Headpiece-assisted DNA data storage in solution and solid†
Chunjie
Hu‡
,
Qingya
Wen‡
,
Qiuyang
Lai
 ,
Ziyi
Xie
,
Ziyi
Xie
 ,
Kaiyue
Zhang
,
Lu
Zhou
and
Zhi-bei
Qu
,
Kaiyue
Zhang
,
Lu
Zhou
and
Zhi-bei
Qu
 *
*
Department of Medicinal Chemistry, School of Pharmacy, Fudan University, Shanghai, China. E-mail: quzhibei@fudan.edu.cn
First published on 3rd December 2024
Abstract
A headpiece was introduced in the construction of a DNA-based data storage platform. It was demonstrated that the involvement of the headpiece could largely improve the stability, recovery, resistance to DNA contamination, and accuracy in sequencing and data retrieval.
We are living in an era of information explosion. It is predicted that the data production in the coming 2025 will be over 175 zettabytes.1 It will be a great challenge to store the cosmic level information efficiently and safely using conventional storage media.2,3 Due to multiple advantages including high density, longevity, energy efficiency, and sustainability, DNA data storage has been attracting more and more interest in academia and industry.4,5 It is an upcoming consensus that DNA-based information storage will help to bridge the gap between data production and storage limitations, especially in archival storage or “cold information” storage.6,7
DNA is a fragile biomolecule easy to degrade physically, chemically, or biologically8 from environmental intervention such as high temperature, ultraviolet light, irradiation, pH, reactive species, chemicals and solvents, biotic fluids, bacteria, and enzymes. Decades ago, palaeontologists and paleoanthropologists discovered that DNA can be stably stored in mineralized fossils for thousands of years, retaining the possibility for sequencing.9 It enlightened technologies to encapsulate DNA in an inorganic matrix to largely improve the stability and resistance to external damage.10–12
Various materials have been applied for DNA encapsulation. For example, Grass and his collaborators13 have constructed DNA storage in glass (silica) particles, calcium phosphate, calcium chloride, magnesium chloride and so on,6 achieving high DNA loading, increased DNA stability, and simple sample handling.
The encapsulation strategy largely improved the stability of DNA in particles and salts. During the pre-treatments before encapsulation and ultimate PCR sequencing processes after DNA release, however, the stability of DNA will be a problem. Besides, chemicals and impurities are introduced during the encapsulation and releasing procedures. Further extraction and purification will be needed, involving additional losses of DNA. The release of DNA usually employs corrosive acids like hydrogen fluoride,14 which may damage the DNA molecules.
Moreover, we have noticed an essential problem which was rarely discussed by previous researchers, that is, external DNA contamination.15,16 Most DNA storage approaches assumed their experiments were done in an ideal clean condition, without any potential microorganisms. However, the buffer applied in DNA experiments could be good for culturing bacteria and fungi,17,18 introducing serious DNA contamination. Furthermore, inappropriate experimental operations would lead to human DNA contamination19 in data storage as well. DNA contaminations can extremely affect the accuracy of sequencing and information retrieval.
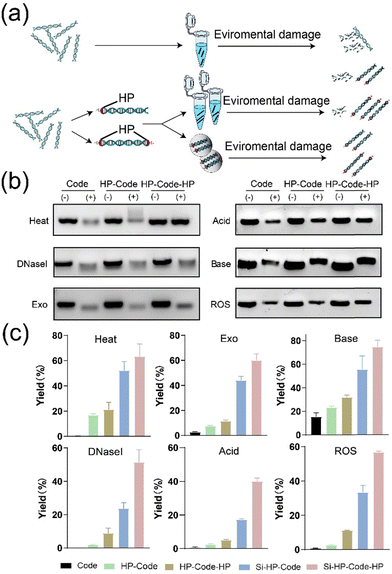
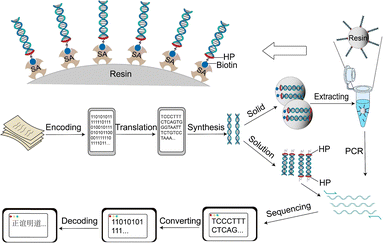
In this work, we attempted to address the several problems mentioned above in DNA data storage. Herein, a headpiece-assisted strategy was proposed to improve the stability, recovery, and resistance to DNA contamination in data storage (Fig. 1).
 | ||
| Fig. 1 Schematic illustration of storing and retrieving data in solution and solid phases in headpiece (HP)-assisted DNA data storage. | ||
A headpiece (HP)20 is a tri-branched polymer that covalently bonds the DNA duplex and provides a handle to a small-molecule ligand if required (Fig. S1 and S2, ESI†). HPs were widely used in DNA encoded libraries (DELs)21,22 for new drug discoveries by pharmaceutical companies such as GSK.23,24 They were verified to stabilize DNA duplexes and to be capable of ligand linkage in large chemical spaces. This work is the first attempt to involve them in DNA data storage, to the best of our knowledge.
Various experiments were constructed to test if the involvement of the headpiece could improve the stability, recovery, and resistance to DNA contamination in data storage as we expected. Considering the DNA duplex has two terminals in both directions, two different types of HP-DNA conjugate were designed (Fig. 2a and Fig. S3, ESI†), with one HP at one terminal (HP-Code), and two HPs at both terminals (HP-Code-HP), respectively. As a proof-of-concept test, we used a 57 bp DNA duplex (namely Code sequence) encoded with “Zheng Yi Ming Dao”, the motto of Shanghai Medical College, Fudan University, in ASCII codes.25
First, gel electrophoresis and realtime fluorescence quantitative PCR (RT-qPCR) experiments26 were applied to test the stabilization of HP-conjugated DNA sequences (Fig. 2b, c and Fig. S4, ESI†). Various environmental treatments including 120 °C heating for 30 min, enzymatic degradation including DNase I and exonuclease, acidic and basic treatments, and ROS production, were employed on nude Code DNA, HP-Code DNA, and HP-Code-HP DNA, respectively. For nude Code sequences without HP protection, strong degradation occurred after 120 °C heating, DNase I and acidic degradations. Weaker but significant degradation could be observed after exonuclease, ROS production and basic treatments. For HP-Code and HP-Code-HP DNAs, stabilizing effects of HP could be found in all environmental treatments, reflecting in the darker bands shown in gel electrophoresis images and higher qPCR yields. In all cases, HP-Code-HP showed better protection effects than HP-Code DNAs. We also demonstrated that the HP protection strategy was capable as a silica encapsulation strategy (Fig. 2c and Fig. S9, ESI†). There were no remarkable DNA losses for HP-Code and HP-Code-HP DNAs encapsulated in silica particles. Next generation sequencing results (Fig. S12 and S13, ESI†) supported that HP conjugation alone and with silica encapsulation would improve DNA stability. It can be concluded that HP could stabilize DNA in solution and solid phases.
As previously described, the tri-branched design of HP provided a handle for small-molecule ligand modification onto DNA duplexes.27 That is, biotin can be easily attached to HP-Code and HP-Code-HP DNAs by facile amine condensation reactions (Fig. S10, ESI†). Thus, biotin–streptavidin affinity interactions28,29 would be easily induced to improve the recovery of encoded DNA in etching-release processes for data storage in silica particles (Fig. 1). It is shown in Fig. S14 (ESI†) that the involvement of HPs is capable for biotinylating and further biotin–streptavidin affinity extraction. And the recovery of encoded DNA could be largely improved, especially for low DNA concentrations less than 10 ng μL−1.
Further characterizations were employed to investigate the capability of HP conjugated DNA in solid phases via inorganic material encapsulation. We chose to follow Grass et al.'s classic silica encapsulation method with glass particles (Fig. 3a). After encapsulation, the silica particles were found to be around 80–200 nm in diameter from transmission electron microscopic (TEM) images and DLS (Fig. S5, ESI†). It was demonstrated from energy-dispersive X-ray spectroscopy (EDX) and scanning-transmission electron microscopy (STEM) high angle annular dark-field (HAADF) element mapping that the DNA strands were successfully encapsulated in silica particles (Fig. 3b and c). This was consistent with the results in ζ-potential, X-ray photoelectron spectra (XPS) and Fourier transform infrared spectrum (FT-IR) (Fig. S6–S8 and Table S1, ESI†). HP conjugated DNA could be undoubtedly encapsulated into silica particles, just like regular DNA strands.
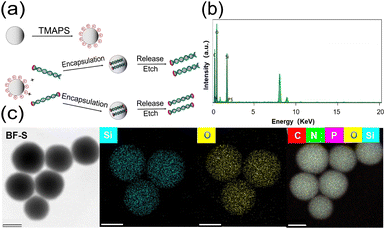 | ||
| Fig. 3 (a) Silica encapsulation and etching-release of encoded DNA. (b) Energy-dispersive X-ray spectra and (c) TEM and HAADF-STEM element mapping images of DNA-silica particles. Scale bar: 100 nm. | ||
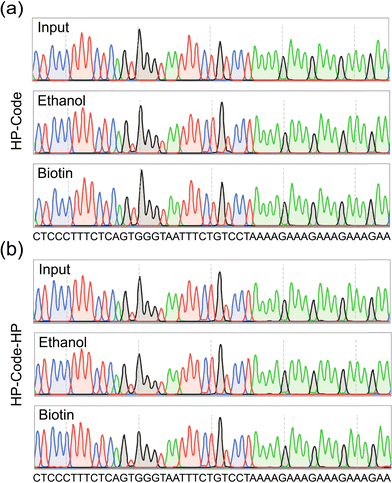
We have verified that HP conjugation can largely improve the stability and recovery for DNA data storage in solution and solid phases. Practical sequencing30 and decoding procedures ought to be tested on HP conjugated DNA codes. HP-Code (Fig. 4a) and HP-Code-HP (Fig. 4b) DNAs in solution, and those released from silica particles using cold ethanol extraction and biotin–streptavidin affinity resin extraction, respectively, were employed for information decoding. It is shown in Fig. 4 that for all cases, including HP-Code and HP-Code-HP, for all conditions like in solution or extracted from etched silica particles using either cold ethanol extraction or biotin–streptavidin affinity extraction, the encoded data could be retrieved and decoded with high accuracy.
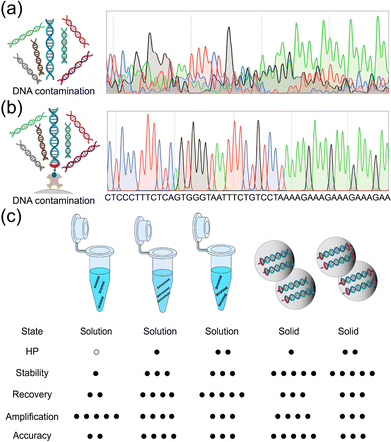
External DNA contamination experiments, where salmon sperm DNA (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was applied as a mimetic of contaminated DNA, were carried out to estimate the resistance of the HP-conjugated DNA storage system. It could be seen that in the presence of external DNA contamination, the retrieving and decoding of the data were largely affected (Fig. 5a). It was impossible to directly retrieve the correct information from the sequencing result. Herein, the biotinylated HP-Code DNA showed powerful resistance to external DNA contamination. Biotin–streptavidin affinity extraction could be used to separate the encoded DNA from untagged contamination DNA strands. And the correct information was successfully retrieved from PCR sequencing at high accuracy (Fig. 5b).
1) was applied as a mimetic of contaminated DNA, were carried out to estimate the resistance of the HP-conjugated DNA storage system. It could be seen that in the presence of external DNA contamination, the retrieving and decoding of the data were largely affected (Fig. 5a). It was impossible to directly retrieve the correct information from the sequencing result. Herein, the biotinylated HP-Code DNA showed powerful resistance to external DNA contamination. Biotin–streptavidin affinity extraction could be used to separate the encoded DNA from untagged contamination DNA strands. And the correct information was successfully retrieved from PCR sequencing at high accuracy (Fig. 5b).
Finally, we summarized the advantages and disadvantages of the four different HP conjugating technologies, those are HP-Code and HP-Code-HP in solution, and HP-Code and HP-Code-HP in the solid state, respectively, and compared those to conventional nude Code DNA storage in solution (Fig. 5c). HP-Code and HP-Code-HP DNAs in solution exhibited improved stabilities, largely improved recoveries, but decreased PCR amplification efficiencies. The ultimate data retrieving accuracies showed improvements as well. The involvement of silica encapsulation greatly improved the stabilities of DNA in the solid state, but the recoveries of DNA and PCR amplification efficiencies declined as well. As to the ultimate data retrieving accuracies, it was demonstrated that silica encapsulation showed positive contributions in data decoding. It was observed that the HP-Code method was better than the HP-Code-HP one. It could be understood as conjugation of HPs at both terminals would hinder DNA unwinding and inhibit PCR amplification.
In summary, HP conjugation design was applied in DNA data storage in solution and solid phases. We verified that HP-conjugated DNA would improve the stability, recovery, resistance to external contamination, and data retrieval accuracy. It provided insights to develop DNA data storage capacitating covalent modification and inorganic encapsulation, expanding the toolbox for practical DNA storage. It will inspire polymer and inorganic chemists to construct future DNA information storage technology with improved performance.
The authors acknowledge support from the National Natural Science Foundation of China (22477017), the National Key R&D Program of China (2022YFC2804800) and the Science & Technology Commission of Shanghai Municipality (No. 23141900900).
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- C. Q. Choi, ASEE Prism, 2020, 29, 22–25 Search PubMed.
- A. Doricchi, C. M. Platnich, A. Gimpel, F. Horn, M. Earle, G. Lanzavecchia, A. L. Cortajarena, L. M. Liz-Marzán, N. Liu, R. Heckel, R. N. Grass, R. Krahne, U. F. Keyser and D. Garoli, ACS Nano, 2022, 16, 17552–17571 CrossRef CAS PubMed.
- L. Ceze, J. Nivala and K. Strauss, Nat. Rev. Genet., 2019, 20, 456–466 CrossRef CAS.
- J. Koch, S. Gantenbein, K. Masania, W. J. Stark, Y. Erlich and R. N. Grass, Nat. Biotechnol., 2020, 38, 39–43 CrossRef CAS.
- C. Geng, S. Liu and X. Jiang, Chem. Sci., 2023, 14, 3973–3981 RSC.
- A. X. Kohll, P. L. Antkowiak, W. D. Chen, B. H. Nguyen, W. J. Stark, L. Ceze, K. Strauss and R. N. Grass, Chem. Commun., 2020, 56, 3613–3616 RSC.
- C. Bancroft, T. Bowler, B. Bloom and C. T. Clelland, Science, 2001, 293, 1763–1765 CrossRef CAS.
- C. Ottoni, B. Bekaert and R. Decorte, Taphonomy of Human Remains: Forensic Analysis of the Dead and the Depositional Environment, 2017, pp. 65–80 Search PubMed.
- L. Orlando, R. Allaby, P. Skoglund, C. Der Sarkissian, P. W. Stockhammer, M. C. Ávila-Arcos, Q. Fu, J. Krause, E. Willerslev, A. C. Stone and C. Warinner, Nat. Rev. Methods Prim., 2021, 1, 14 CrossRef CAS.
- P. L. Antkowiak, J. Koch, P. Rzepka, B. H. Nguyen, K. Strauss, W. J. Stark and R. N. Grass, Chem. Commun., 2022, 58, 3174–3177 RSC.
- R. N. Grass, R. Heckel, M. Puddu, D. Paunescu and W. J. Stark, Angew. Chem., Int. Ed., 2015, 54, 2552–2555 CrossRef CAS PubMed.
- L. M. Wassermann, M. Scheckenbach, A. V. Baptist, V. Glembockyte and A. Heuer-Jungemann, Adv. Mater., 2023, 35, 2212024 CrossRef CAS.
- D. Paunescu, M. Puddu, J. O. B. Soellner, P. R. Stoessel and R. N. Grass, Nat. Protoc., 2013, 8, 2440–2448 CrossRef CAS PubMed.
- M. Puddu, D. Paunescu, W. J. Stark and R. N. Grass, ACS Nano, 2014, 8, 2677–2685 CrossRef CAS PubMed.
- A. Glassing, S. E. Dowd, S. Galandiuk, B. Davis and R. J. Chiodini, Gut Pathog., 2016, 8, 24 CrossRef.
- B. Llamas, G. Valverde, L. Fehren-Schmitz, L. S. Weyrich, A. Cooper and W. Haak, STAR Sci. Technol. Archaeol. Res, 2017, 3, 1–14 Search PubMed.
- B. C. Millar, X. Jiru, J. E. Moore and J. A. P. Earle, J. Microbiol. Methods, 2000, 42, 139–147 CrossRef CAS PubMed.
- R. S. Tanner, Manual of Environmental Microbiology, 2007, pp. 69–78 Search PubMed.
- G. Jun, M. Flickinger, K. N. Hetrick, J. M. Romm, K. F. Doheny, G. R. Abecasis, M. Boehnke and H. M. Kang, Am. J. Hum. Genet., 2012, 91, 839–848 CrossRef CAS PubMed.
- G. Zhao, S. Zhong, G. Zhang, Y. Li and Y. Li, Angew. Chem., Int. Ed., 2022, 61, e202115157 CrossRef CAS.
- L. Jiang, S. Liu, X. Jia, Q. Gong, X. Wen, W. Lu, J. Yang, X. Wu, X. Wang, Y. Suo, Y. Li, M. Uesugi, Z. Qu, M. Tan, X. Lu and L. Zhou, J. Am. Chem. Soc., 2023, 145, 25283–25292 CrossRef CAS.
- A. A. Peterson and D. R. Liu, Nat. Rev. Drug Discovery, 2023, 22, 699–722 CrossRef CAS.
- P. A. Harris, B. W. King, D. Bandyopadhyay, S. B. Berger, N. Campobasso, C. A. Capriotti, J. A. Cox, L. Dare, X. Dong, J. N. Finger, L. C. Grady, S. J. Hoffman, J. U. Jeong, J. Kang, V. Kasparcova, A. S. Lakdawala, R. Lehr, D. E. McNulty, R. Nagilla, M. T. Ouellette, C. S. Pao, A. R. Rendina, M. C. Schaeffer, J. D. Summerfield, B. A. Swift, R. D. Totoritis, P. Ward, A. Zhang, D. Zhang, R. W. Marquis, J. Bertin and P. J. Gough, J. Med. Chem., 2016, 59, 2163–2178 CrossRef CAS PubMed.
- C. C. Arico-Muendel, MedChemComm, 2016, 7, 1898–1909 RSC.
- K. T. Lua, Comput. Stand. Interfaces, 1990, 10, 117–124 CrossRef.
- P. Y. Lee, J. Costumbrado, C. Y. Hsu and Y. H. Kim, J. Vis. Exp., 2012, e3923 Search PubMed.
- Y. Ding, G. J. Franklin, J. L. DeLorey, P. A. Centrella, S. Mataruse, M. A. Clark, S. R. Skinner and S. Belyanskaya, ACS Comb. Sci., 2016, 18, 625–629 CrossRef CAS.
- Z. Qu, Y. Zhang, Z. Dai, Y. Hao, Y. Zhang, J. Shen, F. Wang, Q. Li, C. Fan and X. Liu, Angew. Chem., Int. Ed., 2021, 60, 16693–16699 CrossRef CAS.
- L. J. Smith, R. C. Braylan, J. E. Nutkis, K. B. Edmundson, J. R. Downing and E. K. Wakeland, Anal. Biochem., 1987, 160, 135–138 CrossRef CAS PubMed.
- Z. Chen, W. Zhou, S. Qiao, L. Kang, H. Duan, X. S. Xie and Y. Huang, Nat. Biotechnol., 2017, 35, 1170–1178 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc05109b |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |