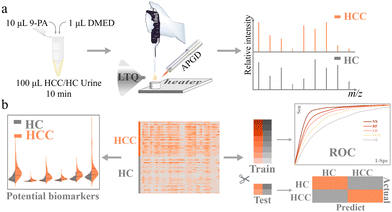
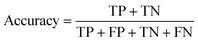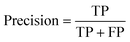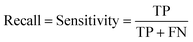Screening of hepatocellular carcinoma via machine learning based on atmospheric pressure glow discharge mass spectrometry†
Jinghan
Fan
ab,
Xiao
Wang
ab,
Yile
Yu
ab,
Yuze
Li
 *c and
Zongxiu
Nie
*c and
Zongxiu
Nie
 *ab
*ab
aBeijing National Laboratory for Molecular Sciences, Key Laboratory of Analytical Chemistry for Living Biosystems, Institute of Chemistry, Chinese Academy of Sciences, Beijing 100190, China. E-mail: znie@iccas.ac.cn; Fax: +86-10-62652123; Tel: +86-10-62652123
bUniversity of Chinese Academy of Sciences, Beijing 100049, China
cState Key Laboratory of High-efficiency Utilization of Coal and Green Chemical Engineering, College of Chemistry and Chemical Engineering, Ningxia University, Yinchuan 750021, China. E-mail: liyuze@nxu.edu.cn
First published on 23rd November 2022
Abstract
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors with a high mortality rate. The diagnosis of HCC is currently based on alpha-fetoprotein detection, imaging examinations, and liver biopsy, which are expensive or invasive. Here, we developed a cost-effective, time-saving, and painless method for the screening of HCC via machine learning based on atmospheric pressure glow discharge mass spectrometry (APGD-MS). Ninety urine samples from HCC patients and healthy control (HC) participants were analyzed. The relative quantification data were utilized to train machine learning models. Neural network was chosen as the best classifier with a classification accuracy of 94%. Besides, the levels of eleven urinary carbonyl metabolites were found to be significantly different between HCC and HC, including glycolic acid, pyroglutamic acid, acetic acid, etc. The possible reasons for the regulation were tentatively proposed. This method realizes the screening of HCC via potential urine metabolic biomarkers based on APGD-MS, bringing a hopeful point-of-care diagnosis of HCC in a patient-friendly manner.
Introduction
Liver is the biggest visceral organ in human body and plays an important role in processing and storing nutrients, producing cholesterols and proteins. Besides, Liver can secrete bile that performs multiple metabolic functions. According to Global Cancer Statistics 2020, hepatocellular carcinoma (HCC) is the fifth most commonly occurring cancer in males and ninth in females.1 The infection of hepatitis virus, excessive intake of aflatoxin, alcoholic intemperance, obesity, etc. all increase the risk of HCC.2Pathological examination is the gold standard of HCC diagnosis. Through the biopsy of liver tissue, HCC can be diagnosed precisely.3 However, there are inherent risks of the biopsy procedure like seeding and bleeding,4 so patients need to undergo the determination of the level of maternal serum alpha-fetoprotein (AFP) or have the imaging examination before biopsy to screen for the possibility of HCC in the first place.5 Nevertheless, AFP detection possesses a high false positive rate6,7 while imaging methods are limited by interobserver variability, side effects of contrast agents, and high cost. Therefore, a cost-effective, time-saving, and patient-friendly method for the screening of HCC is urgently needed.
Urine is the final product of the metabolic system and can be acquired readily.8 It has distinctive advantages of having a simple matrix, being highly accessible, basically sterile, and suitable for long-term storage in large volumes.9 In light of these, it has been a trend to use urine as samples in metabolomic research.10 Through studying the levels of small metabolites (such as carboxyl, aldehyde, and ketone) in the urine of HCC patients and HCs, samples could be identified as HCC or HC through the machine learning method.11,12
Mass spectrometry (MS) is a powerful analytical method since it is capable of providing molecular weight and structure information. Chromatography MS is preferred in analyses of small metabolites in body fluids because it has the superior features of high separation efficiency and strong quantitative ability. At the same time, there are still a few defects that cannot be ignored: sophisticated pretreatments are needed before analysis, operations are complicated, and the introduction of chromatography makes the analysis time relatively long, typically longer than 1 hour.13–15
Carboxyl compounds represent an important kind of metabolite in urine. About 36% of metabolites in urine contain one or more carboxyl groups. The detection of carboxylic compounds in urine, however, imposes strict requirements on measurement performance.16 Firstly, carboxylic acids are weak acids with low ionization efficiency, leading to weak signals in the mass spectra. Secondly, small carboxylic compounds cannot produce characteristic product ions in collision-induced dissociation (CID), which causes difficulty in identification. Thirdly, the mass of urinary carboxylic compounds is always low, yet some mass spectrometers discriminate against small molecules. Hence, in order to improve the ionization efficiency and sensitivity of carboxylic compounds, derivatization reagents are introduced. According to Zhang's study, 32 reagents can be used to derivatize carboxyl of which amine is the most common one.16 Although those derivatization reagents greatly improve the detection results, more sophisticated pretreatments and longer analysis time are also involved in the procedure.17–20
Among all the derivatization reagents, N,N-dimethylethylenediamine (DMED) is one of the most widely used ones in carboxyl-containing metabolite derivatization.21,22 Herein, based on APGD-MS, using DMED, we realized the rapid separation and analysis of carbonyl (mainly carboxylic, also aldehydic and ketonic) compounds in urine. Through machine learning models, we successfully classified HCC and HC cohorts with high precision and sensitivity and obtained 11 potential urinary biomarkers of HCC. Fig. 1 shows the workflow of this cost-effective, time-saving, and pain-free approach to HCC screening.
Experimental
Materials and reagents
The derivatization reagent DMED was purchased from Aladdin Reagent (Beijing, China). 9-Phenylacridine (9-PA) was purchased from Tokyo Chemical Industry Co., Ltd (Tokyo, Japan), and the saturated aqueous solution of 9-PA was used as the internal standard. Triethylamine (TEA) was purchased from Concord Technology Co., Ltd (Tianjin, China). Activating reagent 2-chloro-1-methylpyridinium iodide (CMPI) was purchased from Innochem Technology Co., Ltd (Beijing, China). Creatine and creatinine were purchased from J&K Scientific Ltd (Beijing, China). L-Arginine was purchased from Sinopharm Chemical Reagent Co., Ltd (Shanghai, China). Water used in all experiments is Wahaha Purified water.Sample collection
Urine samples of HCC patients and HC participants were obtained from the First Affiliated Hospital of Gannan Medical University and stored at −80 °C until analysis. In this study, 43 HCC and 47 HC urine samples were enrolled, and the characteristics of those donors are shown in Fig. S1 and Table S1.† The quality control (QC) sample was a mixture of urine from 10 HC samples which had been selected randomly.All experiments were performed in accordance with the guidelines of the Hospital Scientific Research Ethics Committee of the First Affiliated Hospital of Gannan Medical University, and approved by the ethics committee at the First Affiliated Hospital of Gannan Medical University. Informed consent was obtained from human participants of this study.
