3.2% microplastic loadings, however, this was not significant (p = 0.243), and there was no effect on DOC concentration (p = 0.72).PHBV microplastic effects on plant growth
Plant biomass and nutrient contents are shown in Table 1. Shoot biomass decreased at higher microplastic loading (p < 0.001), and biomass was significantly lower for 1.6% and 3.2% loadings (p = 0.021 and p = 0.002, respectively). Root biomass was also significantly reduced (p < 0.001), which was significant at lower plastic loadings (0.6%, p = 0.047) compared to shoot biomass. In addition to biomass changes, there were also effects on the nutrient content of the plant material. C and N content were both reduced with higher microplastic loading (p < 0.001 and p = 0.011, respectively). Shoot P content was also reduced for the 1.6% and 3.2% treatments (p = 0.034 and p = 0.047, respectively), although there was no effect on shoot K (p = 0.747) or S content (p = 0.123). For belowground biomass, N content was lower for the 3.2% treatment (p = 0.002), while root K content increased above 0.6% microplastics compared to the control (p = 0.048). There was no effect of microplastics on root C (p = 0.174), P (p = 0.191) or S (p = 0.177). The plant height and SPAD were determined across the experimental period (Fig. S2 and S3† respectively). Plant height significantly varied across the growth period (p < 0.001), and decreased with increasing microplastic concentration, although at 0.06% microplastic loading, the plant height was comparable to the control. SPAD was also significantly affected by microplastic loading (p < 0.001), with lower chlorophyll content at higher plastic loading, although the SPAD for higher microplastic concentrations (0.6%, 1.6% and 3.2%) converged with the control and 0.06% treatment at the end of the experimental period.| Plant property | p value | Microplastic concentration (% w/w) | ||||
|---|---|---|---|---|---|---|
| 0% | 0.06% | 0.6% | 1.6% | 3.2% | ||
| Shoot biomass (g per pot) | <0.001 | 3.50 ± 0.33a | 3.55 ± 0.69a | 2.04 ± 0.62ab | 1.29 ± 0.21b | 0.66 ± 0.2b |
| Shoot C content (%) | 0.001 | 44.0 ± 0.2a | 43.8 ± 0.3ab | 42.6 ± 0.4bc | 42.9 ± 0.3abc | 42.3 ± 0.1c |
| Shoot N content (%) | 0.020 | 1.33 ± 0.07a | 1.23 ± 0.13ab | 1.23 ± 0.15ab | 1.08 ± 0.08b | 0.78 ± 0.06b |
Shoot C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N ratio N ratio |
0.006 | 33.5 ± 2.2b | 36.7 ± 3.2b | 37.8 ± 5.1b | 40.6 ± 2.9ab | 55.4 ± 4.1a |
| Shoot K content (mg g−1) | 0.75 | 29.4 ± 2.1 | 26.7 ± 1.6 | 29.7 ± 1.8 | 27.0 ± 1.8 | 28.1 ± 2.3 |
| Shoot P content (mg g−1) | 0.047 | 3.70 ± 0.10a | 3.93 ± 1.58a | 3.87 ± 0.87a | 2.90 ± 1.28b | 2.93 ± 0.38b |
| Shoot S content (mg g−1) | 0.12 | 1.00 ± 0.07 | 0.97 ± 0.11 | 0.96 ± 0.15 | 0.85 ± 0.01 | 0.63 ± 0.06 |
| Root biomass (g per pot) | <0.001 | 2.53 ± 0.2ab | 2.87 ± 0.75a | 1.5 ± 0.27bc | 1.01 ± 0.07c | 0.77 ± 0.09c |
| Root C content (%) | 0.17 | 37.3 ± 1.6 | 32.8 ± 1.7 | 34.9 ± 1.1 | 32.8 ± 1.2 | 34 ± 1.6 |
| Root N content (%) | 0.002 | 0.79 ± 0.02a | 0.70 ± 0.03a | 0.77 ± 0.03a | 0.72 ± 0.03ab | 0.60 ± 0.04b |
Root C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N ratio N ratio |
0.003 | 47.4 ± 2.0a | 47.1 ± 2.1b | 45.4 ± 1.3b | 45.9 ± 1.1b | 56.7 ± 2.8b |
| Root K content (mg g−1) | 0.008 | 20.2 ± 1.3b | 19.4 ± 1.6b | 25.7 ± 1.5ab | 25.8 ± 1.5ab | 26.9 ± 1.9b |
| Root P content (mg g−1) | 0.19 | 2.79 ± 0.31 | 2.19 ± 0.30 | 3.43 ± 0.38 | 3.05 ± 0.29 | 2.95 ± 0.33 |
| Root S content (mg kg−1) | 0.18 | 2.61 ± 0.13 | 2.09 ± 0.18 | 3.02 ± 0.30 | 2.92 ± 0.39 | 2.51 ± 0.21 |
Soil enzyme potential activities
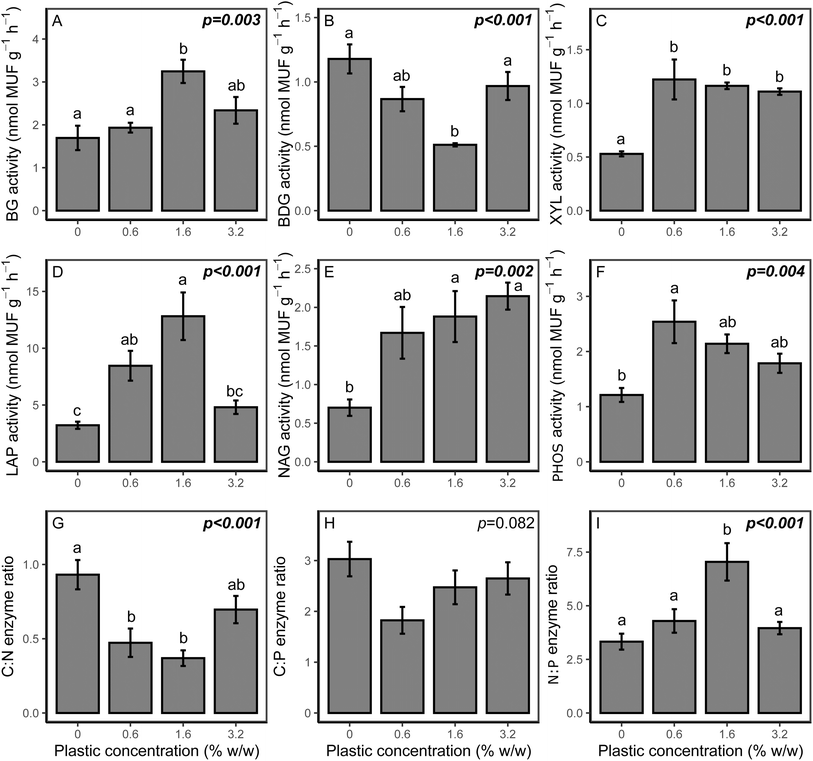
Potential soil enzyme activities and ratios are presented in Fig. 4. For C cycling enzymes, all analysed enzymes were affected by the addition of microplastic. For XYL, potential activity was significantly increased for all PHBV concentrations (p < 0.001) compared to the control. Potential BG activity was significantly higher for the 1.6% and 3.2% treatments (p = 0.004 and p = 0.031, respectively), but did not vary relative to the control for 0.6% w/w PHBV. Potential BDG activity was lower than the control at 1.6% microplastic content (p = 0.008), although other microplastic treatments were comparable to the control. For N cycling enzymes, both LAP and NAG potential activities were higher in the microplastic treatments than the control (p < 0.001 and p = 0.002, respectively), except LAP activity for the 3.2% microplastic loading, which was comparable to the control. PHOS activity was also elevated compared to the control (p = 0.004), which was significant at 0.6% concentration (p = 0.007). Due to differing changes in potential activities of enzymes associated with C, N and P cycling, the relative cycling of C, N and P varied with microplastic addition, as indicated by enzyme ratios of C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N, and N
N, and N![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P. The enzyme C
P. The enzyme C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N ratio was lower for all microplastic treatments (p < 0.001), although there was no effect on the enzyme C
N ratio was lower for all microplastic treatments (p < 0.001), although there was no effect on the enzyme C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ratio (p = 0.082). The enzyme N
P ratio (p = 0.082). The enzyme N![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ratio varied with microplastic addition, although this was only significantly higher for the 1.6% treatment (p = 0.001).
P ratio varied with microplastic addition, although this was only significantly higher for the 1.6% treatment (p = 0.001).
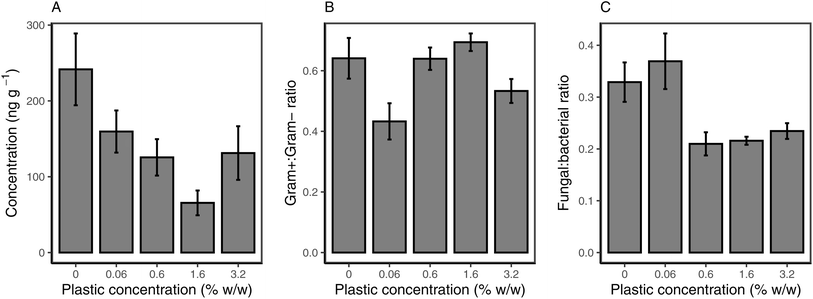
Microbial community biomass via PLFAs
The total PLFA biomass was significantly influenced by microplastic addition (p = 0.043), with all microplastic treatments showing lower PLFA concentration compared to the control. The total PLFA concentration was lowest for 1.6% w/w loading, while it was comparable in the 0.06%, 0.6% and 3.2% w/w (Fig. 5a). The lower concentration of specific PLFAs was also observed for Gram+ bacteria (p = 0.047; Fig. S4c†). Within Gram+ bacteria, the relative decrease in PLFA biomarkers due to microplastic addition was higher for actinobacteria than for Firmicutes (Fig. S4a and b†). Gram− bacteria showed the same trend as Gram+ bacteria (p = 0.037), with the lowest concentration observed at 1.6% PHBV loading (Fig. S4d†). For fungal-derived PLFAs (Fig. S4e†), there was no effect in the 0.06% treatment, however, the concentration of PLFAs was significantly lower relative to the control for all other microplastic treatments (p = 0.026). Fungal PLFAs were lowest in the 0.6% (p = 0.011) and 1.6% (p = 0.007) treatments, compared to the control. There was no effect of microplastic addition on the ratio of Gram+ to Gram− bacteria (p = 0.45; Fig. 5b), however, the ratio of fungal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) bacterial-derived PLFAs was significantly lower at microplastic concentrations above 0.6% (p = 0.014), indicating a larger decrease in fungal communities than in bacterial communities (Fig. 5b).
bacterial-derived PLFAs was significantly lower at microplastic concentrations above 0.6% (p = 0.014), indicating a larger decrease in fungal communities than in bacterial communities (Fig. 5b).
Microbial relative abundance and diversity via 16S rRNA gene amplicon sequencing
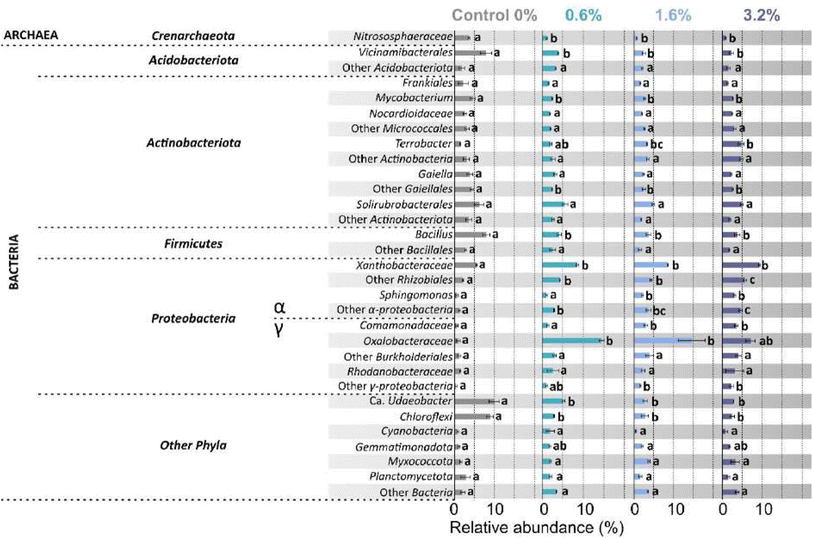
The rarefaction curve (Fig. S5†) showed that samples contained sufficient read counts to capture an accurate representation of the microbial community and diversity within the soil. The total read count per sample increased with increasing PHBV addition, with an average read count of 4085 for control and 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 754 for 3.2% w/w (expressed as counts after removal of chloroplasts and mitochondria). This did not translate into a difference in the Shannon diversity or the Simpson diversity (Table 2). However, species richness significantly varied between treatments (p = 0.033), indicating the number of taxa present increased with increasing PHBV in soil (Table 2). The dendrogram (Fig. S6†) showed a clear distinction between soil control and PHBV treatments, with additional separation between 0.6% w/w and higher concentrations, indicating increased differentiation of the microbial community from the control with increasing PHBV concentration. The analysis of relative abundance of the most abundant groups revealed that 16 out of 31 groups showed no significant change in abundance after addition of PHBV to the soil, whilst 8 groups increased in abundance with increasing PHBV concentration, and 7 groups decreased in abundance with PHBV addition to the soil (Fig. 6). Increase in relative abundance with PHBV addition compared to control was observed for the following groups: Terrabacter (3.2× increase), Xanthobacteraceae (1.7× increase), other Rhizobiales (2.7× increase), Sphingomonas (5× increase), other Alphaproteobacteria (3.7× increase), Comamonadacaeae (6× increase), Oxalobacteraceae (15.6× increase), and other Gammaproteobacteria (6.8× increase). A decrease in relative abundance with PHBV addition to the soil compared to the control was observed for the following groups: Nitrososphaeraceae (5× decrease), Vicinamibacterales (3.5× decrease), Mycobacterium (1.8× decrease), other Gaiellales (1.8× decrease), Bacillus (2.1× decrease), Ca. Udaeobacter (3.5× decrease), and Chloroflexi (3.5× decrease).
754 for 3.2% w/w (expressed as counts after removal of chloroplasts and mitochondria). This did not translate into a difference in the Shannon diversity or the Simpson diversity (Table 2). However, species richness significantly varied between treatments (p = 0.033), indicating the number of taxa present increased with increasing PHBV in soil (Table 2). The dendrogram (Fig. S6†) showed a clear distinction between soil control and PHBV treatments, with additional separation between 0.6% w/w and higher concentrations, indicating increased differentiation of the microbial community from the control with increasing PHBV concentration. The analysis of relative abundance of the most abundant groups revealed that 16 out of 31 groups showed no significant change in abundance after addition of PHBV to the soil, whilst 8 groups increased in abundance with increasing PHBV concentration, and 7 groups decreased in abundance with PHBV addition to the soil (Fig. 6). Increase in relative abundance with PHBV addition compared to control was observed for the following groups: Terrabacter (3.2× increase), Xanthobacteraceae (1.7× increase), other Rhizobiales (2.7× increase), Sphingomonas (5× increase), other Alphaproteobacteria (3.7× increase), Comamonadacaeae (6× increase), Oxalobacteraceae (15.6× increase), and other Gammaproteobacteria (6.8× increase). A decrease in relative abundance with PHBV addition to the soil compared to the control was observed for the following groups: Nitrososphaeraceae (5× decrease), Vicinamibacterales (3.5× decrease), Mycobacterium (1.8× decrease), other Gaiellales (1.8× decrease), Bacillus (2.1× decrease), Ca. Udaeobacter (3.5× decrease), and Chloroflexi (3.5× decrease).
| Treatment | p | Control | 0.60% | 1.60% | 3.20% |
|---|---|---|---|---|---|
| Species richness | 0.033 | 41.6 ± 4.06a | 59.6 ± 3.18ab | 70.0 ± 2.5b | 73.0 ± 7b |
| Shannon index | 0.35 | 3.05 ± 0.07a | 3.12 ± 0.04a | 3.13 ± 0.1a | 3.20 ± 0.07a |
| Simpson index | 0.41 | 0.93 ± 0.004a | 0.92 ± 0.004a | 0.91 ± 0.01a | 0.93 ± 0.004a |
Discussion
PHBV degradation
This study directly quantified the degradation of PHBV microplastics at increasing concentrations alongside their effect on soil nutrient availability, microbial community function and structure, and plant growth. The degradation of PHBV via PHB depolymerase has been previously shown to be expressed in a wide range of soil environments.49,50 The degradation of PHBV microplastic presented here indicated higher degradation of this polymer when introduced as microplastics, relative to previous PHBV and PHB macroplastic studies based on mass loss.7,18,20,51 Biodegradation at 0.06% (realistic) and 0.6% (hotspot) w/w were comparable to degradation observed for PHBV film by indirect CO2 monitoring over the same duration, although this was higher than observed by direct mass loss.19 However, biodegradation of microplastics is suggested to proceed faster than macroplastic, which is first abiotically fragmented to microplastic,11,52 and has a lower surface area to volume ratio for microbial colonisation and degradation. Combined with evidence of changes to C cycling in the soil presented herein, through enhanced potential activity of C cycling enzymes with microplastic inclusion, we suggest direct measures of degradation of biodegradable plastics, such as Py/GC-MS, are required to accurately quantify biodegradation. This approach captures remaining microplastics and nanoplastics,26 and reduces interference in quantifying biodegradation from indirect effects on soil nutrient cycling.The capacity of soil to degrade PHBV was not linear with increasing microplastic concentrations, likely due to nutrient limitation and the finite ability of exoenzyme activity and rate of production.53 This has implications for standardised tests for biodegradation (ISO 17556:2019)21 which use plastic loadings over 1% w/w soil. Standardisation of biodegradation tests is key to ensuring biodegradable products on commercial markets meet required biodegradability standards. However, degradation studies in real-world settings already indicate that while products may pass required standardised tests, degradation rates are lower due to varying moisture, temperature, and nutrient conditions.7,20,53 Our study highlights that the degree of biodegradation is also related to the microplastic loading in soil, therefore, realistic concentrations of plastics in soil should be included in assessments of biodegradation in order to reflect degradation rates in real-world settings. Even the lowest concentration used herein (0.06%), which would be equivalent to one season of plastic mulch film application fragmented into microplastic, may be an overestimation of biodegradable microplastic entering the soil system at any one time. However, with growing concern around biodegradable plastics as a source of microplastics10 and potential hotspots of biodegradable microplastics in soil due to abiotic fragmentation (e.g. 0.6% and above), it is important to consider how the inclusion of microplastics at differing concentrations may regulate biodegradation.
Effects of PHBV on the soil microbial community
There was considerable release of C from PHBV biodegradation, equivalent to up to 52% of the size of the soil DOC pool for the 3.2% w/w treatment (Fig. S7†). However, there was no concurrent increase in DOC, or microbial biomass, suggesting PHBV-derived C released during biodegradation was mineralised to CO2, and that the microbial community exhibited low carbon use efficiency for PHBV. The rapid assimilation of breakdown intermediates parallels that seen for native SOM.54 As hypothesised, increased potential enzyme activity for C, N and P acquiring activity indicated greater investment by the microbial community in acquiring nutrients,55 suggesting a smaller, but potentially more active soil microbial community. Higher BG and LAP activities were previously observed in conjunction with hotspots of nutrient cycling around PHBV microplastics, although these findings were accompanied by increased microbial biomass C.13 The increased enzyme activity associated with SOM turnover also observed in this study, alongside a smaller microbial community, indicated negative effects on microbial carbon use efficiency due to exogenous C input as PHBV microplastic. While PHBV is a bioplastic, and potentially viewed as C neutral,56 increases in turnover of native soil organic matter to acquire nutrients will have implications for the stability of C in SOM pools.57,58The soil microbial community tends to be C limited,59 and labile C would be released during PHBV degradation, yet microbial biomass decreased with PHBV addition. Increased availability of other nutrients (N, P) contradicts limitation of the microbial community by other nutrients through increased enzyme investment, therefore other abiotic factors may be responsible for the observed decreases in the microbial community. Given the decreased fungal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) bacterial ratio, these changes in soil properties impacted the fungal community more than the bacterial community. The bacterial groups which increased in relative abundance, from 16S rRNA analysis, included α- and γ-proteobacteria, which have been observed as the first colonisers of microplastics in soil60 and marine settings,61,62 including PHBV.63 Similar increases in proteobacteria groups were also observed by Zhou et al. (2021)13 with 10% w/w PHBV. Alongside proteobacteria, Terrabacter sp. also increased with PHBV inclusion, which has previously been identified as a degrader of PHB in soils, although other degrading bacteria for PHB have also been identified in different soil types.64 Other groups which can produce PHA depolymerases, such as Bacillus sp.13,65 decreased, highlighting differing responses with soil type and/or pre-existing soil microbial communities. Decreases in Nitrososphaeracaes, which is ammonium oxidising, Chloroflexi and Acidobacteria, such as Vicinamibacterales, likely arose due to changes in N cycling in the soil.66
bacterial ratio, these changes in soil properties impacted the fungal community more than the bacterial community. The bacterial groups which increased in relative abundance, from 16S rRNA analysis, included α- and γ-proteobacteria, which have been observed as the first colonisers of microplastics in soil60 and marine settings,61,62 including PHBV.63 Similar increases in proteobacteria groups were also observed by Zhou et al. (2021)13 with 10% w/w PHBV. Alongside proteobacteria, Terrabacter sp. also increased with PHBV inclusion, which has previously been identified as a degrader of PHB in soils, although other degrading bacteria for PHB have also been identified in different soil types.64 Other groups which can produce PHA depolymerases, such as Bacillus sp.13,65 decreased, highlighting differing responses with soil type and/or pre-existing soil microbial communities. Decreases in Nitrososphaeracaes, which is ammonium oxidising, Chloroflexi and Acidobacteria, such as Vicinamibacterales, likely arose due to changes in N cycling in the soil.66
The inclusion of PHBV into soil also affected its abiotic properties, which are suggested to feedback on the soil microbial community and plant development. The hydrophobicity of the soil increased with higher microplastic loading. While the bulk moisture of the soil was not affected, due to the constant watering regime, the transport and diffusion of soil water and nutrients will be influenced by the hydrophobic regions around microplastics. Reductions in saturated hydraulic conductivity with 2% poly(butylene adipate-co-terephthalate) (PBAT)/poly(lactic acid) (PLA) microplastics have previously been observed.17 In the soil, increased investment in N cycling enzymes relative to C and P enzymes suggests higher competition for available N and mining of soil macromolecular N to meet the microbial demand.67 The degradability of microplastics is also correlated with the priming effect on SOM,68 and introducing more degradable microplastic may strengthen the effect of SOM mining. N limitation may result in the observed plateau in PHBV biodegradation at higher microplastic loadings, which exhibited higher hydrophobicity affecting soil nutrient transport. Significant correlation of biodegradation of PHBV with soil N content has been observed across a range of soil types.69 In this soil, with a low N content of 0.24 g kg−1, N limitation to biodegradation was observed for microplastic loadings above 0.6% w/w, above what inputs would be expected for one growing season use of plastic. More work is required to investigate the microplastic interface with soil pore water, and effects of the hydrophobicity of microplastics on nutrient transport.
Effect of PHBV on plant growth
Increased soil extractable NH4+ and NO3− was observed with microplastic addition, yet there were reductions in microbial biomass, plant biomass and plant N content. While it was hypothesised that plant growth may be reduced due to microbial nutrient immobilisation, instead, we suggest that reduced transport of soil nutrients may reduce plant uptake and create nutrient poor regions resulting in N limitation. Plant N limitation was indicated by reduced chlorophyll content (via SPAD) and N content, and the observation of red anthocyanins also indicated both N and P deficiency.70 P deficiency resulted in reduced shoot P observed at 1.6% and 3.2% w/w, alongside increased potential soil PHOS activity. There was no increase in investment in P enzymes relative to C and N cycling enzymes, however, indicating N availability was more limiting of biodegradation and plant growth than P. There is growing evidence of toxicity of PHBV and PHB, and their degradation products for higher plants such as Sorghum saccharatum, Lepidium sativum and Sinapis alba.71,72 There have been no direct phytotoxicity tests for Zea mays L., although changes in the shoot metabolome of Zea mays L. has previously suggested indirect stress (i.e. nutrient and/or water deficiency), as a result of increasing microplastic content in soil.15 Therefore, reductions in plant growth and biomass observed herein may also be influenced by stress effects of microplastics on soil biogeochemical cycling and the subsequent changes in plant functioning and metabolic response.15,73 Dose-dependent reductions in plant growth have been consistently observed for a wide range of biodegradable microplastics.15,74–77 This study also reveals potential changes in the quality of the crop, with reduced N and P contents, which may have implications for nutritional value of the crop as it enters the food chain. This potential impact on food quality and food security of high microplastic loadings warrants further investigation. However, potential reductions in biomass or nutritional content was not observed at low, realistic (0.06%) plastic loadings, as there was no effect on plant growth or nutrient content. This highlights that while studies using high plastic loadings are important to explore potential effects of hot spots, or accumulation of microplastics, they should also include real-world scenarios (e.g. additives, aged microplastics), which are relevant on a wider scale. This is especially important for defining risk limits for agricultural soils for biodegradable plastic and prevent negative messaging which is not underpinned by fact.Conclusions
Here, we demonstrated that, under increasing PHBV addition, bioplastic degradation is limited both by the acquisition of other nutrients and the effect of hydrophobic regions which may limit the mass flow of water and diffusion of soil nutrients. We show specifically that:(1) The inclusion of bioplastic altered the size and structure of the microbial community, and increased investment in extracellular enzymes to support nutrient acquisition, while biodegradation was nutrient limited.
(2) Despite the smaller, more active microbial community, C inputs from biodegradation were not translated to microbial biomass and indicated reduced carbon use efficiency.
(3) There may be negative impacts of hotspots of biodegradable microplastic inputs into agricultural soils, both for soil carbon stability and crop yield and quality.
(4) At low, and realistic plastic loadings (i.e. input from a plastic mulch film over one growing season), there were minimal effects on the soil microbial community and plant nutrient uptake.
(5) There is a pressing need for longer-term studies reflecting a gradual release of microplastics during fragmentation of biodegradable plastics and to explore the effect of aging/degradation of microplastics. Such studies must include realistic plastic loadings to achieve real-world insights into the effects of biodegradable microplastics on soil. This is also essential for standard assays quantifying biodegradability in soil.
In summary, our results emphasise the importance of combining quantification of biodegradation of microplastics alongside soil microbial function and diversity and plant growth at realistic environmental loadings to assess effects of both biodegradation and the microplastics themselves on soil health.
Data availability
The data supporting this article have been included as part of the ESI.† 16S RNA data for this article are available at the SRA database at https://dataview.ncbi.nlm.nih.gov/object/PRJNA1128446?reviewer=7fu4e3aqurepselvfu0oj9vjnv (PRJNA1128446).Author contributions
M. K. Reay: conceptualization, methodology, formal analysis, investigation, data curation, supervision, writing – original draft, writing – review & editing, visualization. M. Graf: conceptualization, methodology, formal analysis, investigation, data curation, writing – review & editing, visualization. L. M. Greenfield: conceptualization, methodology, investigation, (writing – review & editing). R. Bargiela: data curation, (writing – review & editing). C. Onyije: investigation. C. E. M. Lloyd: writing – review & editing, funding acquisition. I. D. Bull: supervision, writing – review & editing. R. P. Evershed: writing – review & editing, funding acquisition. P. N. Golyshin: writing – review & editing. D. R. Chadwick: conceptualisation, writing – review & editing, supervision, funding acquisition. D. L. Jones: conceptualisation, writing – review & editing, supervision, funding acquisition.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This study funded by the UK Natural Environment Research Council Global Challenges Research Fund programme on Reducing the Impacts of Plastic Waste in Developing Countries (NE/V005871/1). M. K. R., C. O., C. E. M. L., R. P. E. and I. D. B. thank the NERC for partial funding of the National Environmental Isotope Facility (NEIF; contract no. NE/V003917/1) and funding from the European Research Council under the European Union's Seventh Framework Programme (FP/2007–2013) and European Research Council Grant Agreement number 340923 for funding GC-MS capabilities. C. E. M. L. was funded through a The Royal Society Dorothy Hodgkin Fellowship (DHF\R1\191142). We thank Llinos Hughes, Elin Thompson and Bethany Fleming for help in conducting this experiment, and H. L. Whelton and B. Peel for technical support for pyrolysis.References
- A. A. Horton, A. Walton, D. J. Spurgeon, E. Lahive and C. Svendsen, Sci. Total Environ., 2017, 586, 127–141 CrossRef CAS PubMed.
- A. Chamas, H. Moon, J. Zheng, Y. Qiu, T. Tabassum, J. H. Jang, M. Abu-Omar, S. L. Scott and S. Suh, ACS Sustain. Chem. Eng., 2020, 8, 3494–3511 CrossRef CAS.
- E. Bugnicourt, P. Cinelli, A. Lazzeri and V. Alvarez, Express Polym. Lett., 2014, 8, 791–808 CrossRef.
- A. Rivera-Briso and Á. Serrano-Aroca, Polymers, 2018, 10, 732 CrossRef PubMed.
- K. E. Mazur, P. Jakubowska, A. Gaweł and S. Kuciel, Sustainable Mater. Technol., 2022, 31, e00390 CrossRef CAS.
- A. A. Shah, F. Hasan, A. Hameed and S. Ahmed, Ann. Microbiol., 2007, 57, 583–588 CrossRef CAS.
- J. Mergaert, A. Webb, C. Anderson, A. Wouters and J. Swings, Appl. Environ. Microbiol., 1993, 59, 3233–3238 CrossRef CAS.
- R.-J. Mueller, Process Biochem., 2006, 41, 2124–2128 CrossRef CAS.
- I. E. Napper and R. C. Thompson, Environ. Sci. Technol., 2019, 53, 4775–4783 CrossRef CAS PubMed.
- X.-F. Wei, A. J. Capezza, Y. Cui, L. Li, A. Hakonen, B. Liu and M. S. Hedenqvist, Water Res., 2022, 211, 118068 CrossRef CAS PubMed.
- J. Liao and Q. Chen, J. Hazard. Mater., 2021, 418, 126329 CrossRef CAS.
- L. Wei and A. G. McDonald, Polym. Degrad. Stab., 2016, 126, 93–100 CrossRef CAS.
- J. Zhou, H. Gui, C. C. Banfield, Y. Wen, H. Zang, M. A. Dippold, A. Charlton and D. L. Jones, Soil Biol. Biochem., 2021, 156, 108211 CrossRef CAS.
- M. Brtnicky, V. Pecina, J. Holatko, T. Hammerschmiedt, A. Mustafa, A. Kintl, J. Fojt, T. Baltazar and J. Kucerik, Chem. Biol. Technol. Agric., 2022, 9, 75 CrossRef CAS.
- R. W. Brown, D. R. Chadwick, H. Zang, M. Graf, X. Liu, K. Wang, L. M. Greenfield and D. L. Jones, J. Hazard. Mater., 2023, 441, 129959 CrossRef CAS.
- L. M. Greenfield, M. Graf, S. Rengaraj, R. Bargiela, G. Williams, P. N. Golyshin, D. R. Chadwick and D. L. Jones, Agric. Ecosyst. Environ., 2022, 336, 108023 CrossRef CAS.
- Y. Qi, N. Beriot, G. Gort, E. H. Lwanga, H. Gooren, X. Yang and V. Geissen, Environ. Pollut., 2020, 266, 115097 CrossRef CAS.
- Z. Zaidi, D. Mawad and A. Crosky, Front. Mater., 2019, 6, 275 CrossRef.
- S. Muniyasamy, O. Ofosu, M. J. John and R. D. Anandjiwala, J. Renewable Mater., 2016, 4, 133–145 CrossRef CAS.
- J. Kim, N. S. Gupta, L. B. Bezek, J. Linn, K. K. Bejagam, S. Banerjee, J. H. Dumont, S. Y. Nam, H. W. Kang, C. H. Park, G. Pilania, C. N. Iverson, B. L. Marrone and K.-S. Lee, Int. J. Mol. Sci., 2023, 24, 7638 CrossRef CAS PubMed.
- ISO, ISO 17556:2019(en) Plastics—Determination of the Ultimate Aerobic Biodegradability of Plastic Materials in Soil by Measuring the Oxygen Demand in a Respirometer or the Amount of Carbon Dioxide Evolved, 2019 Search PubMed.
- Y. Huo, F. A. Dijkstra, M. Possell, A. Z. Dong and B. Singh, Soil Res., 2023, 61, 755–765 CrossRef CAS.
- T. F. Nelson, R. Baumgartner, M. Jaggi, S. M. Bernasconi, G. Battagliarin, C. Sinkel, A. Künkel, H.-P. E. Kohler, K. McNeill and M. Sander, Nat. Commun., 2022, 13, 5691 CrossRef CAS PubMed.
- L. Tian, Y. Ma and R. Ji, Methods Enzymol., 2021, 121–136 CAS.
- T. Obrador-Viel, V. Zadjelovic, B. Nogales, R. Bosch and J. A. Christie-Oleza, Microb. Biotechnol., 2024, 17(4), e14457 CrossRef.
- M. E. Seeley and J. M. Lynch, Anal. Bioanal. Chem., 2023, 415, 2873–2890 CrossRef CAS.
- M. Fischer and B. M. Scholz-Böttcher, Anal. Methods, 2019, 11, 2489–2497 RSC.
- Z. Steinmetz, A. Kintzi, K. Muñoz and G. E. Schaumann, J. Anal. Appl. Pyrolysis, 2020, 147, 104803 CrossRef CAS.
- F. Watteau, M.-F. Dignac, A. Bouchard, A. Revallier and S. Houot, Front. Sustain. Food Syst., 2018, 2, 81 CrossRef.
- E. D. Okoffo, C. M. Chan, C. Rauert, S. Kaserzon and K. V. Thomas, Environ. Sci. Technol., 2022, 2022, 13774–13785 CrossRef PubMed.
- E. D. Okoffo, F. Ribeiro, J. W. O'Brien, S. O'Brien, B. J. Tscharke, M. Gallen, S. Samanipour, J. F. Mueller and K. V. Thomas, Sci. Total Environ., 2020, 715, 136924 CrossRef CAS.
- E. D. Okoffo and K. V. Thomas, J. Hazard. Mater., 2024, 464, 133013 CrossRef CAS PubMed.
- IUSS Working Group WRB, World Reference Base for Soil Resources 2014, Update 2015. International Soil Classification System for Naming Soils and Creating Legends for Soil Maps. World Soil Resources Reports No. 106, Rome, 2015 Search PubMed.
- M. Graf, L. M. Greenfield, M. K. Reay, R. Bargiela, G. B. Williams, C. Onyije, C. E. M. Lloyd, I. D. Bull, R. P. Evershed, P. N. Golyshin, D. R. Chadwick and D. L. Jones, J. Hazard. Mater., 2023, 458, 131932 CrossRef CAS PubMed.
- AHDB, Nutrient Management Guide (RB209). Section 3 Grass and Forage Crops, 2023 Search PubMed.
- D. L. Rowell, Soil Science: Methods and Applications, Longman Group UK Ltd, London, 1994 Search PubMed.
- S. H. Doerr, Earth Surf. Process. Landforms, 1998, 23, 663–668 CrossRef CAS.
- R. L. Mulvaney, in Methods of Soil Analysis, Part 3, Chemical Methods, ed. D. L. Sparks, A. L. Page, P. A. Helmke, R. H. Loeppert, P. N. Soltanpoor, M. A. Tabatabai, C. T. Johnston and M. E. Sumner, SSSA, Madison, WI USA, 1996, pp. 1123–1184 Search PubMed.
- K. M. Miranda, M. G. Espey and D. A. Wink, Nitric Oxide, 2001, 5, 62–71 CrossRef CAS.
- M. D. R. Vaz, A. C. Edwards, C. A. Shand and M. S. Cresser, Eur. J. Soil Sci., 1994, 45, 353–359 CrossRef.
- M.-C. Marx, M. Wood and S. C. Jarvis, Soil Biol. Biochem., 2001, 33, 1633–1640 CrossRef CAS.
- D. W. Fadrosh, B. Ma, P. Gajer, N. Sengamalay, S. Ott, R. M. Brotman and J. Ravel, Microbiome, 2014, 2, 6 CrossRef PubMed.
- E. Bolyen, J. R. Rideout, M. R. Dillon, N. A. Bokulich, C. C. Abnet, G. A. Al-Ghalith, H. Alexander, E. J. Alm, M. Arumugam, F. Asnicar, Y. Bai, J. E. Bisanz, K. Bittinger, A. Brejnrod, C. J. Brislawn, C. T. Brown, B. J. Callahan, A. M. Caraballo-Rodríguez, J. Chase, E. K. Cope, R. Da Silva, C. Diener, P. C. Dorrestein, G. M. Douglas, D. M. Durall, C. Duvallet, C. F. Edwardson, M. Ernst, M. Estaki, J. Fouquier, J. M. Gauglitz, S. M. Gibbons, D. L. Gibson, A. Gonzalez, K. Gorlick, J. Guo, B. Hillmann, S. Holmes, H. Holste, C. Huttenhower, G. A. Huttley, S. Janssen, A. K. Jarmusch, L. Jiang, B. D. Kaehler, K. Bin Kang, C. R. Keefe, P. Keim, S. T. Kelley, D. Knights, I. Koester, T. Kosciolek, J. Kreps, M. G. I. Langille, J. Lee, R. Ley, Y.-X. Liu, E. Loftfield, C. Lozupone, M. Maher, C. Marotz, B. D. Martin, D. McDonald, L. J. McIver, A. V. Melnik, J. L. Metcalf, S. C. Morgan, J. T. Morton, A. T. Naimey, J. A. Navas-Molina, L. F. Nothias, S. B. Orchanian, T. Pearson, S. L. Peoples, D. Petras, M. L. Preuss, E. Pruesse, L. B. Rasmussen, A. Rivers, M. S. Robeson, P. Rosenthal, N. Segata, M. Shaffer, A. Shiffer, R. Sinha, S. J. Song, J. R. Spear, A. D. Swafford, L. R. Thompson, P. J. Torres, P. Trinh, A. Tripathi, P. J. Turnbaugh, S. Ul-Hasan, J. J. J. van der Hooft, F. Vargas, Y. Vázquez-Baeza, E. Vogtmann, M. von Hippel, W. Walters, Y. Wan, M. Wang, J. Warren, K. C. Weber, C. H. D. Williamson, A. D. Willis, Z. Z. Xu, J. R. Zaneveld, Y. Zhang, Q. Zhu, R. Knight and J. G. Caporaso, Nat. Biotechnol., 2019, 37, 852–857 CrossRef CAS.
- A. Frostegard, A. Tunlid and E. Baath, Appl. Environ. Microbiol., 1993, 59, 3605–3617 CrossRef CAS PubMed.
- R. G. Joergensen, Biol. Fertil. Soils, 2022, 58, 1–6 CrossRef CAS.
- R Core Team, R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria, 2024 Search PubMed.
- A. Kuznetsova, P. B. Brockhoff and R. H. B. Christensen, J. Stat. Software, 2017, 82(13) DOI:10.18637/jss.v082.i13.
- S. Volant, P. Lechat, P. Woringer, L. Motreff, P. Campagne, C. Malabat, S. Kennedy and A. Ghozlane, BMC Bioinf., 2020, 21, 345 CrossRef.
- R. Handrick, S. Reinhardt, P. Kimmig and D. Jendrossek, J. Bacteriol., 2004, 186, 7243–7253 CrossRef CAS PubMed.
- D. Jendrossek and R. Handrick, Annu. Rev. Microbiol., 2002, 56, 403–432 CrossRef CAS PubMed.
- A. S. Al Hosni, J. K. Pittman and G. D. Robson, Waste Manage., 2019, 97, 105–114 CrossRef CAS PubMed.
- Y. Yang, Z. Li, C. Yan, D. Chadwick, D. L. Jones, E. Liu, Q. Liu, R. Bai and W. He, Sci. Total Environ., 2022, 814, 152572 CrossRef CAS PubMed.
- V. Guliyev, B. Tanunchai, M. Udovenko, O. Menyailo, B. Glaser, W. Purahong, F. Buscot and E. Blagodatskaya, Polymers, 2023, 15, 660 CrossRef CAS.
- P. A. W. van Hees, D. L. Jones, R. Finlay, D. L. Godbold and U. S. Lundström, Soil Biol. Biochem., 2005, 37, 1–13 CrossRef CAS.
- R. L. Sinsabaugh, B. H. Hill and J. J. F. Shah, Nature, 2009, 462, 795–798 CrossRef CAS PubMed.
- T. Garrison, A. Murawski and R. Quirino, Polymers, 2016, 8, 262 CrossRef PubMed.
- J. Shi, Z. Wang, Y. Peng, Z. Fan, Z. Zhang, X. Wang, K. Zhu, J. Shang and J. Wang, Environ. Sci. Technol., 2023, 57, 13588–13600 CrossRef CAS.
- K. Mason-Jones, N. Schmücker and Y. Kuzyakov, Soil Biol. Biochem., 2018, 124, 38–46 CrossRef CAS.
- J. L. Soong, L. Fuchslueger, S. Marañon-Jimenez, M. S. Torn, I. A. Janssens, J. Penuelas and A. Richter, Global Change Biol., 2020, 26, 1953–1961 CrossRef PubMed.
- J. Rüthi, B. M. Rast, W. Qi, C. Perez-Mon, L. Pardi-Comensoli, I. Brunner and B. Frey, J. Hazard. Mater., 2023, 441, 129941 CrossRef.
- A. Marín, P. Feijoo, R. de Llanos, B. Carbonetto, P. González-Torres, J. Tena-Medialdea, J. R. García-March, J. Gámez-Pérez and L. Cabedo, Microorganisms, 2023, 11, 1461 CrossRef.
- S. Oberbeckmann, M. G. J. Löder and M. Labrenz, Environ. Chem., 2015, 12, 551 CrossRef CAS.
- C. Odobel, C. Dussud, L. Philip, G. Derippe, M. Lauters, B. Eyheraguibel, G. Burgaud, A. Ter Halle, A.-L. Meistertzheim, S. Bruzaud, V. Barbe and J.-F. Ghiglione, Front. Microbiol., 2021, 12, 734782 CrossRef PubMed.
- S. V. Prudnikova, E. G. Kiselev, A. V. Demidenko, I. V. Nemtsev, E. I. Shishatskaya, S. Thomas and T. G. Volova, Giant, 2024, 18, 100288 CrossRef CAS.
- A. N. Boyandin, S. V. Prudnikova, M. L. Filipenko, E. A. Khrapov, A. D. Vasil'ev and T. G. Volova, Appl. Biochem. Microbiol., 2012, 48, 28–36 CrossRef CAS.
- A. Ho, D. P. Di Lonardo and P. L. E. Bodelier, FEMS Microbiol. Ecol., 2017, fix006 CrossRef.
- Y. Kuzyakov, Soil Biol. Biochem., 2010, 42, 1363–1371 CrossRef CAS.
- G. Zhang, D. Liu, J. Lin, A. Kumar, K. Jia, X. Tian, Z. Yu and B. Zhu, Soil Biol. Biochem., 2023, 180, 109006 CrossRef CAS.
- A. Hoshino, H. Sawada, M. Yokota, M. Tsuji, K. Fukuda and M. Kimura, Soil Sci. Plant Nutr., 2001, 47, 35–43 CrossRef.
- A. Henry, S. Chopra, D. G. Clark and J. P. Lynch, Funct. Plant Biol., 2012, 39, 255 CrossRef CAS PubMed.
- E. Liwarska-Bizukojc, Chemosphere, 2022, 289, 133132 CrossRef CAS.
- E. Liwarska-Bizukojc, Polymers, 2023, 15, 438 CrossRef CAS PubMed.
- Y. Sun, C. Duan, N. Cao, C. Ding, Y. Huang and J. Wang, J. Hazard. Mater., 2022, 424, 127282 CrossRef CAS PubMed.
- E. Liwarska-Bizukojc, Polymers, 2023, 15, 438 CrossRef CAS PubMed.
- A. G. Uzamurera, Z.-Y. Zhao, P.-Y. Wang, Y.-X. Wei, F. Mo, R. Zhou, W.-L. Wang, F. Ullah, A. Khan, X.-B. Xiong, M.-Y. Li, K. Wesly, W.-Y. Wang, H.-Y. Tao and Y.-C. Xiong, Chemosphere, 2023, 329, 138602 CrossRef CAS PubMed.
- H. Sun, Y. Shi, P. Zhao, G. Long, C. Li, J. Wang, D. Qiu, C. Lu, Y. Ding, L. Liu and S. He, Sci. Total Environ., 2023, 868, 161557 CrossRef CAS PubMed.
- M. K. Reay, L. M. Greenfield, M. Graf, C. E. M. Lloyd, R. P. Evershed, D. R. Chadwick and D. L. Jones, J. Hazard. Mater., 2023, 447, 130825 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4va00311j |
| ‡ These authors have contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |