Protein corona potentiates the recovery of nanoparticle-induced disrupted tight junctions in endothelial cells†
Muhammad Daniyal
Ghouri
 ab,
Ayesha
Tariq
ab,
Ayesha
Tariq
 bc,
Jabran
Saleem
ab,
Abdul
Muhaymin
ab,
Rong
Cai
bc,
Jabran
Saleem
ab,
Abdul
Muhaymin
ab,
Rong
Cai
 *ab and
Chunying
Chen
*ab and
Chunying
Chen
 *ab
*ab
aNew Cornerstone Science Laboratory, CAS Key Laboratory for Biomedical Effects of Nanomaterials and Nanosafety & CAS Center for Excellence in Nanoscience, National Center for Nanoscience and Technology of China, Chinese Academy of Sciences (CAS), Beijing 100190, China. E-mail: chenchy@nanoctr.cn; cair@nanoctr.cn
bUniversity of Chinese Academy of Sciences, Beijing 100049, China
cCAS Key Laboratory of Nanosystem and Hierarchical Fabrication National Center for Nanoscience and Technology, Beijing 100190, China
First published on 8th November 2024
Abstract
Nanoparticle interactions with biological systems are intricate processes influenced by various factors, among which the formation of protein corona plays a pivotal role. This research investigates a novel aspect of nanoprotein corona–cell interactions, focusing on the impact of the protein corona on the recovery of disrupted tight junctions in endothelial cells. We demonstrate that the protein corona formed on the surface of star-shaped nanoparticles induces the aggregates of ZO-1, which is quite important for the barriers’ integrity. Our research emphasizes that the APOA1 pre-coating on the nanoparticles reduces the ZO-1 expression of endothelial cells offering a promising strategy for overcoming the bio barriers. These findings contribute to our understanding of the interplay between nanoparticles, protein corona, and endothelial cell junctions, offering insights for developing innovative therapeutic approaches targeting the blood–brain barrier integrity. Our study holds promise for the future of nanomedicine, nano drug delivery systems and development of strategies to mitigate potential adverse effects.
New conceptsNanoparticle interactions with biological systems are complex phenomena influenced by various factors, notably the formation of a protein corona. We firstly demonstrate that the shape-dependent protein corona formed on the surface of gold nanostars induces aggregates of cell-junction proteins and find that a key protein, apolipoprotein A1 (APOA1), in corona decreases the expression of ZO-1 in endothelial cells, which is quite important for the bio barriers’ integrity. Therefore, the APOA1 pre-coating on the nanoparticles offers a promising strategy for overcoming the bio barriers. While immunoglobulin (IgG) rich protein corona present on the surface of spherical nanoparticles potentiates the recovery of disrupted tight junctions. Note that several nanoparticles induce an endothelial cell leakage but all findings overlook the effect induced by protein corona which is inevitably present between nanoparticles and endothelial cells. Our work offers additional insight into the intricate interplay between nanoparticles, protein corona, and endothelial cell junction dynamics, advancing our understanding of nanomedicine and protein corona interactions. This study endorses that the biological effects of protein corona are shape dependent. Furthermore, our findings highlight the potential of leveraging protein corona-coated nanoparticles to enhance the efficacy of nanomaterial-based biomedical applications, thereby addressing critical challenges in materials science and advancing the frontiers of biomedical research. |
Introduction
Endothelial cells (ECs) are the most essential and integral part of vascular integrity, and administer the complex regulation of molecular traffic across blood vessel walls. Central to this regulatory function are endothelial cell–cell junctions, comprised of specialized protein complexes that maintain the impermeability of EC barriers. The significance of controlled transversion of endothelial barriers lies in the nascent field of nanomedicine, where the promise of targeted therapeutic and diagnostic interventions hinges on our ability to navigate these cellular fortifications.1–3Nanoparticles (NPs) have become an intrinsic component of various aspects of modern medicine, finding applications in various domains such as drug delivery, medical imaging, and diagnostics.4 The field of nanomedicine typically involves the formulation of NPs with targeting ligands aimed at specific surface receptors, thereby enhancing the recognition and receptor-mediated internalization of the nanomedicine. The objective of these targeting vehicles towards ECs is to facilitate their recognition of EC-specific surface receptors, enabling their internalization and transport across the EC barrier.5–8 However, the process of targeting EC surface receptors, which is imperative for transcellular transport to occur, renders the nanomedicine highly susceptible to EC cellular processing, particularly endo/lysosomal digestion.5,9
In the context of medical applications, gold nanoparticles (Au NPs) are particularly intriguing owing to their biocompatibility, biodegradability, substantial surface-to-volume ratio, and adjustable structures. More specifically, Au NPs have displayed immense potential for utilization as carriers of drugs and biomolecules, particularly in targeted therapies and theragnostic for ECs.10–12 More recently, it has been demonstrated that the shape of nanoparticles can significantly influence their performance in terms of traversing the EC barrier, their circulation time in the blood, cellular internalization, bio-distribution, and residence time within cells. For instance, non-spherical particles have been reported to exhibit longer circulation times, reduced phagocytosis by macrophages, and lower cellular uptake compared to their spherical counterparts.4,13 Furthermore, studies have indicated that NP charges can be fine-tuned to induce or prevent endothelial leakiness through deliberate design.14 Another study has showcased the effectiveness of Au NPs with sizes ranging from 10 to 30 nm in inducing “nanoparticle-induced endothelial leakiness” (NanoEL).15 These investigations collectively hint at the potential of utilizing the Au NP morphology as a powerful tool for engineering the next generation of theragnostic and drug delivery systems, to overcome the EC barrier.
While the synergy between NPs and ECs has garnered substantial attention, the role of the protein corona (PC) in this dynamic remains a tantalizing gap in scientific studies. Once in the biological system, NPs are inevitably exposed to the immune system, which may nullify their efficacy before reaching the intended location. This defense mechanism is a natural response known as the foreign body reaction, a process that commences with the random attachment of proteins from biological fluids onto the surface of the nanoparticles.16,17 To develop efficacious therapies, it is of utmost importance to comprehend the intricate interactions between nanoparticles and proteins, as well as how the composition of the PC subsequently formed is influenced by key nanoparticle parameters. This knowledge is crucial as it ultimately governs the reduction in the dosage of nanomedicine that effectively reaches the disease site. Given that the PC essentially determines the biological fate of nanomaterials, extensive research has been conducted to unravel the factors that shape corona formation, such as particle size, surface charge, surface chemistry, and the composition of the biological media.18–21
Although previous research has explored the direct influence of nanoparticles on tight junctions, the role of the PC in modulating these effects remains relatively unexplored. This study aims to bridge this knowledge gap by investigating how the PC, formed upon exposure to nanoparticles, influences the recovery of tight junctions in endothelial cells following disruption. Our investigation into nanoparticle-induced disruptions in endothelial cell junctions highlights the potential role of PC in mitigating these effects. The presence of protein corona appears to enhance the expression of tight junction proteins like zonula occludens-1 (ZO-1), suggesting a potential therapeutic avenue for leveraging immunoglobulin (IgG), apolipoprotein A1 (APOA1), and protein corona-coated nanoparticles in restoring endothelial barrier integrity.
Materials and methods
DLS and zeta-potential
DLS (dynamic light scattering for hydrodynamic size) and zeta-potential measurements were performed by using Malvern ZS XPLORER software on the data taken from ZetasizerPro Malvern Analytical Technology, UK. All the nanoparticles and complexes were prepared in distilled water. All measurements were taken in triplicate to avoid possible error.Synthesis of gold nanoparticles
Au nanospheres (NSps) were synthesized by reducing chloroauric acid trihydrate (Sigma-Aldrich) solution (HAuCl4·3H2O) in distilled water using sodium tris-citrate (Na3C6H5O7) (Sigma-Aldrich) at 100 °C, as detailed in the previous study of our group,22 in a round bottom flask with continuous stirring using Teflon-coated magnetic bars. Refluxing is not used to prevent temperature gradients in the liquid. Afterward, a calculated preheated citrate concentration is added to achieve 40 nm particles. The liquid is then allowed to cool to room temperature and extracted after a defined time, usually 15 minutes. Whereas, Au nanostars (NSts) were synthesized through the reduction of HAuCl4 in HEPES buffer, employing a surfactant-free method as reported by Yan et al.23 In summary, 100 μL of a 40 mM solution of HAuCl4·3H2O was supplemented with 20 mL of HEPES buffer (140 mM, pH 7.4) and allowed to stand for 1 hour to yield the biocompatible NSts.Cell culture and differentiation
bEnd.3 cells (mouse pheochromocytoma, American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium (DMEM, Wisent) supplemented with 10% FBS and 1% antibiotics (100 μg mL−1 penicillin and 100 μg mL−1 streptomycin; Invitrogen, Carlsbad, CA, USA) at 37 °C under a humidified atmosphere containing 5% CO2.Protein corona formation on the surface of NPs
800 μL of 25% of mouse plasma (MP) (200 μL of mouse plasma and 600 μL of 1 × PBS) was prepared from an aliquot by diluting it in 1 × PBS and incubating it in the water bath until the temperature reached 37 °C. 200 μL of NSps (50 μg mL−1) and NSts (50 μg mL−1) were added dropwise with constant stirring at 37 °C. The solution was kept for 1 hour at 37 °C with continuous stirring. After incubation, the NSps-MP and NSts-MP complexes (NSp@PC and NSt@PC respectively) were centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 rpm for 10 minutes to remove unbound proteins and washed with 1 × PBS three times.
800 rpm for 10 minutes to remove unbound proteins and washed with 1 × PBS three times.
Liquid chromatography-mass spectrometry (LC-MS)
After the formation of the protein corona, the sample was taken in a 1.5 mL centrifuge tube and lysed with DB lysis buffer (8 M urea, 100 mM TEAB, pH 8.5). The lysate was reduced with 10 mM DTT for 1 h at 56 °C and subsequently alkylated with sufficient iodoacetamide for 1 h at room temperature in the dark. Trypsin and 100 mM TEAB buffer were added into each sample and the sample was mixed and digested at 37 °C for 16 h. Formic acid was mixed with the digested sample, pH was adjusted to be below 3, and then the sample was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min at room temperature. The supernatant was slowly loaded to the C18 desalting column, washed with washing buffer (0.1% formic acid and 3% acetonitrile) 3 times, and then elution buffer (0.1% formic acid and 70% acetonitrile) was added. The eluents of each sample were collected and lyophilized.
000g for 5 min at room temperature. The supernatant was slowly loaded to the C18 desalting column, washed with washing buffer (0.1% formic acid and 3% acetonitrile) 3 times, and then elution buffer (0.1% formic acid and 70% acetonitrile) was added. The eluents of each sample were collected and lyophilized.
Mobile phase A (100% water and 0.1% formic acid) and B (80% acetonitrile and 0.1% formic acid) solutions were prepared. The lyophilized powder was dissolved in 10 μL of solution A, centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 20 min at 4 °C, and 1 μg of the supernatant was injected into a home-made C18 Nano-Trap column (4.5 cm × 75 μm, 3 μm). Peptides were separated in a home-made analytical column (15 cm × 150 μm, 1.9 μm), using a 75 min linear gradient elution starting at 5% buffer B followed by a stepwise increase to 26% in 60 min, 90% in 5 min and remained there for 10 min. The separated peptides were analyzed by a Q ExactiveTM series mass spectrometer (Thermo Fisher), with an ion source of Nanospray Flex™ (ESI), a spray voltage of 2.1 kV and an ion transport capillary temperature of 320 °C. A full scan range from m/z 350 to 1500 with a resolution of 60
000g for 20 min at 4 °C, and 1 μg of the supernatant was injected into a home-made C18 Nano-Trap column (4.5 cm × 75 μm, 3 μm). Peptides were separated in a home-made analytical column (15 cm × 150 μm, 1.9 μm), using a 75 min linear gradient elution starting at 5% buffer B followed by a stepwise increase to 26% in 60 min, 90% in 5 min and remained there for 10 min. The separated peptides were analyzed by a Q ExactiveTM series mass spectrometer (Thermo Fisher), with an ion source of Nanospray Flex™ (ESI), a spray voltage of 2.1 kV and an ion transport capillary temperature of 320 °C. A full scan range from m/z 350 to 1500 with a resolution of 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (at m/z 200), an automatic gain control (AGC) target value was 3 × 106 and a maximum ion injection time was 20 ms. The top 40 precursors of the highest abundant in the full scan were selected and fragmented by higher energy collisional dissociation (HCD) and analyzed in MS/MS, where the resolution was 15
000 (at m/z 200), an automatic gain control (AGC) target value was 3 × 106 and a maximum ion injection time was 20 ms. The top 40 precursors of the highest abundant in the full scan were selected and fragmented by higher energy collisional dissociation (HCD) and analyzed in MS/MS, where the resolution was 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (at m/z 200), the automatic gain control (AGC) target value was 1 × 105, the maximum ion injection time was 45 ms, a normalized collision energy was set as 27%, an intensity threshold was 2.2 × 104, and the dynamic exclusion parameter was 20 s. The all resulting spectra were searched against the mouse specific database by the search engine using MaxQuant. The search parameters are set as follows: mass tolerance for the precursor ion was 10 ppm and mass tolerance for the product ion was 20 ppm. Carbamidomethyl was specified as the fixed modification, oxidation of methionine (M) was specified as the dynamic modification, and acetylation was specified as the N-terminal modification. A maximum of 2 missed cleavage sites were allowed. The proteome was analyzed by Scale Biomedicine Technology Co., LTD (Beijing, China).
000 (at m/z 200), the automatic gain control (AGC) target value was 1 × 105, the maximum ion injection time was 45 ms, a normalized collision energy was set as 27%, an intensity threshold was 2.2 × 104, and the dynamic exclusion parameter was 20 s. The all resulting spectra were searched against the mouse specific database by the search engine using MaxQuant. The search parameters are set as follows: mass tolerance for the precursor ion was 10 ppm and mass tolerance for the product ion was 20 ppm. Carbamidomethyl was specified as the fixed modification, oxidation of methionine (M) was specified as the dynamic modification, and acetylation was specified as the N-terminal modification. A maximum of 2 missed cleavage sites were allowed. The proteome was analyzed by Scale Biomedicine Technology Co., LTD (Beijing, China).
Pre-coating ApoA1 on the surface of NPs
APOA1 (MedChemExpress) was reconstituted and diluted in the buffer provided by the manufacturer. 10 μg mL−1 APOA1 was diluted in TE buffer and added in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio to 5 μg mL−1 washed citrate-capped NSps and NSts at a physiological pH of 7.4. The NSp-ApoA1 and NSt-ApoA1 mixtures were incubated under constant shaking for 1 h at 37 °C to allow spontaneous nonspecific adsorption of each protein around the NSps and NSts. Afterward, both complexes were centrifuged at 13
1 volume ratio to 5 μg mL−1 washed citrate-capped NSps and NSts at a physiological pH of 7.4. The NSp-ApoA1 and NSt-ApoA1 mixtures were incubated under constant shaking for 1 h at 37 °C to allow spontaneous nonspecific adsorption of each protein around the NSps and NSts. Afterward, both complexes were centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 rpm for 10 minutes, washed with 1 × PBS three times and resuspended in DMEM for cell exposure.
800 rpm for 10 minutes, washed with 1 × PBS three times and resuspended in DMEM for cell exposure.
Pre-coating IgG on the surface of NPs
IgG with a molecular weight of 155 kDa was reconstituted as per the instructions provided by the manufacturer (Solarbio). The reconstituted IgG was diluted in TE buffer and added in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio to washed citrate-capped NSps and NSts at a physiological pH of 7.4. The NSp-IgG and NSt-IgG, respectively, were incubated under constant shaking for 1 h at 37 °C to allow spontaneous nonspecific adsorption of IgG around the NSps and NSts. Afterward, both complexes were centrifuged at 13
1 volume ratio to washed citrate-capped NSps and NSts at a physiological pH of 7.4. The NSp-IgG and NSt-IgG, respectively, were incubated under constant shaking for 1 h at 37 °C to allow spontaneous nonspecific adsorption of IgG around the NSps and NSts. Afterward, both complexes were centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 rpm for 10 minutes, washed with 1 × PBS three times and resuspended in DMEM for cell exposure.
800 rpm for 10 minutes, washed with 1 × PBS three times and resuspended in DMEM for cell exposure.
Immunofluorescence staining
bEnd.3 cells (Procell) were cultured on coverslips in 24-well plates. After treatment with the nanoparticles for 24 hours, the wells were washed 3 times with 1 × PBS. The cells were fixed with 4% paraformaldehyde (Biosharp) for 15 minutes. The fixed cells were washed 3 times with 1 × PBS each for 5 minutes. The cells were then permeabilized with 0.1% Triton X-100 (Solarbio) in 1 × PBS for 10 minutes. Then the samples were incubated in a blocking buffer (5% BSA in 1 × PBS) for 1 hour to block nonspecific binding sites and afterward, washed with 1 × PBS. The primary antibody was diluted in the blocking buffer according to the manufacturer's (Proteintech) instructions and the cells were exposed to the primary antibody overnight at 4 °C. The wells were washed three times with 1 × PBS for 5 minutes each to remove unbound primary antibodies, incubated with the secondary antibody (Thermo Fisher Scientific) for 1 hour at room temperature, protected from light, and washed three times with 1 × PBS for 5 minutes each to remove unbound secondary antibodies. The cells were then exposed to phalloidin (Thermo Fisher Scientific) to stain actins for 30 minutes at 37 °C. The slides were rinsed with 1 × PBS and stained with DAPI (Solarbio). The images were captured with a fluorescence microscope (Leica, Germany).Western blot analysis
Western blot was done for the quantitative analysis of ZO-1. The cells were inoculated in a 6-well plate at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ×
× ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 105 cells per mL density. When they grew to 80%, they were exposed to NSps, NSts, NSp@PC, and NSt@PC. Then the cells were harvested after incubation for 24 h, washed with ice-cold 1 × PBS 3 times, lysed in the radioimmunoprecipitation assay (RIPA) buffer, and repeatedly blown on the ice for 5
105 cells per mL density. When they grew to 80%, they were exposed to NSps, NSts, NSp@PC, and NSt@PC. Then the cells were harvested after incubation for 24 h, washed with ice-cold 1 × PBS 3 times, lysed in the radioimmunoprecipitation assay (RIPA) buffer, and repeatedly blown on the ice for 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) min. After that, the lysate was collected, cleaved by ultrasonication for 15 seconds, and centrifuged at 4 °C, 12
min. After that, the lysate was collected, cleaved by ultrasonication for 15 seconds, and centrifuged at 4 °C, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 20 min. Then the supernatant was taken and 5 × SDS sample buffer was added, and boiled at 95 °C for 10
000 × g for 20 min. Then the supernatant was taken and 5 × SDS sample buffer was added, and boiled at 95 °C for 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) min. The total protein concentration was detected using the BCA protein assay kit (Thermo Scientific). The same amounts (20–40 μg) of protein were loaded on 8–12% SDS-PAGE gel and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, MA, USA). The membranes were blocked in 10% skim milk and then incubated with the primary antibodies overnight at 4 °C. After that the membranes were exposed to the second antibodies at room temperature for 2 h. The blots were washed with TBST three times and visualized using an enhanced chemiluminescence (ECL) solution (Abcam). Then the protein bands were visualized with an ECL plus detection system (Millipore, USA). To visualize beta-actin the blots were stripped using a striping buffer for 20 minutes and washed three times with TBST, and the membranes were again blocked with 10% skimmed milk. Afterward, the same steps were followed for probing and visualizing the beta-actin bands.
min. The total protein concentration was detected using the BCA protein assay kit (Thermo Scientific). The same amounts (20–40 μg) of protein were loaded on 8–12% SDS-PAGE gel and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, MA, USA). The membranes were blocked in 10% skim milk and then incubated with the primary antibodies overnight at 4 °C. After that the membranes were exposed to the second antibodies at room temperature for 2 h. The blots were washed with TBST three times and visualized using an enhanced chemiluminescence (ECL) solution (Abcam). Then the protein bands were visualized with an ECL plus detection system (Millipore, USA). To visualize beta-actin the blots were stripped using a striping buffer for 20 minutes and washed three times with TBST, and the membranes were again blocked with 10% skimmed milk. Afterward, the same steps were followed for probing and visualizing the beta-actin bands.
Statistical analysis
The data were presented as mean values ± standard deviation (SD). Each experiment was conducted in triplicate to ensure accuracy and reliability. Statistical analyses were carried out using GraphPad Prism 6.0 (GraphPad Software), with significance determined using Student's t-test. A p-value of less than 0.05 was considered statistically significant. Significance levels were denoted as follows: *p < 0.01 and **p < 0.001.Results and discussion
Physiochemical properties of protein corona coated nanoparticles with different shapes
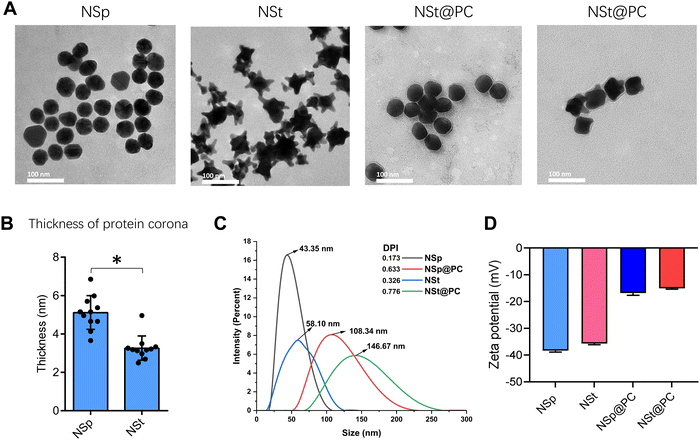
We used two different shapes of Au NPs of comparable sizes, sphere- and star-shaped (NSps and NSts, respectively). The transmission electron microscopy (TEM) images in Fig. 1A show that NSps have a spherical morphology with particle diameters of 40.36 ± 1.15 nm while NSts have a nearly identical size with a diameter of 43.41 ± 2.13 nm. The size difference between the sample groups was also observed through dynamic light scattering (DLS) analysis. The hydrodynamic diameter in ultrapure water was found to be slightly increased from the TEM-derived nanoparticle size. The NSps were found to be in the range of 43.35 ± 1.07 nm while NSts were in the range of 58.10 ± 3.27 nm as shown in Fig. 1C. In addition, we used TEM to visualize the protein corona, the thickness was calculated, as shown in Fig. 1B, as 4.86 ± 1.12 nm for NSps and 3.20 ± 0.66 for NSts. Moreover, the average zeta potentials displayed by NSps and NSts, as shown in Fig. 1D, were determined to be negatively charged with values of −39.02 mV ± 0.28 and −35.57 mV ± 0.34 in ultrapure water, likely due to the presence of a carboxylic group from citrate capping.To understand the effect of PC on the endothelium, we exposed NSps and NSts to MP, and the change in the size and zeta potential was recorded. Both spherical and star-shaped Au-NPs had a significant increase in the hydrodynamic size. The size of NSp@PC was increased to 108.34 ± 5.61 nm while NSt@PC was in the range of 146.67 nm ± 10.29 nm. NSp@PC and NSt@PC also had a similar but significant decrease in the negative charge as observed in zeta potential analysis. The change in the size and zeta potential implies that there is a significant number of proteins attached to both types of NPs. To further elucidate the behavior of the nanoparticles and protein complexes within cellular environments, we measured the zeta potential and hydrodynamic size alterations (Fig. S1A and B, ESI†) for NSp, NSt, NSp@PC and NSt@PC in DMEM (cell culture media without FBS). These results (Table S1, ESI†) demonstrate that the zeta potential and hydrodynamic size of the nanoparticles vary significantly between distilled water and DMEM. The decreased absolute values of the zeta potential in DMEM reflect the influence of the ionic strength and composition of the cell culture media, which can lead to the adsorption of proteins and biomolecules onto the nanoparticle surface, thereby reducing the surface charge. The observed increase in the nanoparticle size in DMEM suggests potential aggregation or the formation of nanoparticle–protein conjugates, which is in line with the behavior of nanoparticles in complex biological environments.24
Shape-dependent protein corona analysis
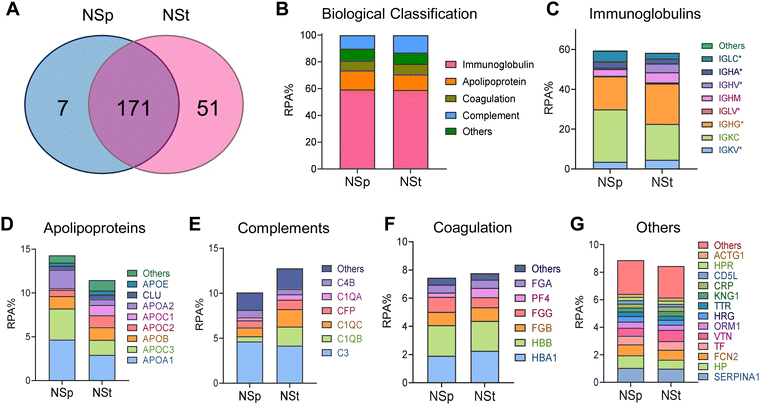
To investigate and distinguish the impact of relatively different protein coronas due to shape on the cell junctions, we incubated the NSps and NSts dropwise in MP for 1 hour with continuous agitation using a rotating shaker. The samples were then centrifuged and washed three times with PBS to remove any unbound proteins. LC-MS was employed to identify and quantitatively determine the individual proteins forming the corona (Fig. 2). We further grouped the constituents of the protein corona in terms of their functions into immunoglobulins, complements, coagulation proteins, apolipoproteins, and others (Fig. 2C–G). The relative protein abundance percentage (RPA%) in NSps and NSts was compared in each group in Fig. 2B. Overall, the types of adsorbed proteins do not appear to be significantly different for both types of nanoparticles, but the amount of protein adsorption varied between the spherical and star-shaped particles. NSts appeared to have 51 unique proteins while NPs had only 7 unique proteins. Immunoglobulins were the most abundant proteins in the corona adsorbed from mouse plasma. Both nanoparticles attracted nearly similar amounts of immunoglobulins i.e., 59% for spheres and 57% for stars (Fig. 2C). On the one hand, spheres attracted a greater number of apolipoproteins (14%) as compared to stars (11%). Furthermore, stars attracted more complement proteins (13%) as compared to spheres (10%).In apolipoproteins, Fig. 2D, APOA1 and APOA2 were the most abundant in spheres (8% or more), which also explains the stability of spheres even after the formation of the protein corona. Due to its amphipathic properties, APOA1 can integrate into the lipid monolayer and provide structural stability. APOA1 also has high affinity towards the NP surface as observed by Ho and colleagues.25 Tsai et al. observed different binding affinities between the four proteins (APOA1, HSA, IgG, and FBG) to NPs.26 Here, APOA1 bound most strongly having the dissociation constant (KD) of 0.12 ± 0.07 μM while IgG with KD = 10.13 ± 3.28 μM bound most weakly to Au-NPs.
In complement proteins (Fig. 2E), C1QA, C1QB, and C1QC were the most abundant on stars (7% or more), it may cause rapid opsonization of star-shaped NPs or may be responsible for complement activation after protein corona formation. All other protein groups composed nearly identical fractions of the protein corona formed on stars and spheres and did not exceed 20% of the total PC.
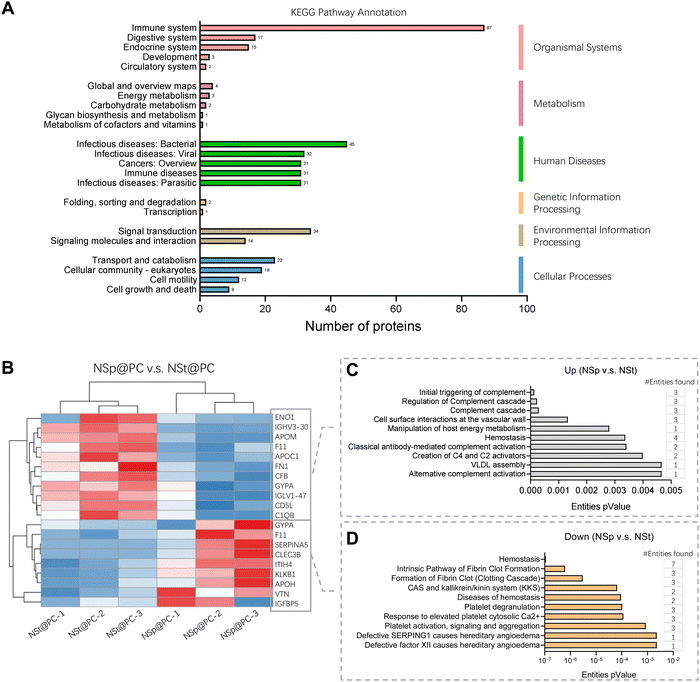
The detailed values for the top 20 most abundant coronal proteins are presented in Tables 1 and 2, respectively. The gene list enrichment analysis of the protein corona revealed an enrichment of immunological KEGG pathways such as immune response and infectious diseases (Fig. 3A), which may be caused by the enrichment of immunoglobulins and complements in the protein coronas. The KEGG pathways were also enriched in the cellular community including focal adhesion, adhering junctions, tight junctions, gap junctions, and signal molecules and interactions including those involving cell adhesion molecules. This indicates that the protein corona likely affects the cell–cell interactions.
| Protein ID | Protein name | Protein name (abbr) | RPA(%) | |
|---|---|---|---|---|
| 1 | P01834 | Ig kappa chain C region | IGKC | 26.32 |
| 2 | P01859 | Ig gamma-2 chain C region | IGHG2 | 12.81 |
| 3 | P0DOY3 | Ig lambda-6 chain C region | IGLC6 | 4.98 |
| 4 | P01024 | Complement C3 | C3 | 4.64 |
| 5 | P02647 | Apolipoprotein A-I | APOA1 | 4.08 |
| 6 | P01871 | Ig mu chain C region | IGHM | 3.50 |
| 7 | P0DOX5 | Ig gamma-1 chain C region | IGHG1 | 3.37 |
| 8 | P01876 | Ig alpha-1 chain C region | IGHA1 | 3.09 |
| 9 | P02656 | Apolipoprotein C-III | APOC3 | 2.92 |
| 10 | P68871 | Hemoglobin subunit beta | HBB | 2.18 |
| 11 | P02652 | Apolipoprotein A-II | APOA2 | 2.12 |
| 12 | P69905 | Hemoglobin subunit alpha | HBA1 | 1.91 |
| 13 | P04114 | Apolipoprotein B-100 | APOB | 1.40 |
| 14 | P02679 | Fibrinogen gamma chain | FGG | 1.25 |
| 15 | P04004 | Vitronectin | VTN | 1.02 |
| 16 | A0A0C4DH72 | Immunoglobulin kappa variable 1–6 | IGKV1-6 | 0.95 |
| 17 | P01009 | Alpha-1-antitrypsin | SERPINA1 | 0.94 |
| 18 | P02675 | Fibrinogen beta chain | FGB | 0.93 |
| 19 | P0C0L5 | Complement C4-B | C4B | 0.82 |
| 20 | P27918 | Properdin | CFP | 0.77 |
| Protein ID | Protein name | Protein name (abbr) | RPA% | |
|---|---|---|---|---|
| 1 | P01834 | Ig kappa chain C region | IGKC | 18.07 |
| 2 | P01859 | Ig gamma-2 chain C region | IGHG2 | 16.39 |
| 3 | P01871 | Ig mu chain C region | IGHM | 5.13 |
| 4 | P01024 | Complement C3 | C3 | 4.18 |
| 5 | P01780 | Ig heavy chain V-III region | IGHV3-21 | 3.44 |
| 6 | P0DOX5 | Ig gamma-1 chain C region | IGHG1 | 3.02 |
| 7 | P02647 | Apolipoprotein A-I | APOA1 | 2.93 |
| 8 | P0DOY3 | Ig lambda-6 chain C region | IGLC6 | 2.61 |
| 9 | P01876 | Ig alpha-1 chain C region | IGHA1 | 2.33 |
| 10 | P69905 | Hemoglobin subunit alpha | HBA1 | 2.26 |
| 11 | P68871 | Hemoglobin subunit beta | HBB | 2.12 |
| 12 | P02746 | Complement C1q subcomponent subunit B | C1QB | 2.10 |
| 13 | P02747 | Complement C1q subcomponent subunit C | C1QC | 1.93 |
| 14 | P02656 | Apolipoprotein C-III | APOC3 | 1.71 |
| 15 | P04114 | Apolipoprotein B-100 | APOB | 1.44 |
| 16 | P02655 | Apolipoprotein C-II | APOC2 | 1.35 |
| 17 | P01599 | Ig kappa chain V-I region | IGKV1-17 | 1.31 |
| 18 | P02654 | Apolipoprotein C-I | APOC1 | 1.18 |
| 19 | P27918 | Properdin | CFP | 1.05 |
| 20 | P01009 | Alpha-1-antitrypsin | SERPINA1 | 1.05 |
Heatmap and hierarchical clustering of the corona proteomic profiles of NSp@PC vs. NSt@PC with a significant difference (Fig. 3B) and the corresponding Reactome pathway annotations (Fig. 3B and C). Compared to NSt@PC, NSp@PC was found to have more protein abundance on ‘cellular surface interactions at the vascular wall’, which includes FN1, GYPA and IGLV, indicating that immunoglobulins may also affect the interaction of the nanoparticles with the vascular wall.
Variations in the expression of TJ proteins in endothelial cells when exposed to nanoparticles for contrasting conditions of protein corona
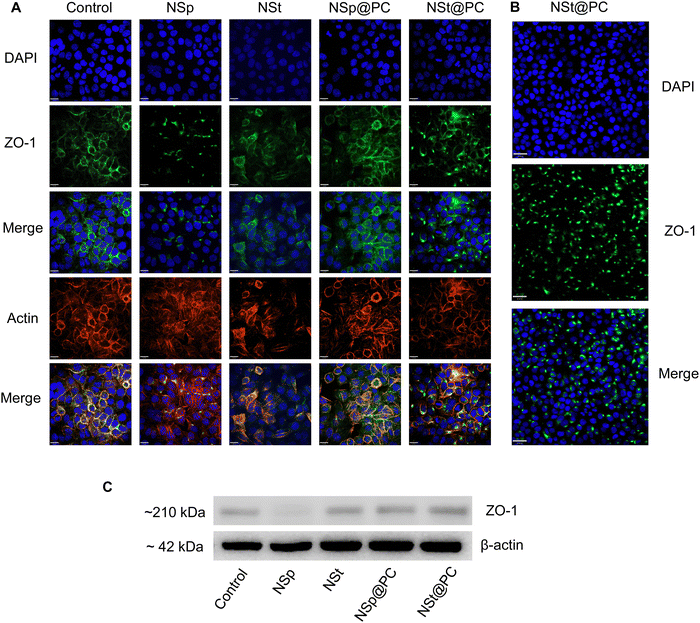
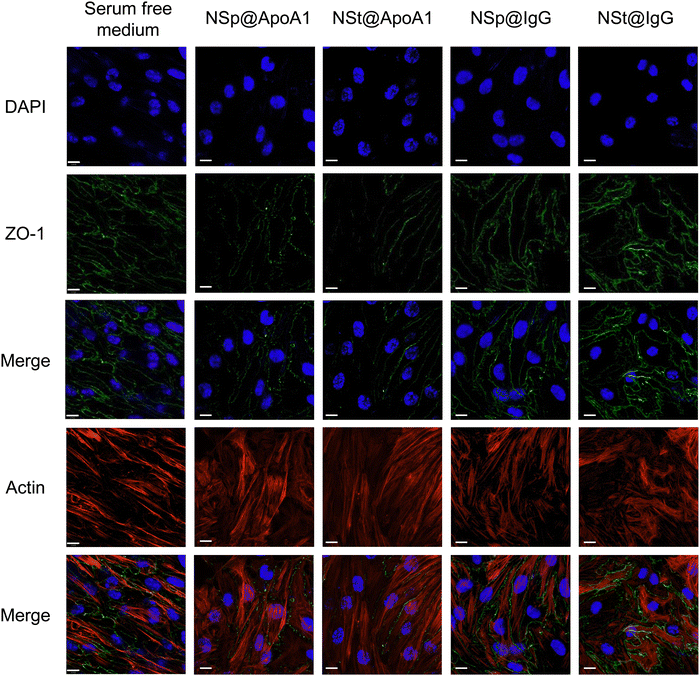
We investigated the influence of protein corona-coated nanoparticles on the expression of tight junction (TJ) proteins in endothelial cells, focusing specifically on ZO-1, a key regulator of cellular junction processes. ZO-1 plays a vital role in promoting cell–cell adhesion by interacting with cytoskeletal scaffold proteins, thereby enhancing the integrity of endothelial cells.27Using our library of nanoparticles (NSps, NSts, NSp@PC, and NSt@PC), we examined the contrast between bare and protein corona-coated nanomaterials on the permeability and integrity of brain endothelial cell monolayers. Following treatment of immortalized murine brain microvascular endothelial cells (bEnd.3), which contribute to the blood–brain barrier in mice, with equivalent concentrations of NSp and NSt for 24 hours, noticeable gaps were observed between endothelial cells. This increased leakiness observed in the bare nanoparticles’ groups was further corroborated by decreased expression of ZO-1 immunofluorescence staining and western blot analysis (Fig. 4A, C and Fig. S2, ESI†). While a relatively higher charge of NSp compared to NSt may have some influence, it is important to note that the marginal disparity in charge (approximately 3 mV) is not likely to be the sole factor. The combined effect of nanoparticle shape and size also play crucial roles in influencing the endothelial cell leakiness. In contrast, treatment with NSp@PC and NSt@PC did not result in any discernible gaps between the cells.
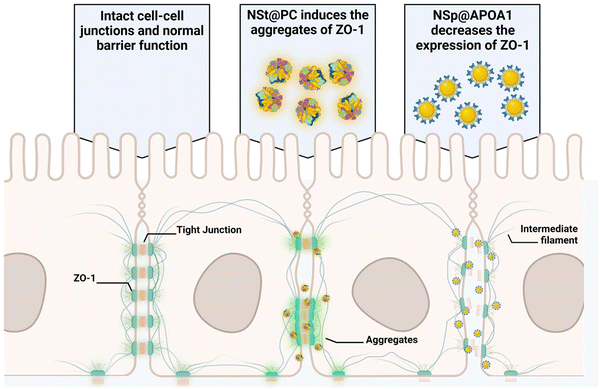
Pre-coating NSp and NSt nanoparticles with MP resulted in significant alterations in the surface dynamics of the nanoparticles. Specifically, the formation of a protein corona around the nanoparticles changed their surface charge, hydrodynamic size, and overall surface chemistry, which in turn influenced their interaction with cellular membranes and proteins. The protein corona modifies how nanoparticles interact with cells, potentially reducing cytotoxicity and improving biocompatibility, leading to distinct changes in their behavior. Interestingly, the presence of protein corona resulted in an upregulation of ZO-1 expression compared to the NSp group, particularly evident in NSp@PC, suggesting that the protein corona promotes the expression of tight junction proteins. This theorizes that PC formed on spherical nanoparticles has the potential to recover the disrupted cell–cell junctions, whereas NSt@PC nanoparticles induced the aggregates of ZO-1, as shown in Fig. 4B. The underlying mechanism causing ZO-1 aggregation in the presence of NSt@PC is not yet fully understood, and further investigation is needed to elucidate this phenomenon. We plan to explore this in future studies to gain a deeper understanding of the interactions at play.
These results highlight the shape-dependent biological effects of the protein corona. Moreover, it is evident that the presence of charge polarization across the cell membrane surface, possibly originating from the discrete distribution of various proteins from the luminal surface to the cell junction, may play a role in modulating cellular behavior and function in response to protein corona coated nanoparticles.28
APOA1 pre-coating nanoparticles inhibit the expression of ZO-1
The protein corona formed on NSp and NSt surfaces primarily consists of IgG, as evidenced by the predominant presence of IgG in the corona, as shown in Fig. 2B. We hypnotized that this profusion of IgG is likely to be the primary contributor to the effects observed during subsequent interactions of these pre-coated nanoparticles with cell–cell junctions. Interestingly, our LC-MS analysis also revealed the presence of several apolipoproteins in the corona, with APOA1 being the most abundant as depicted in Fig. 2D. Apolipoproteins take part in lipid transport and metabolic functions within the central nervous system (CNS) and across the blood–brain barrier (BBB).29,30 APOA1 plays a significant role in the delivery of doxorubicin loaded in surfactant-coated poly(butyl-cyanoacrylate) nanoparticles to the brain.31 Additionally, it is involved in the interaction of lipid–drug conjugate (LDC) nanoparticles with brain vessel endothelial cells,32 potentially enhancing scavenger receptor class B type I (SR-BI) dependent transcytosis.33 Thus, APOA1 pre-coating is supposed to be a useful strategy to facilitate nanoparticle transport across the BBB and further transport to deeper regions of the brain. This conclusion is consistent with our results, though, the mechanism is different.Recognizing the importance of these proteins, we went on to investigate their individual effects on ECs, as shown in Fig. 5. APOA1 downregulated the expression of ZO-1 compared to the control. The expression of ZO-1 was least observed in NS@ApoA1 and NP@ApoA1. The specific effects of APOA1 on ZO-1 expression could be influenced by several factors including the anti-inflammatory properties of APOA1. Additionally, cellular interactions may play a role, considering APOA1's involvement in lipid metabolism. The presence of APOA1 on nanoparticles might influence the cellular lipid composition or metabolism, indirectly affecting ZO-1 expression and hence the BBB integrity. Our investigation of APOA1 provides pivotal evidence that APOA1 can open up the path for drugs to transverse the BBB. Further mechanistic insights into APOA1's anti-inflammatory effects are essential, especially concerning endothelial cell junction disruption. On the other hand, IgG-coated nanoparticles resulted in comparatively increased expression of ZO-1. IgG, a component of the immune system, plays an important role in modulating cellular responses within the BBB endothelium.34 The upregulated expression of ZO-1 by IgG pre-coated nanomaterials is consistent with the protein corona-coated nanostars and spherical particles. The role of IgG in mediating the targeting and cellular internalization of nanoparticles for drug delivery applications has already been reported, this study specifically explores its effect on the endothelium junctions.35 These findings also support our hypothesis that protein corona formed on the surface of spherical nanoparticles aids the recovery of compromised cell junctions (Fig. 6).
Conclusion
Our study highlights the profound impact of nanoparticle surface chemistry on protein adsorption, EC leakiness, and barrier function. We observed that the physiological and chemical properties of nanoparticles influence the adsorption of different proteins, with the resultant protein corona layers modulating cytotoxicity responses and EC barrier interactions. Importantly, our investigation into nanoparticle-induced disruptions in endothelial cell junctions sheds light on the intriguing potential of APOA1.While anti-inflammatory properties of APOA1 may theoretically contribute to maintaining the BBB integrity by reducing inflammation-associated disruptions, its involvement in lipid metabolism and transport could also play a significant role. In particular, APOA1's role in lipid transport and regulation could impact the BBB integrity by influencing the cellular lipid composition and membrane properties. Changes in lipid metabolism may compromise the structural integrity of endothelial cells that form the BBB, thereby affecting tight junctions and permeability.
Moreover, our study underscores the role of the protein corona in potentiating the recovery of disrupted cell junctions. The presence of protein corona, particularly enriched with IgG, appears to enhance ZO-1 expression and promote tighter endothelial cell junctions. This finding opens new avenues for leveraging IgG, APOA1, and protein corona-coated nanoparticles in therapeutic strategies to restore the integrity of endothelial barriers compromised by nanoparticle interactions.
In summary, our study offers valuable insights into the complex interplay between nanoparticles, protein corona, and endothelial cell junctions, advancing our understanding of nanomedicine and protein corona interactions. These findings pave the way for developing innovative therapeutic approaches targeting the BBB integrity and hold promise for the future of nanomedicine.
Author contributions
C. Y. Chen and R. Cai conceived and designed the study. M. D. Ghouri conducted the experiments and analyzed the data. A. Tariq, J. Saleen, and A. Muhaymin contributed to the manuscript writing and provided critical revisions and intellectual input. All authors reviewed and approved the final manuscript.Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request. The raw data supporting the findings of this study, including the western blot assay results, and zeta potential and size measurements are made available to researchers who wish to examine the ESI† in more detail.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Key R&D Program of China (2021YE0112600 and 2021YEA1200900), the National Natural Science Foundation of China (32071402, 22388101, 22027810), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB36000000) and the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (CIFMS2019-I2M-5-018). It was also supported by the Beijing Nova Program (20220484006).Notes and references
- Y. Cong, D. Baimanov, Y. Zhou, C. Chen and L. Wang, Adv. Drug Delivery Rev., 2022, 191, 114615 CrossRef CAS PubMed.
- X. Wang, X. Cui, J. Wu, L. Bao, Z. Tan and C. Chen, Sci. Adv., 2023, 9, 2252 CrossRef.
- Q. Wang, Q. Liang, J. Dou, H. Zhou, C. Zeng, H. Pan, Y. Shen, Q. Li, Y. Liu and D. T. Leong, Nat. Nanotechnol., 2023, 1–11 Search PubMed.
- M. D. Ghouri, J. Saleem, J. Ren, J. Liu, A. Umer, R. Cai and C. Chen, Adv. Mater. Interfaces, 2022, 9, 2101391 CrossRef CAS.
- Q. Wang, H. Cheng, H. Peng, H. Zhou, P. Y. Li and R. Langer, Adv. Drug Delivery Rev., 2015, 91, 125–140 CrossRef CAS PubMed.
- D. Iannotta, A. W. Kijas, A. E. Rowan and J. Wolfram, Nat. Nanotechnol., 2024, 19, 13–20 CrossRef CAS.
- M. I. Teixeira, C. M. Lopes, M. H. Amaral and P. C. Costa, Colloids Surf., B, 2023, 221, 112999 CrossRef CAS PubMed.
- M. Hu, X. Li, Z. You, R. Cai and C. Chen, Adv. Mater., 2023, 35, 2303266 Search PubMed.
- S. Han, R. da Costa Marques, J. Simon, A. Kaltbeitzel, K. Koynov, K. Landfester, V. Mailänder and I. Lieberwirth, Nat. Commun., 2023, 14, 295 CrossRef CAS.
- Y. H. Song, R. De and K. T. Lee, Adv. Colloid Interface Sci., 2023, 320, 103008 CrossRef CAS.
- T. O. Lilius, K. N. Mortensen, C. Deville, T. J. Lohela, F. F. Stæger, B. Sigurdsson, E. M. Fiordaliso, M. Rosenholm, C. Kamphuis, F. J. Beekman, A. I. Jensen and M. Nedergaard, J. Controlled Release, 2023, 355, 135–148 CrossRef CAS.
- F. U. Rehman, F. Iftikhar, C. Zhao and Z. Sajid, Gold nanoparticles for treatment of cerebral diseases, Gold Nanoparticles for Drug Delivery, Academic Press, 2024, pp. 251–276 Search PubMed.
- W. Zhang, D. Zhu, Z. Tong, B. Peng, X. Cheng, L. Esser and N. H. Voelcker, Pharmaceutics, 2023, 15, 2271 CrossRef CAS PubMed.
- W. J. F. Vanbilloen, J. S. Rechberger, J. B. Anderson, L. F. Nonnenbroich, L. Zhang and D. J. Daniels, Pharmaceutics, 2023, 15, 1804 CrossRef CAS PubMed.
- M. I. Setyawati, C. Y. Tay, B. H. Bay and D. T. Leong, ACS Nano, 2017, 11, 5020–5030 CrossRef CAS PubMed.
- R. Cai and C. Chen, Adv. Mater., 2019, 31, 1805740 CrossRef CAS PubMed.
- R. Cai, J. Ren, Y. Ji, Y. Wang, Y. Liu, Z. Chen, Z. Farhadi Sabet, X. Wu, I. Lynch and C. Chen, ACS Appl. Mater. Interfaces, 2020, 12, 1997–2008 CrossRef CAS PubMed.
- D. Baimanov, R. Cai and C. Chen, Bioconjugate Chem., 2019, 30, 1923–1937 CrossRef CAS PubMed.
- J. Ren, N. Andrikopoulos, K. Velonia, H. Tang, R. Cai, F. Ding, P. C. Ke and C. Chen, J. Am. Chem. Soc., 2022, 144, 9184–9205 CrossRef CAS.
- J. Ren, R. Cai, J. Wang, M. Daniyal, D. Baimanov, Y. Liu, D. Yin, Y. Liu, Q. Miao, Y. Zhao and C. Chen, Nano Lett., 2019, 19, 4692–4701 CrossRef CAS PubMed.
- A. Umer, M. D. Ghouri, T. Muyizere, R. M. Aqib, A. Muhaymin, R. Cai and C. Chen, Precis. Chem., 2023, 1, 341–356 CrossRef CAS PubMed.
- X. Wang, M. Wang, R. Lei, S. F. Zhu, Y. Zhao and C. Chen, ACS Nano, 2017, 11, 4606–4616 CrossRef CAS PubMed.
- R. Yan, J. Chen, J. Wang, J. Rao, X. Du, Y. Liu, L. Zhang, L. Qiu, B. Liu, Y.-D. Zhao, P. Jiang, C. Chen and Y.-Q. Li, Small, 2018, 14, 1802745 CrossRef PubMed.
- B. Meesaragandla, Y. Komaragiri, R. Schlüter, O. Otto and M. Delcea, Sci. Rep., 2022, 12, 16643 CrossRef PubMed.
- Y. T. Ho, N. A. Azman, F. W. Y. Loh, G. K. T. Ong, G. Engudar, S. A. Kriz and J. C. Y. Kah, Bioconjugate Chem., 2018, 29, 3923–3934 CrossRef CAS PubMed.
- D.-H. Tsai, F. W. DelRio, A. M. Keene, K. M. Tyner, R. I. MacCuspie, T. J. Cho, M. R. Zachariah and V. A. Hackley, Langmuir, 2011, 27, 2464–2477 CrossRef CAS PubMed.
- A. Traweger, S. Toepfer, R. N. Wagner, J. Zweimueller-Mayer, R. Gehwolf, C. Lehner, H. Tempfer, I. Krizbai, I. Wilhelm, H.-C. Bauer and H. Bauer, Tissue Barriers, 2013, 1, e25039 CrossRef PubMed.
- M. Mahmoudi, I. Lynch, M. R. Ejtehadi, M. P. Monopoli, F. B. Bombelli and S. Laurent, Chem. Rev., 2011, 111, 5610–5637 CrossRef CAS PubMed.
- A. C. Raulin, Y. A. Martens and G. Bu, Annu. Rev. Biochem., 2022, 91, 731–759 CrossRef CAS PubMed.
- E. M. Rhea and W. A. Banks, Pharm. Res., 2021, 38, 1469–1475 CrossRef CAS PubMed.
- B. Petri, A. Bootz, A. Khalansky, T. Hekmatara, R. Müller, R. Uhl, J. Kreuter and S. Gelperina, J. Controlled Release, 2007, 117, 51–58 CrossRef CAS PubMed.
- A. Gessner, C. Olbrich, W. Schröder, O. Kayser and R. Müller, Int. J. Pharm., 2001, 214, 87–91 CrossRef CAS PubMed.
- S. M. Armstrong, M. G. Sugiyama, K. Y. Fung, Y. Gao, C. Wang, A. S. Levy, P. Azizi, M. Roufaiel, S.-N. Zhu and D. Neculai, Cardiovasc. Res., 2015, 108, 268–277 CrossRef CAS.
- J. K. Ryu and J. G. McLarnon, J. Cell. Mol. Med., 2009, 13, 2911–2925 CrossRef CAS PubMed.
- A. Syed, A. Baker, M. Mohany, A. M. Elgorban, M. S. Khan and S. S. Al-Rejaie, Artif. Cells, Nanomed., Biotechnol., 2023, 51, 384–396 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nh00178h |
| This journal is © The Royal Society of Chemistry 2025 |