Instrumentation
MS experiments were mainly performed on LTQ XL (Thermo Scientific, San Jose, CA). The LTQ capillary temperature was 300 °C and the tube lens voltage was 100 V. The homemade desorption, separation, and ionization (DSI) platform based on atmospheric pressure glow discharge (APGD) ion source was placed in front of the LTQ as shown in Fig. S2.† The temperature of the heater was set as 230 °C. A high voltage (CE1500002T, Rainworm Electronics (Shanghai) Co., Ltd Shanghai, China) was applied to the needle electrode in current-controlled mode (max voltage = 1500 V, current = 8 mA). The argon flow rate was set as 0.4 L min−1. The angle formed by the ion source and the MS inlet was 135 degrees. The analysis time for each sample was 3 minutes. More details about the DSI platform and its usage can be found in our previous work.23 High-resolution MS was performed on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Scientific, San Jose, CA, US). All the MS systems were operated in the positive ion mode. The evaporation procedure of extracting carboxyl compounds was performed on a concentration vacuum centrifuge (Beijing JM Technology Co., Ltd, Beijing, China). The evaporation temperature was set as 40 °C.Comparison of derivatization protocols
Scheme S1† shows the derivatization reactions of carbonyl compounds with DMED. The typical general derivatization method was performed according to previous works.13,24,25 Briefly, 100 μL of urine sample was mixed with 300 μL of ethyl acetate and 10 μL of 0.5% (v/v) formic acid aqueous solution, and vortexed for 30 s. Samples were centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g and 4 °C for 3 min and the ethyl acetate layer was collected. The extraction procedure was repeated twice. The supernatants were dried under vacuum. Then, 200 μL of acetonitrile (ACN), 15 μL of 20 mol mL−1 CMPI, and 30 μL of 20 μmol mL−1 TEA were added to the sediment and mixed. The mixture was incubated at 40 °C for 5 min. Afterwards, 30 μL of 20 μmol mL−1 DMED was added and incubated at 40 °C for 1 h. The resulting solution was dried under vacuum and redissolved in 100 μL of ACN/water (1/9, v/v) followed by MS analysis. The simplified method was optimized according to the work of our team.23 Details of the derivatization protocols are shown in Fig. S3.† Briefly, 1 μL of DMED was added to 100 μL of urine sample and vortexed for 30 s. The mixed sample was derived for 10 min at room temperature. The prepared solution was then analyzed by high-resolution MS.
000 g and 4 °C for 3 min and the ethyl acetate layer was collected. The extraction procedure was repeated twice. The supernatants were dried under vacuum. Then, 200 μL of acetonitrile (ACN), 15 μL of 20 mol mL−1 CMPI, and 30 μL of 20 μmol mL−1 TEA were added to the sediment and mixed. The mixture was incubated at 40 °C for 5 min. Afterwards, 30 μL of 20 μmol mL−1 DMED was added and incubated at 40 °C for 1 h. The resulting solution was dried under vacuum and redissolved in 100 μL of ACN/water (1/9, v/v) followed by MS analysis. The simplified method was optimized according to the work of our team.23 Details of the derivatization protocols are shown in Fig. S3.† Briefly, 1 μL of DMED was added to 100 μL of urine sample and vortexed for 30 s. The mixed sample was derived for 10 min at room temperature. The prepared solution was then analyzed by high-resolution MS.
Data analysis
The raw data were extracted using Thermo Xcalibur Qual Browser 4.0 and JupyterLab 2.26 in Anaconda. Orthogonal partial least-squares discriminant analysis (OPLS-DA) and variable importance of projection (VIP) score plots were performed on SIMCA 14.1. The correlation heatmap was obtained using Metaboanalyst 5.0 at https://www.metaboanalyst.ca. Machine learning analyses were conducted using Orange 3.29.3. Other analyses were processed on Origin 2021. Details of the data processing procedure are shown in Fig. S4.†Equations
For machine learning works, the area under the curve (AUC), accuracy, F1, precision, and recall were used to evaluate the classifiers. They could be calculated with true positive (TP), false positive (FP), true negative (TN), and false negative (FN) as shown in the following equation:Results and discussion
Optimization of derivatization protocol
In the general derivatization protocol of carboxyl compounds, activating reagents and an alkaline environment are necessary to accelerate the reaction, which, however, brings more sophisticated pretreatments and longer analysis time.17–20To simplify this procedure, we employed a DSI platform to activate the derivatization. Since this platform could provide a high temperature of 230 °C, carboxyl (also aldehydic and ketonic) compounds could directly obtain energy and be activated without activating reagents and the alkaline environment. This simplification shortened the analysis time from 4–5 hours to 10 minutes and avoided the usage of exogenous reagents.
For the purpose of balancing signals, cost of time, and economy, the adding volume of DMED and derivatization time were optimized. One hundred microliters of QC sample were mixed with 2.5, 1, 0.5, 0.25, 0.1, or 0.05 μL DMED respectively (made up to 2.5 μL with water) and derived for 1, 5, 10, 30, and 60 minutes under room temperature before APGD-MS analyses. In order to quantify the level of urinary compounds, the mass peak intensity of creatinine (Icreatinine + 2 × Idimer creatinine) was chosen as an internal standard in the optimization experiments because of its superior stability under room temperature.26
As shown in Fig. S5a,† with the prolonging of the derivatization time, the relative peak intensity of DMED showed a downtrend. It can be caused by the continuous derivation of high-level compounds or the self-reaction of DMED (Fig. S5b†). With the adding volume of DMED increased, the relative peak intensity of DMED showed an uptrend, which indicated that excess DMED was not entirely involved in the derivatization. Comparing the 0.25 μL and 0.1 μL lines, the relative peak intensity of DMED remained flat after 10 min, but were both a half lower than the 0.5 μL line. It implied that 0.25 μL of DMED can basically realize the derivatization of low-quantity compounds in 100 μL of urine.
The relative intensities of 41 mass peaks under different derivatization conditions were compared. The level of a quarter of the metabolites was relatively low, like 5-aminosalicylic acid, so the derivatization could be finished with only 0.25 μL of DMED (Fig. S6a†). For other metabolites with higher levels, such as cysteine and proline, the derivatization could be finished with at the most 1 μL of DMED (Fig. S6b and c†). Accordingly, we can surmise that when the added volume of DMED was 1.0 μL, the metabolites could be derived sufficiently. Although the added volume of DMED varied, the relative intensities of most of the derivatives reached a peak in 5–10 min. Therefore, a derivatization time of 10 minutes was considered the optimum.
In conclusion, the derivatization protocol was simplified as follows: 1.0 μL of DMED was added to 100 μL of urine sample. The mixture was vortexed for 30 s and incubated at room temperature for 10 minutes (Fig. S3b†).
Comparison of different protocols
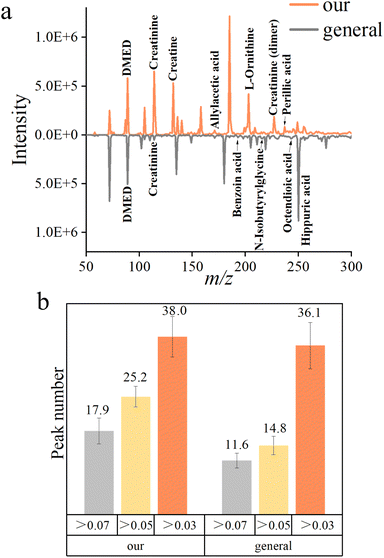
The general protocols of DMED derivatization are always accompanied by activation reagents and the alkaline environment. Along with the above, pretreatments like extraction, purification, etc. greatly increased the processing time.In order to compare the quality of the mass spectra acquired by different protocols, the average mass spectra of both methods in 3 minutes are shown in Fig. 2a. The simplified derivatization protocol was able to detect more peaks (Fig. 2b). This result was verified by the high-resolution MS analyses (Tables S2 and S3†).23 It was interesting that among the compounds identified in the general protocol, the number of small molecules was low while large molecules were numerous. We assumed it may be caused by the volatilization of light carbonyl compounds in the evaporation procedure.
Relative quantification of metabolites
9-PA was chosen as the internal standard (IS) for the relative quantification of metabolites in urine because it neither presented in urine nor interfered with metabolites and showed stable intensity in mass spectra (Fig. S7†). Creatinine was used to evaluate the relatively quantitative ability of the IS. Since creatinine is the most abundant metabolite in urine, it is usually regarded as the standard of the concentration of urine.27 It came out in a good linear relationship, which can be used for reliable relative quantification (Fig. S8a†). Besides, we gradually diluted the solutions of creatinine, creatine, and L-arginine and found the limit of detection (LOD) of this method to be 10−10, 10−12, and 10−11 mol L−1 respectively (Fig. S8b†). In addition, the repeatability of the whole process was tested, and the RSD of typical metabolites was about 20%, which indicated the acceptable repeatability of this method (Fig. S8c and d†).Classification by machine learning
Ninety urine samples (43 HCC and 47 HC) were analyzed by the aforementioned method in Fig. S3b.† Each sample was tested three times in parallel, and a total of 270 spectra were obtained. Mass spectra of urine were collected in positive ion mode over the mass range of 50–300 Da. All the data were normalized using the peak intensity of 9-PA. A heatmap of all the mass spectra data matrices is shown in Fig. S9,† and obvious differences can be observed.Twenty percent of the data sets was randomly picked from each cohort and set as the test set, the other 80% was the training set. Five supervised classifiers RF (Random Forest), LR (Logistic Regression), NN (Neural Network), SVM (Support Vector Machines), and GB (Gradient Boosting) were implanted to classify data sets of two cohorts. Evaluation results were used to measure the classification efficiency.
As shown in Fig. 3a and b, NN gave the best result with an AUC score of 0.990 and classification accuracy of 94.4% for the training cohort. It was chosen as the classifier for the screening of HCC. The evaluation results of NN for the test set are listed in Fig. 3c, and the prediction accuracy was 92.0%. It indicated that combined with machine learning, the analysis of urine carbonyl metabolites acquired by APGD-MS could be a reliable method for HCC screening.
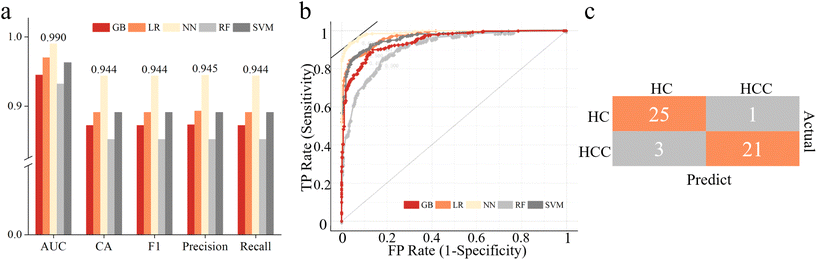 | ||
| Fig. 3 Screening results of machine learning models. (a) Evaluation results of 5 different models. (b) ROC curves of 5 different models. (c) Confusion matrix of test set. | ||
Metabolic alterations and metabolic pathways
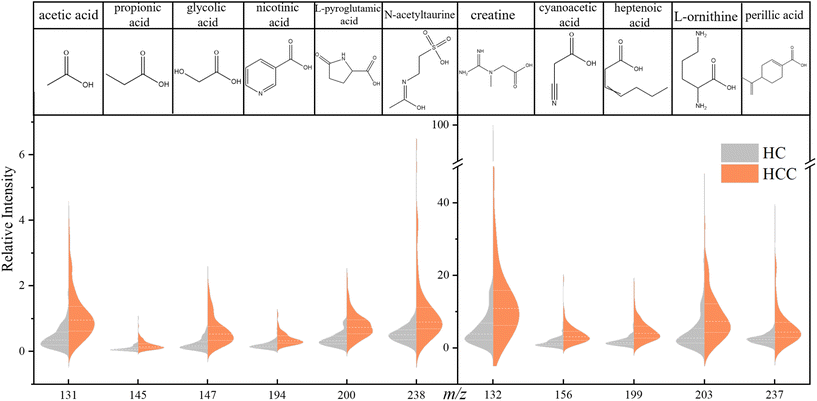
In order to explore the differences in carbonyl compounds between HC and HCC, we obtained an OPLS-DA plot as shown in Fig. S10.† It turned out that these two cohorts can be differentiated unequivocally. Based on this fact, differential metabolites were further selected and studied.A significant difference was defined as fold change (FC) > 2 or FC < 0.5, p-value < 0.05, and variable importance of projection (VIP) value > 1. Under this criteria, 11 compounds can be considered to be the differential metabolites of HCC: acetic acid, creatine, propionic acid, glycolic acid, cyanoacetic acid, nicotinic acid, heptenoic acid, L-pyroglutamic acid, L-ornithine, perillic acid, and N-acetyltaurine. The violin plots of these 11 metabolites are shown in Fig. 4. It can be seen that the level of these 11 acids was generally higher in the HCC cohort, and the most significantly enriched among them was glycolic acid, L-pyroglutamic acid, acetic acid, etc.
In order to explore the latent relationship between urinary carbonyl compounds, the Pearson correlation heatmap and pathway map were obtained based on the relative intensity of 41 peaks (Fig. 5). It can be found that acetaldehyde, cysteine, L-ornithine, etc. were negatively correlated with creatinine, proline, etc. Acetic acid, creatine and glycolic acid presented strong positive correlations with each other. It was inferred that close connections between metabolic pathways of these compounds may exist, yet the specific mechanism needs to be further investigated. In addition, the possible reasons for some altered metabolites were speculated as follows.
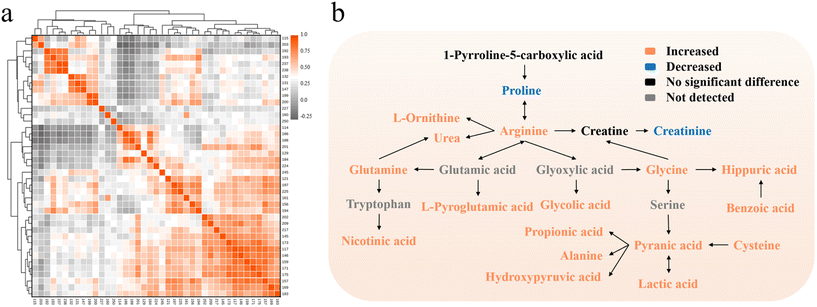 | ||
| Fig. 5 (a) Correlation heatmap of 41 metabolites. (b) Pathway map of the differential metabolites between HCC and HC. | ||
Liver is a vital place for acetate metabolism, but the diffuse degeneration and necrosis of liver tissue caused by HCC would affect the normal functioning of liver. In cancer cells, the energy supply mode of glycolysis increases while the tricarboxylic acid (TCA) cycle decreases. But acetic acid is metabolized to carbon dioxide and water through TCA. Additionally, acetate is one of the main products of β-oxidation of fatty acid in liver whereas HCC will cause the increase of that and lead to the increased level of acetate.28 Therefore, the acetate metabolism of hepatocytes would reduce but the resultant of acetate would increase so that the excess acetate may be excreted from human body through urine, which leads to the upregulation of urinary acetic acid.
Liver is also the major organ for amino acid metabolism, and liver injury would result in its downregulation. According to the research of Leeda-Arporn, HCC would cause the upregulation of glutamic acid levels in serum.29 Since the changing trend of metabolites in serum is basically simultaneous with that in urine and pyroglutamic acid is the product of glutamic acid after dehydration as well as cyclization, the pyroglutamic acid level in urine would increase.
The metabolic cycle of oxalic acid exists in human hepatocytes: mitochondria metabolize glycine to oxalate, glycolate, and glyoxylate and excrete into the cytoplasm.30 In normal cells, peroxisomes can be involved in the metabolic cycle, which uptake glycolate and glyoxylate, and transform them into oxalate for further use. Nonetheless, peroxisomes are not available for glycolic acid metabolism in HCC cells. This abnormality will cause a high level of glycolic acid and force it to be excreted into urine, resulting in the upregulation of urinary glycolic acid level.
Creatine kinase is the central controller of cellular energy homeostasis, and the mitochondrial isoenzyme of creatine kinase in mitochondria plays a pivotal role in cellular energy metabolism. Overexpression of mitochondrial isoenzyme of creatine kinase has been found in HCC tissue, which makes it a biomarker of HCC.31 It may also accelerate the mutual transformation of creatine and creatine phosphate, present the upregulation of their levels and finally bring on the increase of urinary creatine levels.
Using these 11 potential HCC biomarkers as features, we repeated the machine learning classification (Fig. S11a and b†). It suggested that NN was still the optimal model for HCC screening with an AUC score of 0.936 and classification accuracy of 84.4% for the training cohort. The evaluation results for the test set are listed in Fig. S11c,† and the accuracy was 90.0%. While the classification effect and prediction accuracy slightly vary from using all 41 mass peaks, they were still acceptable. It indicated that these 11 metabolites can be considered to be the potential urinary biomarkers of HCC, realizing the rapid screening of HCC.
Conclusions
In conclusion, we simplified the derivatization protocol on the basis of an APGD-based platform from hours to minutes and realized the qualitative and semi-quantitative analyses of carboxylic, aldehydic, and ketonic compounds in urine. Accordingly, we developed a rapid screening method for HCC using the machine learning model and selected 11 potential urinary biomarkers. The prediction accuracy of the test set using all detected metabolites or these 11 potential biomarkers reached 92.2% and 90.0% respectively. This method has the advantages of being cost-effective, time-saving, painless, and highly accurate. With further improvement, a more convenient and automatic screening system could be established. If the screening can be involved before the pathological examination, it would be a promising hint for further diagnosis.Author contributions
J. F.: conceptualization, methodology, formal analysis, investigation, data curation, and writing – original draft; X. W.: formal analysis, writing – review and editing; Y. Y.: writing – review and editing; Y. L.: conceptualization, methodology, writing – review and editing; Z. N.: conceptualization, supervision, writing – review and editing. All the authors have read and agreed to the manuscript.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by grants from the National Natural Sciences Foundation of China (Grant No. 21625504, 21827807, and 21790390/21790392) and Chinese Academy of Sciences.References
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, CA Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- S. K. Clinton, E. L. Giovannucci and S. D. Hursting, J. Nutr., 2020, 150, 663–671 CrossRef PubMed.
- J. Ahmad, H. X. Barnhart, M. Bonacini, M. Ghabril, P. H. Hayashi, J. A. Odin, D. C. Rockey, S. Rossi, J. Serrano, H. L. Tillmann and D. E. Kleiner, J. Hepatol., 2022, 76, 1070–1078 CrossRef CAS PubMed.
- F. P. Russo, A. Imondi, E. N. Lynch and F. Farinati, Dig. Liver Dis., 2018, 50, 640–646 CrossRef PubMed.
- K. A. Gebo, G. Chander, M. W. Jenckes, K. G. Ghanem, H. F. Herlong, M. S. Torbenson, S. S. El-Kamary and E. B. Bass, Hepatology, 2002, 36, S84–S92 Search PubMed.
- H. Ma, X. Sun, L. Chen, W. Cheng, X. X. Han, B. Zhao and C. He, Anal. Chem., 2017, 89, 8877–8883 CrossRef CAS PubMed.
- X. Liu, X. Chi, Q. Gong, L. Gao, Y. Niu, X. Chi, M. Cheng, Y. Si, M. Wang, J. Zhong, J. Niu and W. Yang, PLoS One, 2015, 10, e0127518 CrossRef PubMed.
- The Editors of Encyclopaedia., “urine.”, https://www.britannica.com/science/urine.
- C. Burton and Y. Ma, Curr. Med. Chem., 2019, 26, 5–28 CrossRef CAS PubMed.
- J. Yang, R. Wang, L. Huang, M. Zhang, J. Niu, C. Bao, N. Shen, M. Dai, Q. Guo, Q. Wang, Q. Wang, Q. Fu and K. Qian, Angew. Chem., Int. Ed., 2020, 59, 1703–1710 CrossRef CAS PubMed.
- C. E. Zhang, M. Niu, Q. Li, Y. L. Zhao, Z. J. Ma, Y. Xiong, X. P. Dong, R. Y. Li, W. W. Feng, Q. Dong, X. Ma, Y. Zhu, Z. S. Zou, J. L. Cao, J. B. Wang and X. H. Xiao, J. Ethnopharmacol., 2016, 194, 299–306 CrossRef CAS PubMed.
- X. J. Wang, A. H. Zhang and H. Sun, Hepatology, 2012, 57, 2072–2077 CrossRef PubMed.
- Y. He, Y. Luo, H. Chen, J. Chen, Y. Fu, H. Hou and Q. Hu, Anal. Biochem., 2019, 569, 1–9 CrossRef CAS PubMed.
- H. J. Issaq, O. Nativ, T. Waybright, B. Luke, T. D. Veenstra, E. J. Issaq, A. Kravstov and M. Mullerad, J. Urol., 2008, 179, 2422–2426 CrossRef CAS PubMed.
- P. Ramakrishnan, S. Nair and K. Rangiah, J. Chromatogr. A, 2016, 1443, 83–92 CrossRef CAS PubMed.
- T. Y. Zhang, S. Li, Q. F. Zhu, Q. Wang, D. Hussain and Y. Q. Feng, TrAC, Trends Anal. Chem., 2019, 119, 115608 CrossRef CAS.
- Y. Q. Huang, Q. Y. Wang, J. Q. Liu, Y. H. Hao, B. F. Yuan and Y. Q. Feng, Analyst, 2014, 139, 3446–3454 RSC.
- X. Bian, B. Sun, P. Zheng, N. Li and J. L. Wu, Anal. Chim. Acta, 2017, 989, 59–70 CrossRef CAS PubMed.
- M. Zeng and H. Cao, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2018, 1083, 137–145 CrossRef CAS PubMed.
- Y. R. Liu, K. Thomsen, Y. Nie, S. J. Zhang and A. S. Meyer, Green Chem., 2016, 18, 6246–6254 RSC.
- J. Ding, T. Kind, Q.-F. Zhu, Y. Wang, J.-W. Yan, O. Fiehn and Y.-Q. Feng, Anal. Chem., 2020, 92, 5960–5968 CrossRef CAS PubMed.
- Q.-F. Zhu, J.-W. Yan, Y. Gao, J.-W. Zhang, B.-F. Yuan and Y.-Q. Feng, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2017, 1061–1062, 34–40 CrossRef CAS PubMed.
- Y. Li, L. Jiang, Z. Wang, Y. Wang, X. Cao, L. Meng, J. Fan, C. Xiong and Z. Nie, Anal. Chem., 2022, 94, 9894–9902 CrossRef CAS PubMed.
- S. J. Zheng, S. J. Liu, Q. F. Zhu, N. Guo, Y. L. Wang, B. F. Yuan and Y. Q. Feng, Anal. Chem., 2018, 90, 8412–8420 CrossRef CAS PubMed.
- Q. F. Zhu, Z. Zhang, P. Liu, S. J. Zheng, K. Peng, Q. Y. Deng, F. Zheng, B. F. Yuan and Y. Q. Feng, J. Chromatogr. A, 2016, 1460, 100–109 CrossRef CAS PubMed.
- F. W. Spierto, W. H. Hannon, E. W. Gunter and S. J. Smith, Clin. Chim. Acta, 1997, 264, 227–232 CrossRef CAS PubMed.
- M. Luginbuhl and W. Weinmann, Drug Test. Anal., 2017, 9, 1537–1541 CrossRef PubMed.
- H. Yamashita, T. Kaneyuki and K. Tagawa, Biochim. Biophys. Acta, 2001, 1532, 79–87 CrossRef CAS PubMed.
- R. Leela-Arporn, H. Ohta, M. Tamura, N. Nagata, K. Sasaoka, A. Dermlim, K. Nisa, T. Osuga, K. Morishita, N. Sasaki and M. Takiguchi, J. Vet. Intern. Med., 2019, 33, 1653–1659 CrossRef PubMed.
- P. R. Baker, S. D. Cramer, M. Kennedy, D. G. Assimos and R. P. Holmes, Am. J. Physiol., 2004, 287, C1359–C1365 CrossRef CAS PubMed.
- Y. Soroida, R. Ohkawa, H. Nakagawa, Y. Satoh, H. Yoshida, H. Kinoshita, R. Tateishi, R. Masuzaki, K. Enooku, S. Shiina, T. Sato, S. Obi, T. Hoshino, R. Nagatomo, S. Okubo, H. Yokota, K. Koike, Y. Yatomi and H. Ikeda, J. Hepatol., 2012, 57, 330–336 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2an01756c |
| This journal is © The Royal Society of Chemistry 2023 |