Recent advances in SERS assays for detection of multiple extracellular vesicles biomarkers for cancer diagnosis
Chloe
Duffield
 ,
Laura M.
Rey Gomez
,
Laura M.
Rey Gomez
 ,
Simon Chang-Hao
Tsao
and
Yuling
Wang
,
Simon Chang-Hao
Tsao
and
Yuling
Wang
 *
*
School of Natural Sciences, Faculty of science and engineering, Macquarie University, Sydney, NSW 2109, Australia. E-mail: yuling.wang@mq.edu.au
First published on 7th December 2024
Abstract
As the prevalence of cancer is escalating, there is an increased demand for early and sensitive diagnostic tools. A major challenge in early detection is the lack of specific biomarkers, and a readily accessible, sensitive and rapid detection method. To meet these challenges, cancer-derived small extracellular vesicles (sEVs) have been discovered as a new promising cancer biomarker due to the high abundance of sEVs in body fluids and their extensive cargo of biomarkers. Additionally, surface-enhanced Raman scattering (SERS) presents a sensitive, multiplexed, and rapid method that has gained attraction with recent studies showing promising results from patient samples for the multiplex detection of cancer sEVs. Various label-based SERS multiplex assays have been developed in the field of SERS including bead assays, lateral flow immunoassays, microfluidic devices, and artificial intelligence (AI)-based label-free SERS chips, targeting multiple surface proteins to ensure comprehensive multiplex diagnostics. These assays hold promise for enabling early detection, quantification, and subtyping of cancer-derived sEVs for cancer diagnostic applications. This review aims to provide a summary of the recent advances in the field of SERS multiplex assays for detection, quantification, and subtyping of sEVs to facilitate cancer diagnosis. This review further provides unique insights into the use of sEVs as a biomarker and aims to address the issues surrounding their translation from laboratories to clinics.
1. Introduction
The demand for better cancer diagnosis and disease progression monitoring is ever increasing with 19–20 million people being diagnosed each year and 10 million deaths annually.1 With the ever-increasing demand for early diagnostics, small extracellular vesicles (sEVs) have emerged as promising biomarker targets because of their intrinsic role in tumor development, progression,2–7 and intercellular communication within tumor microenvironments.3,4,8–10 Moreover, sEVs which are readily available and abundant in most bodily fluids, carry target biomolecules such as surface proteins, nucleic acids, and carbohydrates.2,6,11,12 Through the molecular subtyping of sEVs, further information can be deduced about proliferation, progression, and tumorigenesis at very early stages of cancer.2–4 However, their heterogenous nature poses a challenge as each sEV population has different surface protein expression profiles. Therefore, profiling of multiple surface biomarkers is often required for accurate capture of sEV populations, thus moving the field of research to multiplex assay types.2–4 Multiplex assays have become increasingly important as the detection of multiple cancer-specific biomarkers ensures a high degree of accuracy for diagnostics. This issue is highly prevalent due to the heterogeneity in sEV populations as expression levels vary widely patient-to-patient. With the lack of a single cancer-specific biomarker to accurately diagnose a type of cancer, the use of multiple biomarkers has proven useful and allows for the consideration of a wide variety of patient cohorts.Raman spectroscopy is an optical technique that provides insights into the vibrational and rotational properties of molecules.13–15 However, since the Raman signals are inherently weak, enhancements are often required for practical applications.13–15 Surface-enhanced Raman scattering (SERS) is a powerful spectroscopic method that significantly amplifies Raman signals by utilizing molecules adsorbed on rough metal surfaces or nanostructures. SERS enhancement primarily occurs through two mechanisms: electromagnetic and chemical. Electromagnetic enhancement results from the resonance with surface plasmons, particularly when target molecules are adsorbed on nanoscale metallic surfaces such as gold (Au) or silver (Ag).16–19 Specific nanostructures, like gold nanostars (AuNS), can create “plasmonic hot spots” – regions of intense electromagnetic field enhancement due to localized surface plasmon resonance.17,20 Chemical enhancement involves charge transfer between the metal surface and attached molecules, often facilitated by Raman reporter molecules, and can increase signal intensity by a factor of 102 to 104.21–24
SERS-based liquid biopsy assays have gained popularity in precision oncology due to several advantages: rapid analysis, non-destructive nature, minimal invasiveness, real-time capabilities, high sensitivity (from both electromagnetic and chemical enhancements), and versatility (applicable to both label-based and label-free detection methods).12,25,26 By leveraging these signal enhancement capabilities, SERS liquid biopsy assays offer a promising approach for highly sensitive, real-time analysis of biomarkers in bodily fluids, with potential applications in early cancer detection and treatment monitoring.
SERS has progressed into a rapidly developing field of research that allows for ultrasensitive biosensing, down to single molecule detection and imaging in a rapid and non-invasive manner.9,16,28–30 While SERS has many benefits, such as multiplexing capabilities and high sensitivity, it also does not have the typical issues often associated with fluorescence, such as photobleaching and overlapping emission spectra between fluorophores.9,31,32 These SERS assays allow for real-time analysis for precision oncology and are highly sensitive due to increased signal generated by electromagnetic and chemical enhancements.19,31,33
Label-free detection typically involves the direct measurement of target analyte with SERS enhancement from a designed platform.31,34 The complexity of patient samples and the need for multiplex detection often lead to problems with specificity due to complex SERS spectra, and a lack of sensitivity when dealing with low concentrations. On the other hand, with label-based techniques, the use of SERS nanotag technology for labelling sEVs has become increasingly attractive to induce high signals that can be quantified.2,16,27,31,35 With slight variations in SERS nanotag synthesis such as structure, Raman reporter molecule, size and bioconjugation technique, issues arise with stability, reproducibility and specificity.22,36–38 Thus, there is no consensus on whether label-based or label-free methods are more suitable for SERS biosensing applications.
This review article aims to provide a comprehensive summary of recent developments and on-going research in the field of multiplex label-based and label-free SERS assays for the detection of cancer-derived sEVs. While SERS bioassay is a hot topic, this review focuses on the unique perspectives of using sEVs as biomarkers and aims to collate recent advances in the field of SERS multiplex assays to address the issues surrounding their translation from laboratories to clinics. We begin with an overview of sEVs as cancer biomarkers, including information on the isolation methods for assay applications. It covers the design of plasmonic nanostructures for label-free and SERS nanotag technology, including a summary on the appropriate selection of nanotags for multiplex assay designs. This review also highlights the recent promising innovations in SERS technologies for the multiplex detection of sEVs, with emphasis on their enhanced sensitivity, specificity, and real-time disease monitoring capabilities.
2. sEV as cancer biomarkers
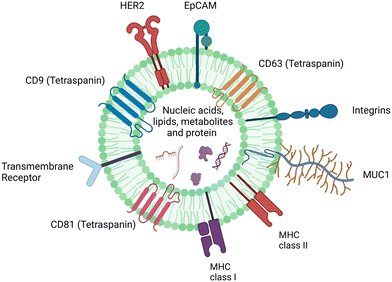
sEVs are denoted by the International Society for Extracellular Vesicles (ISEV) as being extracellular vesicles of less than 200 nm in size. They are released by the cells into extracellular space to then be circulated throughout the body and are detectable in all somatic fluids such as blood, saliva, urine, and cerebrospinal fluid.2,5,11,25,34,39–44 sEVs present as promising biomarkers for cancer diagnostics as they are readily available in bodily fluids, have multiple surface markers for capture and subtyping, and can share characteristics of their parent tumor cells.8,10,11,45,46 In this section, a brief introduction about sEVs as biomarkers in the context of cancer will be discussed to provide insight into considerations for SERS assay designs using cancer-derived sEVs.sEVs have a variety of markers which can be used for subtyping and detection in SERS multiplex assays.7,11,40,41,47Fig. 1 highlights the structure of a single sEV, and the potential membrane components and internal cargo as adapted during their biogenesis.
The biomarkers of sEVs vary greatly and mimic that of their parent cells. Examples include cancer specific proteins (e.g. HER2) to differentiate sEVs from normal and cancer cells, tetraspanins (CD9, CD81, and CD63) which are common protein biomarkers among all sEVs, and miRNA (e.g. miR-21) derived from sEV cargo.9,12,18,48–51 While there are different assays for each biomarker, the most common one is the use of surface proteins as they can be easily captured with antibodies or aptamers.52–54 Single biomarkers have proven ineffective in accurately discriminating between cancer types, as many cancers share similar biomarkers.4,9,22,55 An example of this includes EpCAM, which has been found in multiple cancer types and is also expressed in low amounts in healthy patients.49,50 Further studies by medical professionals are required to establish relevant clinical thresholds that determine the amount of biomarker required to specify a positive result. Therefore, due to the high heterogeneity of sEV populations, multiple biomarkers are often required to both capture sEVs and detect the presence of cancer, as each patient's sEV samples can have varying expression levels.7,45,56–58
3. Label-free SERS detection and subtyping
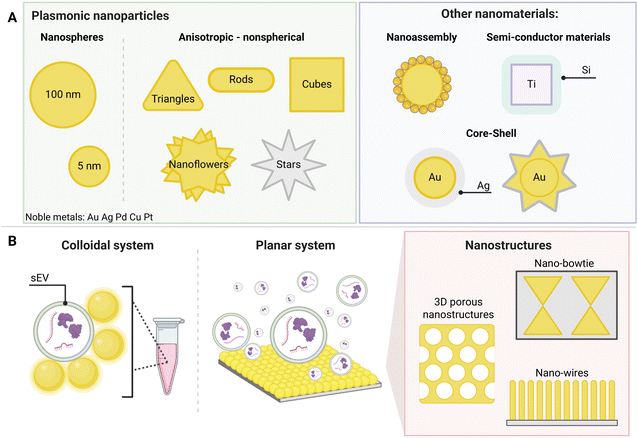
A label-free assay utilizes the unique Raman ‘fingerprint’ from the target biomarker directly, which is enhanced by the SERS substrate as a consequence of electromagnetic enhancements. Through the use of strong optical properties from SERS substrates such as plasmonic nanoparticles and nanostars (Fig. 2), label-free SERS assays for sEVs detection have been developed for ease of use, higher specificity, broader multiplex capabilities, and rapid diagnostics.3,29,30,33,48,59Combined with artificial intelligence (AI), new techniques have emerged as promising approaches for multiplex sEV detection, particularly for cancer diagnosis and subtyping.34,60–62 This section will explore recent multiplex label-free SERS assays for the detection of cancer-derived sEVs with the aim of addressing enhanced sensitivity and specificity capabilities, real-time disease monitoring, their suitability for use in clinical settings and the problems currently faced.
3.1 Sample preparation techniques for label-free assays
Sample preparation for label-free assays is vital as highly clean spectra are needed for AI model training and sample identification, making it very important for assay design.2,7,11,25 Due to the high heterogenicity of sEVs, the isolation method chosen can impact the results with chemical and other contaminants interfering with the SERS spectra.Current isolation methods of sEVs include ultracentrifugation, acoustofluidics, solubility precipitation, size exclusion chromatography (SEC), immunoaffinity-based techniques using antibodies or aptamers, lipid self-assembly nanoprobes, charge dielectrophoresis, isolation kits and ion exchange chromatography.58,63–65 Among those, ultracentrifugation is considered the gold standard, however, it often results in low purity and prolonged processing times.7,65,66 SEC has become the preferred method as higher purities are able to be obtained.11,56,64 Therefore, selecting the right isolation method is essential for ensuring high sample purity and the reliability of label-free assays, ultimately allowing for accurate AI model training and improved sEV detection.
3.2 SERS substrate
Various types of SERS substrates have been developed for biosensing applications to improve detection sensitivity through strong electromagnetic enhancements.48,60,67–69 In label-free assays, SERS substrates can be used in two forms: as nanomaterials arranged in a planar configuration, known as substrate-based or drop-cast assays or where nanoparticles are in colloidal suspensions, referred to as liquid-based assays.SERS substrates have been developed in a variety of designs for detection of biomarkers including the use of gold nanopillars, gold nanoparticles on a glass slide or even hybrid graphene–gold nanopyramids.70–72 Traditionally, SERS substrates for sEV detection are composed of metals like silver (Ag) and gold (Au), as they exhibit favourable plasmonic properties, consequently increasing the Raman signal through surface plasmon resonance.16,30,35 Signal enhancements are required to overcome potential background signals from non-specific molecules on the assay surface. These have been addressed by utilizing layered materials and designing novel nanostructures that generate significantly increased enhancement, thereby mitigating background signal interference.33,34,73 Label-free assays have become specifically targeted for sEVs as the only changes required are the data processing techniques. Generally, the ideal SERS substrate for sEV detection generates the high electromagnetic enhancement, is reproducible, has low background interferences, and demonstrates a high degree of stability.17,28,48,60,68,69,74
3.3 Label-free assays
With the need of high signal enhancement for multiplexing capabilities, unique substrate designs have been implemented for SERS label-free assays. A recent study by Dong et al. (2020) demonstrates an innovative design with the use of different substrates for SERS enhancements, including an Au-coated TiO2 macroporous inverse opal (MIO) structure.48 This assay utilized a ‘beehive’ type structure to induce a ‘slow light effect’ that significantly improved the SERS performance, with an enhancement factor (EF) of 1.25 × 103 (Fig. 3A). It was able to detect cancer owing to the strong Raman signal intensity at 1087 cm−1 arising from the P–O bond within the phosphoproteins of cancer-derived sEVs, irrespective of cancer type.48 A study with 30 prostate cancer patients confirmed a two-fold increase of intensity in 93% of patient samples compared to healthy patients.48 With low-cost and timesaving attributes compared to the gold standard enzyme-linked immunosorbent assay (ELISA) and western blot strategies, the macroporous ‘beehive’ design presents a potentially effective method for rapidly pre-screening clinical applications.5,39,48,75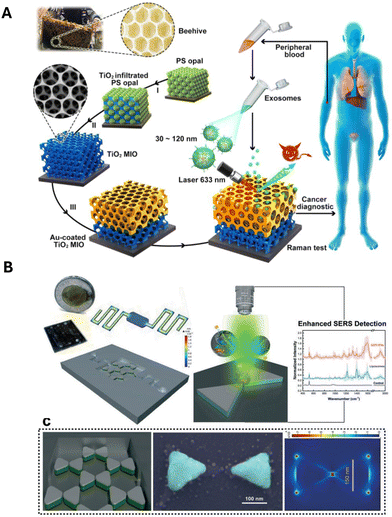 | ||
| Fig. 3 Label-free assays with unique nanostructures as SERS substrate. (A) Beehive structure to enhance sEVs Raman fingerprint. Adapted from ref. 48 with permission from ACS Applied Materials and Interfaces, copyright [2021]. (B) Nano-bowtie structures used as SERS substrate for sEVs detection. Adapted from ref. 3 with permission from Lab on a Chip, copyright [2020]. | ||
Another example of an innovative label-free SERS platform was developed by Jalali et al. (2021), which utilized a microfluidic chip design with nano-bowtie nanoparticles (Fig. 3B).3 These allows for an EF of 9 × 105 due to the plasmonic ‘hot spot’ located in the middle of the bowtie structure. Thus, showing a higher EF than that of the beehive structure. As the sEVs flow through, they are captured in the middle of the nano-bowtie nanoparticles and their intensity is enhanced substantially, allowing for the subsequent multiplexing identification of sEV populations using AI techniques.
3.4 Data analysis techniques for label-free assays
Data analysis is the key element required for label-free assay data interpretation and AI has presented an ideal solution to deconvolute and interpret SERS spectra obtained from sEVs.15,60,76,77 These methods utilize statistical models such as principal component analysis (PCA), least squares discriminant analysis (LDA), discriminant function analysis (DFA), multivariate curve resolution (MCR), graphical models, k-nearest neighbours, random forests and partial least-squares discriminant analysis (PLS-DA), whereby individual samples’ populations of sEV ‘signatures’ can be learned through the identification of specific features, and predictions made for subsequent unknown samples.60,61,76–79 This is summarized in Fig. 4, where AI is often used as the broad-spectrum term that encapsulates machine and deep learning models. Recent advances in this field have taken typical assay designs and utilized AI-based machine learning and neural network models to determine the presence and/or classify a specific type of cancer, in an effort to reduce the complexity of data output for end-users.15,34,61,76,78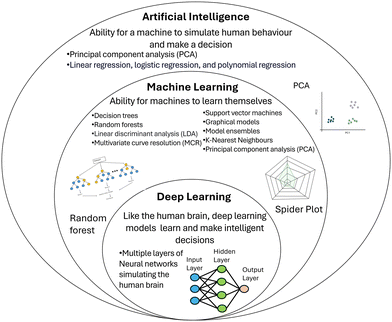 | ||
| Fig. 4 Summary of statistical models used in each artificial intelligence subsect including machine learning and deep learning. These models are often more complex and require extensive datasets. | ||
In a typical machine learning model, AI is trained using common data analysis techniques such as PCA and LDA to build models based on a training set of data. The AI then receives unknown data and assigns it to a group based on the known model from its training set. In neural network models, which are less common and more complex, nodes are used to connect information, similar to the human brain.13,15,80,81 While AI models pose effective tools to limit the data processing requirements involved with SERS, many complications still arise from issues such as overfitting of data, poor data quality, and insufficient sample size.15 In machine learning models, biases can cause incorrect interpretations, particularly when the data doesn't fit the training set data.60,82 Due to the complexity of patient samples, inaccuracies can occur from various factors, such as age, pre- and post-operative status, gender, and other variables.15,76,78,79 To combat this issue, AI models need to be trained with an extensive database whereby features can be extracted and selected to improve overall accuracy. While AI models use conventional data processing techniques during the training stages, the interpretation of the data without the need for specialized knowledge or manual processing makes the output clearer to end-users. They present a potentially effective method to determine the likelihood of cancer and present recommendations such as biopsies or ongoing treatment. As larger training datasets become available, AI has the potential to be a highly effective tool for point-of-care (POC) devices for the detection and subtyping of sEVs.
In a short summary of this section, label-free SERS assays for the detection of multiple sEV biomarkers present an effective method for cancer diagnosis (Fig. 5). Substrate design is fundamental for SERS signals with new designs being implemented such as nano-bowtie microfluidic systems and beehive nanostructures, which allow for high electromagnetic enhancements. In addition, novel data analysis strategies for label-free assays include the use of AI models for feature extraction and final readout. PCA and LDA are typically used in machine learning models, however future research will tend towards deep learning models which are able to achieve more complex data analysis and learning techniques. With the need to ensure utility, AI has come an effective tool for POC detection where complex data analysis is no longer required. Table 1 also provides a detailed summary of recent SERS label-free assays for sEV detection to provide design insights for future developments. Further information is provided on their limits of detection, SERS nanotag, capture probes, and cancer type being detected.
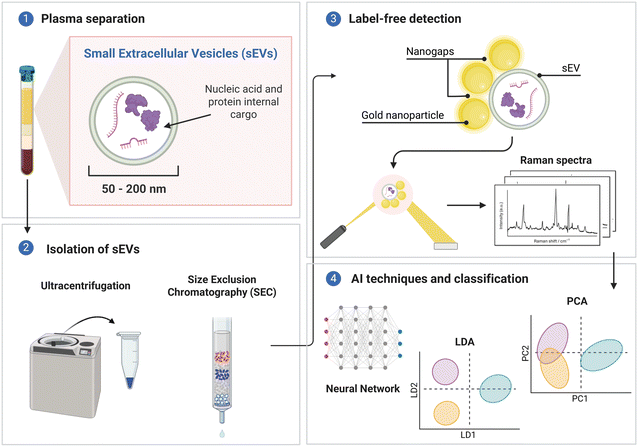 | ||
| Fig. 5 Label-free assay preparation including (1) plasma separation, (2) isolation of sEVs, (3) label-free detection and (4) AI techniques and classification. | ||
| Assay type | sEV isolation method | Accuracy, sensitivity and specificity (%) | Substrate | Detection target | Algorithm/statistical tests used | Cancer type | Ref. |
|---|---|---|---|---|---|---|---|
| AI-SERS array chip | Size exclusion chromatography | 90.2% sensitivity | Gold nanoparticles (spherical) | sEVs from various cancers | Multiple instance learning (MIL) | Lung, colorectal, pancreas, stomach and breast cancer | 82 |
| 94.4% specificity | Neural network algorithm | ||||||
| Size exclusion chromatography | 100% diagnostic accuracy for human patients with different breast cancer subtypes who do not undergo standard surgery | Gold nanostars (AuNS) | Estrogen receptor (ER), progesterone receptor (PR), and human epithelial receptor 2 (HER2) | Principal component analysis with linear discriminant analysis (PCA-LDA) | Breast cancer | 34 | |
| Partial least-squares discriminant analysis (PLS-DA) | |||||||
| Macroporous SERS Probe | Isolation kit | Not reported | Au-coated TiO2 Macroporous inverse opal (MIO) | sEVs from various cancers | t-Test | Colon, liver and lung cancer | 48 |
| Machine learning fuzzy diagnosis | Isolation kit | 91.1% accuracy | Gold nanoparticles (spherical) on the surface of the chip | H8, HeLa, and MCF-7 cell derived sEVs | Savitzky−Golay (SG) algorithm | Breast and cervical cancer | 60 |
| Adaptive iterative reweighted penalized least-squares algorithm (airPLS) | |||||||
| HOOKE intP Raman Analytical Software | |||||||
| Linear discriminant analysis (PCA-LDA) | |||||||
| AI Microfluidic chip | Size exclusion chromatography | 87% accuracy | Singe sEV nanocavity made from MoS2 on Si/SiO2 substrate | EGFRvIII, EGFR and MGMT in glioblastoma cells | Convolutional Neural Network (CNN) Algorithm | Glioblastoma (brain) cancer | 85 |
| Principal component analysis (PCA) | |||||||
| 3D ANN Nanosensor | Ultracentrifugation | 100% accuracy | Titanium nanostructure | Individual extracellular vesicles from breast, lung and colorectal cells | Principal component analysis (PCA) | Breast, lung and colorectal cancer | 2 |
| 100% sensitivity | Multivariate curve resolution (MCR) | ||||||
| 100% specificity | |||||||
| 79% for three tissue of origin and multiclass classification | |||||||
| Nano-bowtie microfluidic device | Size exclusion chromatography | 1.32 × 105 particles per mL | Polystyrene nanoparticles | Glioma sEVs | Principal component analysis (PCA) | Glioblastoma | 3 |
4. SERS label-based assays
The detection and subtyping of sEVs often employs the use of label-based methods to overcome the limitations of label-free methods reported in Section 3. The ‘targeted approaches’ for label-based assays are summarized in Fig. 6, where the use of SERS nanotags/labels is central to their functionalization.22,26,83 In this section, we discuss the SERS nanotag design, provide insights with respect to sEV detection, and highlight current advancement and challenges with label-based SERS assays and their potential solutions.4.1 Sample preparation techniques for label-based assays
In contrast with label-free assays, label-based assays do not require such high sample pre-processing. In almost all cases, the assays aim to reduce the amount of pre-processing for POC applications and either isolate sEVs within the assay or directly apply samples such as plasma, serum, urine, or whole blood. However, for proof-of-concept, similar to that for label-free, the most common methods of isolation include SEC, ultracentrifugation, and isolation kits.4,10,66,844.2 SERS nanotag design
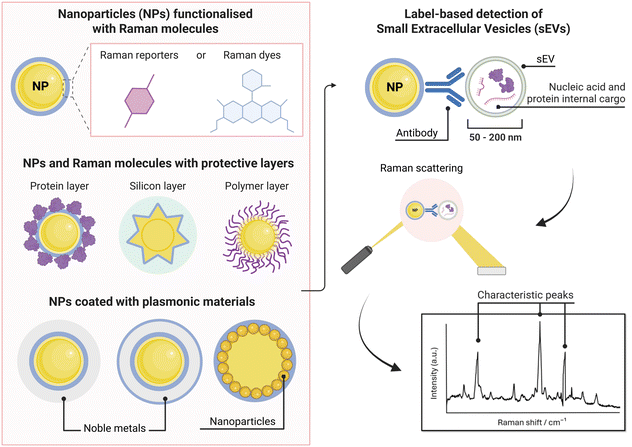
SERS nanotag technology is widely used in label-based applications and typically consists of a nanoparticle, which is coated with a Raman reporter molecule and bioconjugated with antibodies or aptamers for functionalization.16,27,30,35,36,83,86 SERS nanotags can differ based on their nanoparticle shape and plasmonic metal type, protective shells/coatings, and stabilising mechanisms. Examples of different nanoparticle shapes could include star, rod, flower, and spherical.16,17,27,36,87 For signal enhancement and protection purposes, coatings such as silver or silicon layers are often implemented. Further alterations can be made to improve the stability of the SERS nanotags including the addition of polyethylene glycol (PEG), silica shells, polymer coatings, and BSA layers.16,17,27,36,87 Therefore, there are many careful design considerations required for their biosensing applicability.SERS nanotags can achieve high sensitivity and capture specificity; however, a common problem is often instability and signal variability, depending on the type of particle and Raman reporter, which poses problems for quantification.16,30,35,86 Slight variations can affect the signal intensity by orders of magnitude due to localized ‘hot spots’, which are difficult to control.22,27 Thus, the appropriate design of SERS nanotags is vital for biosensing applications.
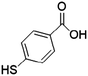
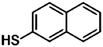
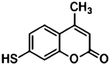
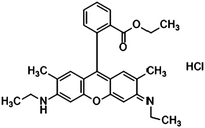
Furthermore, the selection of Raman reporter molecules is crucial due to the potential overlapping Raman peaks. In SERS applications, it is essential to identify ‘characteristic’ peaks that correspond to specific biomarkers to ensure accurate diagnosis.27,88 This is typically done through the identification of the highest characteristic peak in the spectrum. Previous studies in the broader field of biosensing applications have utilised many different Raman reporters, however, achieving multiplexing becomes a challenge when characteristic peaks from multiple Raman reporters overlap in the spectra. Due to this, most studies for multiplex biosensing applications use four or less Raman reporters, with the bulk majority using the three most common; 4-mercaptobenzoic acid (4-MBA), 2,3,5,6-tetrafluoro-4-mercaptobenzoic acid (TFMBA), and 5,5-dithiobis-(2-nitrobenzoic acid) (DTNB) as these three demonstrate clear peak distinctions.16,27,79,88 Four out of twelve studies conducted in the field of SERS multiplex assays for cancer-derived sEVs utilize this or similar combinations. Other Raman reporter molecules used in the multiplex SERS label-based assays for sEV detection are highlighted in Table 2, indicating the main characteristic peak of each.
| Raman reporter | Abbreviation | Molecule structure | Main characteristic peak/cm−1 | Ref. |
|---|---|---|---|---|
| 4-Mercaptobenzoic acid | 4-MBA |
|
1080 | 4, 10, 55, 92 and 104 |
| 4-Nitrothiophenol | 4-NTP |
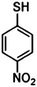
|
1335 | 9, 74 and 95 |
| 4-Aminothiophenol | 4-ATP |
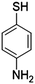
|
1140 | 9 and 84 |
| 5,5′-Dithiobis-(2-nitrobenzoic acid) | DTNB |
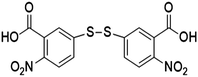
|
1337 | 4, 10, 32, 55 and 92 |
| 2,3,5,6-Tetrafluoro-4-mercaptobenzoic acid | TFMBA |
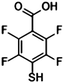
|
1380 | 4, 10 and 55 |
| 4-Mercaptopyridine | 2-Mpy |
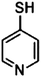
|
1002 | 4 and 9 |
| 2-Naphthalenethiol | 2-NAT |
|
1080 and 1378 | 32 |
| 2,7-Mercapto-4-methylcoumarin | MMC |
|
1170 | 32 |
| Rhodamine X | ROX |
|
1503 | 74 |
With the need for high signal enhancements, another technique has been reported for biosensing applications, which includes the use of Surface-enhanced resonance Raman scattering (SERRS) rather than typically used non-resonant molecules as reported above.21,89,90 Dyes such as NIR4f and NIR5e have been reported as ideal resonant Raman reporters for use with nanostars or nanospheres as they contribute to the signal intensity through molecular electronic resonances as demonstrated by Choi et al. (2023).21 The study used nanostars coated with NIR4f resonant Raman dye and compared them to AuNS with non-resonant 4-nitrothiophenol (4-NTP), showing an increase of 167 times at 785 nm vs. 633 nm excitation.21 However, this comes with the added complexity of their spectra which features fluorescence broadening and multiple peaks that can complicate spectra deconvolution.21 With improvements in spectral deconvolution and AI technologies, the improved signal intensity could be useful in multiplex quantification and detection purposes.32,33,91
4.3 Paper-based assays
Lateral flow immunoassays (LFIAs) are widely used paper-based diagnostic assays, commercially available in many forms, that leverage the capillary flow of porous membranes to transport a sample across the device and promote interaction with bioreceptors to detect target analytes.54,92–94 These assays are cost-effective and easy to use, making them suitable for POC applications.59,92,93,95–97 A typical LFIA is made up of a sample pad, conjugate pad, nitrocellulose membrane (NC), and an absorbent pad (Fig. 7A), which allows for a target analyte to flow through by capillary forces, interact with SERS nanotags and be captured on the test line to produce a signal.59,92,93,95–98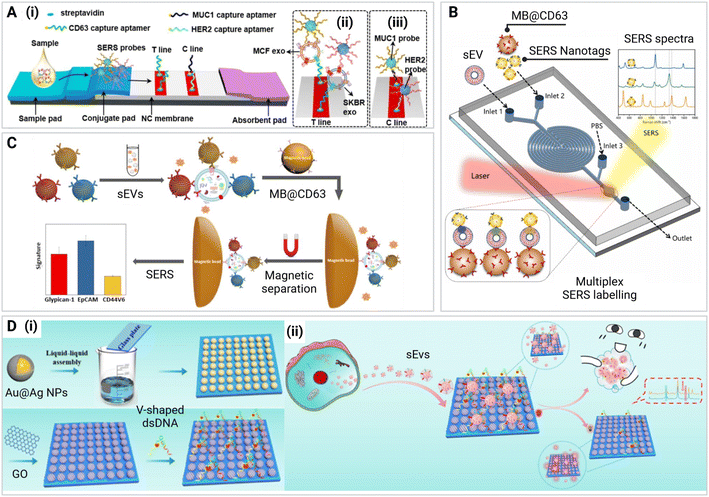 | ||
| Fig. 7 Label-based SERS assays including (A) Paper LFIA for the detection of sEVs. Aptamers were used for capture on a test line and control line. Adapted from ref. 92 with permission from ACS Applied Interfaces copyright [2023]. (B) A microfluidic device using immunomagnetic separation where sEVs are isolated by aptamer-functionalized magnetic beads (MBs) and detected using Raman reporter tagged AuNPs. Adapted from ref. 55 with permission from Sensors and Actuators B: Chemical copyright [2023]. (C) Liquid-based assay using MBs to capture sEVs which are labelled with SERS nanotags in suspension. A magnet is then used to draw the MBs into a pellet before obtaining the SERS reading from the supernatant. Adapted from ref. 10 with permission from ACS Sensors copyright [2020]. (D) Chip/array-based assay showing (i) the liquid-based assembly of a SERS substrate composed of silver-coated gold nanoparticles (Au@Ag NPs) bound onto a glass plate. A graphene oxide (GO) layer is then placed on top and functionalized with V-shaped double-stranded DNA (dsDNA) for (ii) capture and SERS-based detection of cell line derived sEVs. Adapted from ref. 74 with permission from ACS Sensors copyright [2023]. | ||
In a recent publication by Su et al. (2023), a LFIA was developed that utilized aptamers for the multiplex capture of two biomarkers, HER2 and MUC1, expressed on the surface of sEVs.92 To enable SERS-based detection, they designed two SERS nanotags, each incorporating gold nanostars (AuNS) as the SERS substrate and a different Raman reporter molecule – 4-MBA and DTNB for each biomarker.17,92,94,98 The captured serum sEVs were detected at the test line on the LFIA strip. The LFIA demonstrated a limit of detection (LOD) of 3.27 × 106 and 4.80 × 106 particles per mL, respectively.92 To deconvolute the output SERS spectra, multivariate curve resolution-alternating least squares spectral unmixing (MCR-ALS) was used, which demonstrated a 92% and 88% similarity with the individual SERS spectra of the HER2 and MUC1 probes, respectively.92 While the spectral deconvolution approach employed in the SERS-based LFIA enabled the accurate identification and quantification of the two sEV biomarkers, MUC1 and HER2, for diagnostic purposes, the detection of more than two biomarkers is typically required to reliably determine the cancer type and stage.9,11,92 This SERS-based LFIA meets most of the ASSURED criteria for POC devices, being affordable, sensitive, specific, user-friendly, and rapid, as outlined by the World Health Organization (WHO).99 However, the requirement for SERS measurements and the lack of portable Raman devices still poses a challenge in terms of deliverability to end-users.86,100,101
4.4 Microfluidic chips
Microfluidic devices have emerged as a more recent and advanced approach to address the challenges encountered by paper-based assays in the capture, detection, and subtyping of cancer-derived sEVs using SERS technology, and aim to be integrated sample-to-answer platforms.102,103 They typically consist of micron-sized channels and chambers on a substrate such as glass, silicon, or polymers like polydimethylsiloxane (PDMS).102,103 The workflow of microfluidic devices can vary depending on the specific design, but typically involve loading the sample into the inlet, followed by the sample flowing through microchannels to trap sEVs in designated chambers, and then detecting them using SERS.26,66,102,104Compared to LFIAs, microfluidic devices offer several advantages, such as faster analysis, increased multiplex capabilities, potential re-usability, higher sensitivity, lower LOD, small volume sample requirements, potential for automation, and the ability to integrate various laboratory functions like sample processing.66,102–104 Due to this, numerous microfluidic device designs have been recently developed for multiplex SERS-based cancer sEV detection and subtyping.
Recent examples, Han et al. (2022), Lin et al. (2023), and Wang et al. (2021), demonstrate multiplexed microfluidic-based methods for capturing and subtyping sEVs from different cancer types, including osteosarcoma, melanoma, breast, and lung cancer4,55,104. These devices can achieve LODs of 102–103 particles per mL. While the three devices share the common goal of sEV detection and characterization, their specific aims and design considerations differ. Han et al. (2022) and Lin et al. (2023) (Fig. 7B) aim to profile and characterize tumor-derived sEVs based on cancer-specific surface markers.55,104 However, the approaches used in these two studies only utilize one sEV tetraspanin (CD63) thus limiting the capture efficiency of the assay. The accurate detection and characterization of sEV subpopulations requires the use of multiple tetraspanins in combination with multiple cancer-specific surface markers. In contrast, the study by Wang et al. focuses primarily on accurately determining the phenotypic heterogeneity of sEVs using the three most commonly expressed tetraspanin markers (CD63, CD81, and CD9).4 The detection of general sEV surface markers allows for the quantification of captured sEV populations and provides insights into their surface protein expression profiles.4 The design utilizes a ‘nano-mixing’ system with three distinct capture regions, allowing for a broader range of the sEV population to be captured, while also targeting cancer-specific sEVs using CD45 antibodies.4 By integrating these design elements, combining both general sEV markers and cancer-specific biomarkers, more specific, precise, and accurate detection can be achieved. Furthermore, all three devices were tested with patient plasma samples and were all able to capture cancer sEVs with high degree of sensitivity. However, this integrated approach presents practical challenges and complications in SERS spectral deconvolution due to the increased number of Raman reporter molecules required for multiplex detection. Nonetheless, overcoming these hurdles through careful assay design and optimization could potentially lead to more comprehensive and clinically relevant sEV profiling for cancer diagnosis and monitoring.
4.5 Liquid-based bead assays
Bead assays represent another common approach for SERS-based assays, particularly for the multiplex detection, quantification, and subtyping of cancer-derived sEVs.65,68 While bead assays are used regularly for other target biomarkers, only three recent studies by Wang et al. (2018), Ning et al. (2020), and Zhang et al. (2020) have demonstrated the utility of SERS-based bead assays for the multiplexed detection and subtyping of sEVs.9,10,32 These assays typically involve the use of SERS nanotags and magnetic beads (MBs), which, through various interactions, enable the detection of sEVs through subsequent SERS measurements.9,10,32 All methods noted in this section utilized SERS nanotags with characteristic spectral peaks to detect sEVs associated with cancer. The sEVs bind to MBs functionalized with antibodies or aptamers to target specific biomarkers on the surface of the sEVs. The SERS nanotags then bind to the captured sEVs forming a complex, which is then accumulated in a pellet using a magnet that attracts the MBs. The measurement can either be a negative or positive response depending on the assay design. In a negative response assay, the supernatant is measured and the decrease in signal intensity of the distinctive peaks indicate the presence of target sEVs due to the absence of SERS nanotags in suspension. In a positive response assay, the pellet is resuspended and a measurement taken of the SERS nanotag-sEV-MB complexes. A positive repsonse is obtained whereby more sEVs result in a higher SERS signal.In the design by Zhang et al. (2020), SERS nanotags of MBA-EpCAM (1077 cm−1), TFMBA-CD44V6 (1380 cm−1), and DTNB-MIL38 (1340 cm−1) were incubated with sEVs suspended in cell culture media before being further incubated with MBs for capture.10 Using a magnet to separate the captured sEVs from other vesicles and proteins in the solution, a SERS readout was used to determine the surface subtyping in a positive response manner.10 Using this design, the assay was able to effectively phenotype sEVs from pancreatic cancer, colorectal cancer, and bladder cancer cell lines with a LOD of 2.3 × 106 particles per mL.10 To demonstrate clinical relevance, sEVs were also spiked into plasma from healthy donors, showing similar Raman peaks although with lower intensity due to the SERS nanotags being removed by target sEVs on the MBs. In another study using the same assay design, Zhang et al. (2022) (Fig. 7C) compared pancreatic ductal adenocarcinoma (PDAC) patient plasma with healthy donors. The results found that the assay had a sensitivity of 100% (CI: 66.4–100%), a specificity of 100% (CI: 75.3–100%) and AUC of 1.000 for PDAC diagnosis. This design could therefore be applied to other cancer types for use with patient samples.105 Therefore, the bead assay design shows an effective method that can be used in clinical applications for the subtyping of multiple cancer types.
On the other hand, studies conducted by Wang et al. (2018) and Ning et al. (2020), utilize a negative response system.9,32 Ning et al. (2020) focused on prostate cancer detection, comparing healthy samples that exhibited strong peaks at 1002 (2-Mpy), 1140 (4-ATP), and 1335 cm−1 (NTP) with patient samples.9 In the latter, they observed a decrease in the dominant 1002 cm−1 peak, indicating the presence of prostate cancer sEVs. The LOD for each cell line was determined to be 26 particles per μL for sEVs with LNCaP, 72 particles per μL for sEVs with SKBR3 and 35 particles per μL for sEVs with HepG2. However, the study did not report any statistical analysis for the patient samples to determine if the decrease in the SERS peaks was significant. Wang et al. (2018) investigated breast cancer detection using whole blood samples.32 They found that healthy individuals displayed the strongest SERS intensities at 1326 cm−1 (DTNB), 1378 cm−1 (2NAT), and 1170 cm−1 (MMC), suggesting the absence of cancer-related sEVs. In contrast, breast cancer patient samples showed a weakened intensity of the breast cancer biomarker (aptamer H2) signal, indicating the presence of breast cancer sEVs.32 Likewise, colorectal cancer patients showed a decrease in signal for the T84 colorectal cancer biomarker and prostate patients showed a decrease in intensity for LNCaP probes. The LODs for cell line-derived sEVs were determined to be 32 SKBR3 sEVs per μL, 73 sEVs per μL for T84 sEVs and 203 sEVs per μL for LNCaP sEVs. However, the study did not mention any statistical analysis conducted to determine if the decrease in signal was significant.
While all presented assays demonstrate the ability to allow for multiplex detection of sEVs, positive response assays demonstrate superior reliability in minimizing false positive rates as they offer more intuitive readouts for end-users, significantly enhancing result interpretation and understanding as compared to negative response readouts. Bead assays therefore offer several advantages, including multiplex capabilities and cost-effectiveness for POC applications, with positive response liquid-based bead assays being the ideal choice for on-going studies.8,65
4.6 Biosensor chip/array-based assays
Chip-array assays typically utilize a plasmonic immunocapture-based substrate that immobilizes the capture of sEVs, and nanotags SERS readouts.12 Many examples, such as those proposed by Li et al. (2018) and Zhang et al. (2023) employ unique features to capture sEVs on a plasmonic surface, resulting in high sensitivity and specificity.74,84 The reported LODs for Li et al. (2018) and Zhang et al. (2023) are one exosome in 2 mL (0.5 particles per mL) and 1.5 × 102 particles per mL, respectively.74,84 Therefore, the LODs of these assays fall within the range of 102 particles per mL, being lower than those of typical ELISA and western blot gold standard techniques.74,75 Compared to liquid-based bead assays, miniaturized devices, and paper-based designs, the chip/array-based assays demonstrate higher sensitivity, similar to that of microfluidic devices.The assay proposed by Li et al. (2018) presents a polydopamine (PDA) bi-functionalized SERS base for sEV capture with SERS nanotags as labels for quantification.84 The assay demonstrates high sensitivity and specificity, with the ability to accurately differentiate clinical serum samples from pancreatic cancer patients and healthy individuals using migration inhibitory factor (MIF), glypican-1 (GPC1), and EGFR-based PDA SERS methods.84 Thus, demonstrating a clear ability to successfully discriminate between healthy and cancer patients and potentially improve consequent therapeutic outcomes with early detection.
The ratiometric aptamer biosensor proposed by Zhang et al. (2023) features immobilized gold–silver nanoparticles on the surface of a glass plate, which are then coated with graphene oxide (GO) and V-shaped double-stranded DNA complimentary to HER2 and EpCAM surface markers, serving as capture probes for sEVs via competitive reactions (Fig. 7D).74 One advantage of this assay design is the use of ratiometric SERS, which effectively overcomes the influence of external factors such as temperature, salt concentration, and pH value, resulting in improved SERS signal performance.74 Furthermore, the competitive reaction between the V-shaped double-stranded DNA and the sEVs eliminates the need for tetraspanin-based capture, as the assay relies on the EpCAM and HER2 surface markers for sEV capture. However, due to the nature of the V-shaped double-stranded DNA, the assay is only able to accommodate two biomarkers at a time. Thus, for clinical applications, the ability to screen multiple cancer-specific biomarkers is limited in the current design.
To summarise this section, the design of SERS nanotags plays a crucial role in the development of SERS label-based assays, offering significant advantages in terms of sensitivity, specificity, and multiplexing capabilities. One of the key benefits of label-based methods is the lack of need for sample preparation techniques to minimise background signals.
A critical aspect of SERS nanotag design is the selection of appropriate Raman reporters, which are responsible for generating the characteristic spectral peaks used for detection and quantification. Table 2 provides an overview of the most used Raman reporters and their distinctive peaks, serving as a valuable reference for researchers in the field. However, with a need for clear characteristic peaks, current methods of data processing limit the ability to multiplex more than four Raman reporters at a time, leaving room for further research and development in this field.
Overall, multiplexed SERS label-based assays have emerged as an effective method for the detection of cancer-derived sEVs. Recently developed SERS multiplex label-based assays show great promise as a POC devices for cancer detection. These assays have demonstrated the ability to simultaneously detect multiple biomarkers with high sensitivity and specificity. Various assay formats have been explored, including paper-based platforms, microfluidic chips, miniaturized SERS devices, liquid-based bead assays, biosensors, and chip/array-based assays. Each format offers unique advantages in terms of sample handling, multiplexing capability, and ease of use. However, more focus is required on improving SERS multiplexing to incorporate more biomarkers for accurate diagnostics. A detailed summary of recent publications on SERS multiplex assay types for detecting and profiling cancer-derived sEVs is provided in Table 3.
| Assay type | Specific assay name | Target/s | LOD reported | Nanoparticle used | SERS nanotag | Capture probe on substrate | Detection targets | Reporter molecule/s | Cancer Type | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Paper-based assay | Lateral flow assay | Cancer specific markers /tetraspanins | 3.27 × 106 particles per mL for serum SKBR sEVs | Gold nanostars with silica protective layer | AuNS@HER2 (detection aptamer) | CD63 capture aptamer | SKBR and MCF7 exosomes | DTNB and 4-MBA | Breast cancer | 92 |
| 4.80 × 106 particles per mL for serum MCF7 sEVs | AuNS@MUC1 (detection aptamer) | |||||||||
| Microfluidic chips | Cancer specific markers/tetraspanins | 2 particles per μL or 2 × 103 particles per mL | Gold nanoparticles (spherical) | AuNPs@MBA@anti-CD63 | N/A | CD63, VIM and EpCAM protein biomarkers | 4-MBA, TFMBA and 4-Mpy | Osteosarcoma | 104 | |
| AuNPs@TFMBA@anti-CD9 | ||||||||||
| AuNPs@TFMBA@anti-VIM | ||||||||||
| AuNPs@MPY@anti-EpCAM | ||||||||||
| Cancer specific markers/tetraspanins | LODs obtained by CD63-MBA, PD-L1-DTNB, and EGFR-TFMBA SERS nanotags were 5.46 × 103, 2.44 × 103, and 4.46 × 102 particles per mL respectively | Gold nanoparticles (spherical) | AuNP@MBA@Anti-CD63 | Anti-CD63 functionalized MBs | CD63, PD-L1 and EGFR | 4-MBA, DTNB, TFMBA | Lung cancer | 55 | ||
| AuNP@DTNB@Anti-PD-L1 | ||||||||||
| AuNP@TFMBA@Anti-EGFR | ||||||||||
| Cancer specific markers/tetraspanins | 103 sEVs per mL | Gold nanoparticles (Spherical) | AuNP@MBA@Ab | Antibodies of CD63, CD81 and CD9 in the micro-wells | CD81, CD63 and CD9 | 4-MBA, DTNB, TFMBA and MPY | Melanoma, breast and lung cancer | 4 | ||
| AuNP@DTNB@Ab | MCSP | |||||||||
| AuNP@TFMBA@Ab | MCAM | |||||||||
| AuNP@MPY@Ab | ErbB3 | |||||||||
| LNGFR | ||||||||||
| CD45 | ||||||||||
| SK-MEL28, SK-MES-1, NCI-H1703, and NCI-H1650, and MCF7 cell line derived sEVs | ||||||||||
| Liquid-based bead assays | Cancer specific markers | 32 SKBR3 particles per μL | Gold Nanoparticles (spherical) and magnetic B=beads (MBs) | AuNP@SKBR3(aptamer)@DTNB | MB@SiO2@Au@aptamer | SKBR3 (breast) | DTNB, 2NAT and MMC | Breast, prostate and colorectal | 32 | |
| T84 exosomes 73 particles per μL | AuNP@LNCaP(aptamer)@2NAT | Aptamers include: HER2 (breast), PSMA (prostate) and CEA (colorectal) | LNCaP (prostate) | |||||||
| LNCaP sEVs 203 particles per μL | AuNP@T84(aptamer)@MMC | T84 cells (colorectal) | ||||||||
| Cancer specific markers | LNCaP – 26 particles per μL | MBs and Gold–silver–silver core–shell–shell nanotrepangs | GSSNT@2-Mpy@L-PSMA | MB@PSMA(aptamer) | LNCaP, SKBR3 and HepG2 exosomes | 2-Mpy, 4-ATP and 4-NTP | Breast, liver and prostate cancer | 9 | ||
| SKBR – 72 particles per μL | GSSNT@4-ATP@L-Her2 | MB@HER2(aptamer) | ||||||||
| HepG2 – 35 particles per μL | GSSNT@NTP@L-AFP | MB@AFP(aptamer) | ||||||||
| Cancer specific markers/tetraspanins | 2.3 × 106 particles per mL | Gold nanoparticles (spherical) | AuNPs@DTNB | Streptavidin@MB@biotin-CD63 antibody | Glypican-1 | DTNB, 4-MBA and TFMBA | Pancreatic cancer | 10 | ||
| AuNP@TFMBA | Epithelial cell adhesion molecules (EpCAM) | |||||||||
| AuNP@4-MBA | CD44 variant isoform 6 (CD44V6) | |||||||||
| From panc-1 sEVs | ||||||||||
| Biosensor chip/Array-based assays | Polydopamine bi-functionalized SERS immunoassay | Cancer specific markers/tetraspanins | 1 exosome in 2 mL | Polydopamine-encapsulated antibody-reporter-Ag(shell)–Au(core) (PEARL) | AuNP@Ag@pATP@PDA@Antibodies | Anti-MIF | MIF | p-ATP | Pancreatic cancer | 84 |
| Anti-GPC1 | GPC1 | |||||||||
| Anti-CD63 | CD63 | |||||||||
| Anti-EGFR | PANC-01 sEVs | |||||||||
| Ratiometric SERS biosensor | Cancer specific markers | 1.5 × 102 particles per mL | Gold-Silver Nanoparticles | Au@AgNPs/graphene Oxide (GO) substrate@NTP | V-shaped double-stranded DNA | EpCAM | 4-NTP and Rhodamine X | Pancreatic and breast cancer | 74 | |
| HER2 |
5. Conclusion and perspectives
sEVs present a promising biomarker for multiplexed, SERS-based cancer detection.11,43 Current research on SERS label-based and label-free methods allows for early detection, quantification, and subtyping of cancer-derived sEV populations. Various label-free designs have been proposed (e.g. AI-based array chips) which allow for sensitive detection of cancer sEVs. Without the need for a label, label-free assays can ensure multiple different sEV populations to be detected. The simplicity of the system also affords further advantages for overall cost, stability and rapid testing. However, with the need of pure samples and complex AI data analysis, often extensive sample preparation is required to extract sEVs to remove additional background, and extensive AI data training thus, limiting the ability for these assays to be easily translated to clinical settings. Further research should be focused on implementing larger training datasets to ensure accuracy within AI models and prevent overfitting of data.34,60,61,106On the other hand, label-based techniques have emerged as a ‘targeted approach’ to sEV detection. The use of SERS nanotags to specifically label sEVs has improved specificity, added quantification capacities and reduced matrix effects. Due to this, label-based techniques have moved to using limited sample processing to adhere to clinical testing requirements. However, with the need a newly designed SERS nanotag for each target biomarker, multiplexing capabilities have been limited to four or less Raman reporters due to spectral overlap and complex data convolution.22,83,89,92 Further research should be focused on improving SERS nanotag multiplexing capabilities.
Within the field of label-based assays, LFIAs present a method that is rapid, cost-effective and easy for end-users. As compared to microfluidic devices, bead assays and chip/array-based designs – which, although more complex, offer higher sensitivity and re-usability – LFIAs (pregnancy and COVID-19 tests) are well known and easier to translate into clinical settings. Microfluidic devices, bead assays and chip/array-based designs still require further testing and face the challenge of translation to clinical settings whereby clinicians will have to be trained to use such devices. Therefore, LFIAs present at current the most ideal design for rapid translation to clinical settings for cancer diagnostics.
While both label-based and label-free techniques offer promising approaches for detecting sEVs, label-based methods provide enhanced specificity in testing. This increased precision is afforded by the specific targeting of markers on sEVs, such as sEV-specific tetraspanins and cancer-associated proteins. Label-based assays, as compared to label-free offer a significant advantage: the ability to utilize unprocessed clinical samples with high accuracy. This capability has been demonstrated through the successful use of direct plasma and serum samples in label-based assays. Despite the added complexity of the assay requiring SERS nanotags, the improved accuracy and ability to work with raw clinical specimens make label-based techniques the future direction for sEV detection in cancer diagnostics.
Additionally, a greater focus on patient samples, rather than solely replying on cell lines or spiked samples, would allow the devices to be optimized for POC applications and consider the complexity of individual sEV populations per patient.53,92,107,108 The critical need for accessibility and affordability in diagnostic solutions is also addressed with designs being focused on re-usability or cost-effective materials.44,94,99 Therefore, the presented SERS label-based and label-free assays for cancer sEV detection demonstrate enhanced sensitivity and specificity, real-time disease monitoring capabilities, and the potential for facilitation of precision oncology in POC settings.
5.1 Perspective on clinical applications
Clinical applications are a valuable motivating aspect for SERS multiplex sEV detection and subtyping assay designs, with most aiming to assist with current precision oncology methods. However, with the current problems such as stability of SERS nanotags, cost-effectiveness, the need for extensive multiplex capabilities, and the lack of extensive data for AI-based methods, there is still a need for an adequate device to be successfully applied to clinical applications.For the translation from laboratories to clinics, there are still further developments required including:
[1] the need to develop a highly reproducible and stable bioconjugated SERS nanotag for use in assay designs to ensure accuracy and deliverability.21,35,83,89,109
[2] The use of whole blood for easy clinical use and to significantly decrease sample processing times.53,107,110–113
[3] The utilization of all three tetraspanins in order to ensure total sEV capture in label-based assay designs. This would ensure a greater range of the sEV population would be characterized and thus ensure better detection, quantification and subtyping.4,45,56,114
[4] The implementation of a clinical threshold for label-free sEV detection methods to determine clinical relevancy to various cancer types, as some cancer markers are also expressed on healthy cells. For example, clinical thresholds play a crucial role in determining the minimum detectable level of EpCAM necessary for a positive result in SERS-based cancer diagnostics. The exceptional sensitivity of SERS assays, capable of detecting even trace amounts of biomarkers, presents both opportunities and challenges for clinical application. While this high sensitivity potentially enables earlier cancer detection, it also complicates result interpretation, making simple ‘yes’ or ‘no’ diagnostic outputs difficult without specific benchmark detection. This challenge mirrors the established practice in routine pathology testing, where defined thresholds are commonly implemented to ensure accurate diagnoses. To address this, comprehensive clinical studies are essential to establish specific thresholds for each cancer biomarker, including EpCAM. These studies must involve diverse patient populations and correlate SERS-detected biomarker levels with clinical outcomes, accounting for variations in biomarker expression between cancer types and stages. This approach will facilitate the standardization of SERS-based diagnostics across different platforms and clinical settings, ultimately ensuring an easier and more effective transition of diagnostic SERS assays from the laboratory to clinical practice.49,50
[5] The integration of AI-based software for results interpretation to improve user-friendliness. This would limit the sample processing required when used in POC applications.15,34,61,78,79
[6] The fabrication of a patient sample database for AI technologies. This would enable larger data sets to be used to develop more complex and highly accurate AI models.
With the current approaches, two possibilities are presented for clinical applications for the detection and subtyping of cancer sEVs. One is the development of a portable SERS device for LFIA.86,101 However, the tests would have to employ the use of SERS and AI-based technologies for simple readout and recommendations to end-users.60,76 With further developments, the design of a portable testing system could be useful for home-testing or rapid clinical testing in a format that is well-known to most users (lateral flow-based assay similar to those of commonly used in pregnancy and COVID-19 tests). The test could employ the use of a cartridge where the assay can be inserted into the portable Raman. The device could be integrated with a computer or phone to supply an easy-to-read outcome of the assay. In this case, the portable Raman reader would be reusable and easy to use for POC applications. However, the LFIA design would have to incorporate a broad range of surface marker targets to accurately determine cancer type. Another potential design could be a high-throughput automated device that utilizes either a label-based or label-free designs for multiplex detection capabilities of all cancer types. It is also desirable to have a method of easy quantification and an AI-based analysis method that can predict the cancer type and stage based on the output SERS spectra. Therefore, with further improvements to current designs, a practical device could be implemented for multiplex sEV detection and subtyping that is easy to use, sensitive, low-cost, and accurate to ensure early disease detection and monitoring capabilities.
Author contributions
C.D. drafted the manuscript. YW, SCT and LR provided critical insights and discussion toward manuscript conceptualization.Data availability
This review article synthesizes and analyses data from previously published studies. All data referenced in this review are available from the original sources, which are cited in the references section.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by the Cancer Council NSW project grant (RG 22-12). Y. W. acknowledges the funding support from Australian Research Council (ARC) Future Fellowships (FT210100737). L. R. and C. D. acknowledges the funding support from the government funded Research Training Program (RTP) Scholarship for the Doctor of Philosophy and Master of Research in Natural Sciences Program at Macquarie University. Figures were created using BioRender.References
- B. S. Chhikara and K. Parang, Global Cancer Statistics 2022: The Trends Projection Analysis, Chem. Biol. Lett., 2023, 10(1), 451–451 Search PubMed https://scholar.google.com/scholar?q=urn:nbn:sciencein.cbl.2023.v10.451 .
- R. Haldavnekar, K. Venkatakrishnan and B. Tan, Cancer Stem Cell Derived Extracellular Vesicles with Self-Functionalized 3D Nanosensor for Real-Time Cancer Diagnosis: Eliminating the Roadblocks in Liquid Biopsy, ACS Nano, 2022, 16(8), 12226–12243, DOI:10.1021/acsnano.2c02971.
- M. Jalali, I. I. Hosseini, T. AbdelFatah, L. Montermini, S. W. Hogiu, J. Rak and S. Mahshid, Plasmonic Nanobowtiefluidic Device for Sensitive Detection of Glioma Extracellular Vesicles by Raman Spectrometry, Lab. Chip, 2021, 21(5), 855–866, 10.1039/D0LC00957A.
- J. Wang, A. Wuethrich, R. J. Lobb, F. Antaw, A. A. I. Sina, R. E. Lane, Q. Zhou, C. Zieschank, C. Bell, V. F. Bonazzi, L. G. Aoude, S. Everitt, B. Yeo, A. P. Barbour, A. Möller and M. Trau, Characterizing the Heterogeneity of Small Extracellular Vesicle Populations in Multiple Cancer Types via an Ultrasensitive Chip, ACS Sens., 2021, 6(9), 3182–3194, DOI:10.1021/acssensors.1c00358.
- C. Théry, et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines, J. Extracell. Vesicles, 2018, 7(1), 1535750, DOI:10.1080/20013078.2018.1535750.
- M. H. Vasconcelos, H. R. Caires, A. Ābols, C. P. R. Xavier and A. Linē, Extracellular Vesicles as a Novel Source of Biomarkers in Liquid Biopsies for Monitoring Cancer Progression and Drug Resistance, Drug Resistance Updates, 2019, 47, 100647, DOI:10.1016/j.drup.2019.100647.
- T. An, S. Qin, D. Sun, Y. Huang, Y. Hu, S. Li, H. Zhang, B. Li, B. Situ, L. Lie, Y. Wu and L. Zheng, Unique Protein Profiles of Extracellular Vesicles as Diagnostic Biomarkers for Early and Advanced Non-Small Cell Lung Cancer, Proteomics, 2019, 19(12), 1800160, DOI:10.1002/pmic.201800160.
- L. Min, B. Wang, H. Bao, X. Li, L. Zhao, J. Meng and S. Wang, Advanced Nanotechnologies for Extracellular Vesicle-Based Liquid Biopsy, Adv. Sci., 2021, 8(20), 2102789, DOI:10.1002/advs.202102789.
- C.-F. Ning, L. Wang, Y.-F. Tian, B.-C. Yin and B.-C. Ye, Multiple and Sensitive SERS Detection of Cancer-Related Exosomes Based on Gold–Silver Bimetallic Nanotrepangs, Analyst, 2020, 145(7), 2795–2804, 10.1039/C9AN02180A.
- W. Zhang, L. Jiang, R. J. Diefenbach, D. H. Campbell, B. J. Walsh, N. H. Packer and Y. Wang, Enabling Sensitive Phenotypic Profiling of Cancer-Derived Small Extracellular Vesicles Using Surface-Enhanced Raman Spectroscopy Nanotags, ACS Sens., 2020, 5(3), 764–771, DOI:10.1021/acssensors.9b02377.
- K. Abhange, A. Makler, Y. Wen, N. Ramnauth, W. Mao, W. Asghar and Y. Wan, Small Extracellular Vesicles in Cancer, Bioact. Mater., 2021, 6(11), 3705–3743, DOI:10.1016/j.bioactmat.2021.03.015.
- Y. Liang, B. M. Lehrich, S. Zheng and M. Lu, Emerging Methods in Biomarker Identification for Extracellular Vesicle-Based Liquid Biopsy, J. Extracell. Vesicles, 2021, 10(7), e12090, DOI:10.1002/jev2.12090.
- W. Lee, A. T. M. Lenferink, C. Otto and H. L. Offerhaus, Classifying Raman Spectra of Extracellular Vesicles Based on Convolutional Neural Networks for Prostate Cancer Detection, J. Raman Spectrosc., 2020, 51(2), 293–300, DOI:10.1002/jrs.5770.
- M. Fleischmann, P. J. Hendra and A. J. McQuillan, Raman Spectra of Pyridine Adsorbed at a Silver Electrode, Chem. Phys. Lett., 1974, 26(2), 163–166, DOI:10.1016/0009-2614(74)85388-1.
- F. Lussier, V. Thibault, B. Charron, G. Q. Wallace and J.-F. Masson, Deep Learning and Artificial Intelligence Methods for Raman and Surface-Enhanced Raman Scattering, TrAC, Trends Anal. Chem., 2020, 124, 115796, DOI:10.1016/j.trac.2019.115796.
- M. Sánchez-Purrà, B. Roig-Solvas, A. Versiani, C. Rodriguez-Quijada, H. d. Puig, I. Bosch, L. Gehrke and K. Hamad-Schifferli, Design of SERS Nanotags for Multiplexed Lateral Flow Immunoassays, Mol. Syst. Des. Eng., 2017, 2(4), 401–409, 10.1039/C7ME00052A.
- B. Andreiuk, F. Nicolson, L. M. Clark, S. R. Panikkanvalappil, Kenry, M. Rashidian, S. Harmsen and M. F. Kircher, Design and Synthesis of Gold Nanostars-Based SERS Nanotags for Bioimaging Applications, Nanotheranostics, 2022, 6(1), 10–30, DOI:10.7150/ntno.61244.
- S. Gupta, S. Huda, P. K. Kilpatrick and O. D. Velev, Characterization and Optimization of Gold Nanoparticle-Based Silver-Enhanced Immunoassays, Anal. Chem., 2007, 79(10), 3810–3820, DOI:10.1021/ac062341m.
- M. D. Porter, R. J. Lipert, L. M. Siperko, G. Wang and R. Narayanan, SERS as a Bioassay Platform: Fundamentals, Design, and Applications, Chem. Soc. Rev., 2008, 37(5), 1001–1011, 10.1039/B708461G.
- Y. Gao, J. Wang, W. Wang, T. Zhao, Y. Cui, P. Liu, S. Xu and X. Luo, More Symmetrical “Hot Spots” Ensure Stronger Plasmon-Enhanced Fluorescence: From Au Nanorods to Nanostars, Anal. Chem., 2021, 93(4), 2480–2489, DOI:10.1021/acs.analchem.0c04518.
- N. Choi, M. Adanalic, A. Dankov, R. Grzeschik, M. Hechler and S. Schlücker, NIR Dye-Encoded Nanotags for Biosensing: Role of Functional Groups on Sensitivity and Performance in SERRS-Based LFA, J. Raman Spectrosc., 2023, 54(9), 929–939, DOI:10.1002/jrs.6519.
- M. Sánchez-Purrà, B. Roig-Solvas, C. Rodriguez-Quijada, B. M. Leonardo and K. Hamad-Schifferli, Reporter Selection for Nanotags in Multiplexed Surface Enhanced Raman Spectroscopy Assays, ACS Omega, 2018, 3(9), 10733–10742, DOI:10.1021/acsomega.8b01499.
- K. Plakas, L. E. Rosch, M. D. Clark, S. Adbul-Rashed, T. M. Shaffer, S. Harmsen, S. S. Gambhir and M. R. Detty, Design and Evaluation of Raman Reporters for the Raman-Silent Region, Nanotheranostics, 2022, 6(1), 1–9, DOI:10.7150/ntno.58965.
- S. Rodal-Cedeira, A. Vázquez-Arias, G. Bodelón, A. Skorikov, S. Núñez-Sánchez, A. Laporta, L. Polavarapu, S. Bals, L. M. Liz-Marzán, J. Pérez-Juste and I. Pastoriza-Santos, An Expanded Surface-Enhanced Raman Scattering Tags Library by Combinatorial Encapsulation of Reporter Molecules in Metal Nanoshells, ACS Nano, 2020, 14(11), 14655–14664, DOI:10.1021/acsnano.0c04368.
- A. Eguchi, E. Kostallari, A. E. Feldstein and V. H. Shah, Extracellular Vesicles, the Liquid Biopsy of the Future, J. Hepatol., 2019, 70(6), 1292–1294, DOI:10.1016/j.jhep.2019.01.030.
- K. Balaji Shanmugasundaram, J. Li, A. Ibn Sina, A. Wuethrich and M. Trau, Toward Precision Oncology: SERS Microfluidic Systems for Multiplex Biomarker Analysis in Liquid Biopsy, Mater. Adv., 2022, 3(3), 1459–1471, 10.1039/D1MA00848J.
- W. Zhang, L. Jiang, J. A. Piper and Y. Wang, SERS Nanotags and Their Applications in Biosensing and Bioimaging, J. Anal. Test., 2018, 2(1), 26–44, DOI:10.1007/s41664-018-0053-9.
- M. M. Joseph, N. Narayanan, J. B. Nair, V. Karunakaran, A. N. Ramya, P. T. Sujai, G. Saranya, J. S. Arya, V. M. Vijayan and K. K. Maiti, Exploring the Margins of SERS in Practical Domain: An Emerging Diagnostic Modality for Modern Biomedical Applications, Biomaterials, 2018, 181, 140–181, DOI:10.1016/j.biomaterials.2018.07.045.
- J. U. Lee, A. H. Nguyen and S. J. Sim, A Nanoplasmonic Biosensor for Label-Free Multiplex Detection of Cancer Biomarkers, Biosens. Bioelectron., 2015, 74, 341–346, DOI:10.1016/j.bios.2015.06.059.
- C. Duffield, et al., Synthesis and characterization of reporter molecules embedded core-shell nanoparticles as SERS nanotags. DOI:10.1142/S1793545821410078.
- J. Wang, K. M. Koo, Y. Wang and M. Trau, Engineering State-of-the-Art Plasmonic Nanomaterials for SERS-Based Clinical Liquid Biopsy Applications, Adv. Sci., 2019, 6(23), 1900730, DOI:10.1002/advs.201900730.
- Z. Wang, S. Zong, Y. Wang, N. Li, L. Li, J. Lu, Z. Wang, B. Chen and Y. Cui, Screening and Multiple Detection of Cancer Exosomes Using an SERS-Based Method, Nanoscale, 2018, 10(19), 9053–9062, 10.1039/C7NR09162A.
- R. Di Santo, S. Romanò, A. Mazzini, S. Jovanović, G. Nocca, G. Campi, M. Papi, M. De Spirito, F. Di Giacinto and G. Ciasca, Recent Advances in the Label-Free Characterization of Exosomes for Cancer Liquid Biopsy: From Scattering and Spectroscopy to Nanoindentation and Nanodevices, Nanomaterials, 2021, 11(6), 1476, DOI:10.3390/nano11061476.
- Y. Xie, X. Su, Y. Wen, C. Zheng and M. Li, Artificial Intelligent Label-Free SERS Profiling of Serum Exosomes for Breast Cancer Diagnosis and Postoperative Assessment, Nano Lett., 2022, 22(19), 7910–7918, DOI:10.1021/acs.nanolett.2c02928.
- Z. Zhang, R. Guan, J. Li and Y. Sun, Engineering Rational SERS Nanotags for Parallel Detection of Multiple Cancer Circulating Biomarkers, Chemosensors, 2023, 11(2), 110, DOI:10.3390/chemosensors11020110.
- M. Salehi and A. Hamann-Steinmeier, Bioconjugation of SERS Nanotags and Increasing the Reproducibility of Results, BioNanoMaterials, 2017, 18(1–2), 20160011, DOI:10.1515/bnm-2016-0011.
- M. Arruebo, M. Valladares and Á. González-Fernández, Antibody–Conjugated Nanoparticles for Biomedical Applications, J. Nanomater., 2009, 2009(1), 439389, DOI:10.1155/2009/439389.
- L. Zhang, Y. Mazouzi, M. Salmain, B. Liedberg and S. Boujday, Antibody-Gold Nanoparticle Bioconjugates for Biosensors: Synthesis, Characterization and Selected Applications, Biosens. Bioelectron., 2020, 165, 112370, DOI:10.1016/j.bios.2020.112370.
- J. A. Welsh, D. C. I. Goberdhan, L. O'Driscoll, E. I. Buzas, C. Blenkiron, B. Bussolati, H. Cai, D. Di Vizio, T. A. P. Driedonks, U. Erdbrügger, J. M. Falcon-Perez, Q. Fu, A. F. Hill, M. Lenassi, S. K. Lim, M. G. Mahoney, S. Mohanty, A. Möller, R. Nieuwland, T. Ochiya, S. Sahoo, A. C. Torrecilhas, L. Zheng, A. Zijlstra, S. Abuelreich, R. Bagabas, P. Bergese, E. M. Bridges, M. Brucale, D. Burger, R. P. Carney, E. Cocucci, F. Colombo, R. Crescitelli, E. Hanser, A. L. Harris, N. J. Haughey, A. Hendrix, A. R. Ivanov, T. Jovanovic-Talisman, N. A. Kruh-Garcia, V. Ku'ulei-Lyn Faustino, D. Kyburz, C. Lässer, K. M. Lennon, J. Lötvall, A. L. Maddox, E. S. Martens-Uzunova, R. R. Mizenko, L. A. Newman, A. Ridolfi, E. Rohde, T. Rojalin, A. Rowland, A. Saftics, U. S. Sandau, J. A. Saugstad, F. Shekari, S. Swift, D. Ter-Ovanesyan, J. P. Tosar, Z. Useckaite, F. Valle, Z. Varga, E. van der Pol, M. J. C. van Herwijnen, M. H. M. Wauben, A. M. Wehman, S. Williams, A. Zendrini, A. J. Zimmerman, MISEV Consortium, C. Théry and K. W. Witwer, Minimal Information for Studies of Extracellular Vesicles (MISEV2023): From Basic to Advanced Approaches, J. Extracell. Vesicles, 2024, 13(2), e12404, DOI:10.1002/jev2.12404.
- H. Wu, M. Fu, J. Liu, W. Chong, Z. Fang, F. Du, Y. Liu, L. Shang and L. Li, The Role and Application of Small Extracellular Vesicles in Gastric Cancer, Mol. Cancer, 2021, 20(1), 71, DOI:10.1186/s12943-021-01365-z.
- E. I. Buzas, The Roles of Extracellular Vesicles in the Immune System, Nat. Rev. Immunol., 2023, 23(4), 236–250, DOI:10.1038/s41577-022-00763-8.
- S. Samanta, S. Rajasingh, N. Drosos, Z. Zhou, B. Dawn and J. Rajasingh, Exosomes: New Molecular Targets of Diseases, Acta Pharmacol. Sin., 2018, 39(4), 501–513, DOI:10.1038/aps.2017.162.
- D. Badovinac, K. Goričar, H. Zavrtanik, M. Petrič, T. Lavrin, N. Mavec, V. Dolžan, A. Tomažič and M. Lenassi, Plasma Extracellular Vesicle Characteristics Correlate with Tumor Differentiation and Predict Overall Survival in Patients with Pancreatic Ductal Adenocarcinoma Undergoing Surgery with Curative Intent, J. Pers. Med., 2021, 11(2), 77, DOI:10.3390/jpm11020077.
- M. Oliveira-Rodríguez, S. López-Cobo, H. T. Reyburn, A. Costa-García, S. López-Martín, M. Yáñez-Mó, E. Cernuda-Morollón, A. Paschen, M. Valés-Gómez and M. C. Blanco-López, Development of a Rapid Lateral Flow Immunoassay Test for Detection of Exosomes Previously Enriched from Cell Culture Medium and Body Fluids, J. Extracell. Vesicles, 2016, 5, 31803, DOI:10.3402/jev.v5.31803.
- W. Shen, K. Guo, G. B. Adkins, Q. Jiang, Y. Liu, S. Sedano, Y. Duan, W. Yan, S. E. Wang, K. Bergersen, D. Worth, E. H. Wilson and W. Zhong, A Single Extracellular Vesicle (EV) Flow Cytometry Approach to Reveal EV Heterogeneity, Angew. Chem., Int. Ed., 2018, 57(48), 15675–15680, DOI:10.1002/anie.201806901.
- Y. Xue, X. Feng, X. Fan, G. Zhu, J. McLaughlan, W. Zhang and X. Chen, Extracellular Vesicles for the Diagnosis of Cancers, Small Struct., 2022, 3(1), 2100096, DOI:10.1002/sstr.202100096.
- F. Urabe, N. Kosaka, K. Ito, T. Kimura, S. Egawa and T. Ochiya, Extracellular Vesicles as Biomarkers and Therapeutic Targets for Cancer, Am. J. Physiol.: Cell Physiol., 2020, 318(1), C29–C39, DOI:10.1152/ajpcell.00280.2019.
- S. Dong, Y. Wang, Z. Liu, W. Zhang, K. Yi, X. Zhang, X. Zhang, C. Jiang, S. Yang, F. Wang and X. Xiao, Beehive-Inspired Macroporous SERS Probe for Cancer Detection through Capturing and Analyzing Exosomes in Plasma, ACS Appl. Mater. Interfaces, 2020, 12(4), 5136–5146, DOI:10.1021/acsami.9b21333.
- E. Schmelzer and L. M. Reid, EpCAM Expression in Normal, Non-Pathological Tissues, Front. Biosci., Landmark Ed., 2008, 13, 3096–3100, DOI:10.2741/2911.
- L. Huang, Y. Yang, F. Yang, S. Liu, Z. Zhu, Z. Lei and J. Guo, Functions of EpCAM in Physiological Processes and Diseases (Review), Int. J. Mol. Med., 2018, 42(4), 1771–1785, DOI:10.3892/ijmm.2018.3764.
- A. Nanou, L. L. Zeune, F.-C. Bidard, J.-Y. Pierga and L. W. M. M. Terstappen, HER2 Expression on Tumor-Derived Extracellular Vesicles and Circulating Tumor Cells in Metastatic Breast Cancer, Breast Cancer Res., 2020, 22(1), 86, DOI:10.1186/s13058-020-01323-5.
- M. Amorim, G. Fernandes, P. Oliveira, D. Martins-de-Souza, E. Dias-Neto and D. Nunes, The Overexpression of a Single Oncogene (ERBB2/HER2) Alters the Proteomic Landscape of Extracellular Vesicles, Proteomics, 2014, 14(12), 1472–1479, DOI:10.1002/pmic.201300485.
- K. Ekström, R. Crescitelli, H. I. Pétursson, J. Johansson, C. Lässer and R. Olofsson Bagge, Characterization of Surface Markers on Extracellular Vesicles Isolated from Lymphatic Exudate from Patients with Breast Cancer, BMC Cancer, 2022, 22(1), 50, DOI:10.1186/s12885-021-08870-w.
- Z. Li, Y. Gu, S. Ge, Y. Mao, Y. Gu, X. Cao and D. Lu, An Aptamer-Based SERS–LFA Biosensor with Multiple Channels for the Ultrasensitive Simultaneous Detection of Serum VEGF and Osteopontin in Cervical Cancer Patients, New J. Chem., 2022, 46(43), 20629–20642, 10.1039/D2NJ03567G.
- W. Lin, L. Yuan, Z. Gao, G. Cai, C. Liang, M. Fan, X. Xiu, Z. Huang, S. Feng and J. Wang, An Integrated Sample-to-Answer SERS Platform for Multiplex Phenotyping of Extracellular Vesicles, Sens. Actuators, B, 2023, 394, 134355, DOI:10.1016/j.snb.2023.134355.
- M. Okada-Tsuchioka, N. Kajitani, W. Omori, T. Kurashige, S. Boku and M. Takebayashi, Tetraspanin Heterogeneity of Small Extracellular Vesicles in Human Biofluids and Brain Tissue, Biochem. Biophys. Res. Commun., 2022, 627, 146–151, DOI:10.1016/j.bbrc.2022.08.025.
- J. Jablonska, M. Rist, I. Spyra, L. Tengler, M. Domnich, B. Kansy, B. Giebel, B. K. Thakur, N. Rotter, S. Lang and S. Ludwig, Evaluation of Immunoregulatory Biomarkers on Plasma Small Extracellular Vesicles for Disease Progression and Early Therapeutic Response in Head and Neck Cancer, Cells, 2022, 11(5), 902, DOI:10.3390/cells11050902.
- D. Kim, H.-K. Woo, C. Lee, Y. Min, S. Kumar, V. Sunkara, H.-G. Jo, Y. J. Lee, J. Kim, H. K. Ha and Y.-K. Cho, EV-Ident: Identifying Tumor-Specific Extracellular Vesicles by Size Fractionation and Single-Vesicle Analysis, Anal. Chem., 2020, 92(8), 6010–6018, DOI:10.1021/acs.analchem.0c00285.
- M. Oliveira-Rodríguez, E. Serrano-Pertierra, A. C. García, S. López-Martín, M. Yañez-Mo, E. Cernuda-Morollón and M. C. Blanco-López, Point-of-Care Detection of Extracellular Vesicles: Sensitivity Optimization and Multiple-Target Detection, Biosens. Bioelectron., 2017, 87, 38–45, DOI:10.1016/j.bios.2016.08.001.
- X. Diao, X. Li, S. Hou, H. Li, G. Qi and Y. Jin, Machine Learning-Based Label-Free SERS Profiling of Exosomes for Accurate Fuzzy Diagnosis of Cancer and Dynamic Monitoring of Drug Therapeutic Processes, Anal. Chem., 2023, 95(19), 7552–7559, DOI:10.1021/acs.analchem.3c00026.
- D. Vang, M. S. Kelly, M. Sheokand, M. Sharma, L. Esfandiari, R. I. Dima and P. Strobbia, Machine Learning Approaches in Label-Free Small Extracellular Vesicles Analysis with Surface-Enhanced Raman Scattering (SERS) for Cancer Diagnostics, bioRxiv, 2024, 22, 2024.02.19.581099. DOI:10.1101/2024.02.19.581099.
- S. Wang, A. Khan, R. Huang, S. Ye, K. Di, T. Xiong and Z. Li, Recent Advances in Single Extracellular Vesicle Detection Methods, Biosens. Bioelectron., 2020, 154, 112056, DOI:10.1016/j.bios.2020.112056.
- A. Görgens, G. Corso, D. W. Hagey, R. Jawad Wiklander, M. O. Gustafsson, U. Felldin, Y. Lee, R. B. Bostancioglu, H. Sork, X. Liang, W. Zheng, D. K. Mohammad, S. I. van de Wakker, P. Vader, A. M. Zickler, D. R. Mamand, L. Ma, M. N. Holme, M. M. Stevens, O. P. B. Wiklander and S. El Andaloussi, Identification of Storage Conditions Stabilizing Extracellular Vesicles Preparations, J. Extracell. Vesicles, 2022, 11(6), e12238, DOI:10.1002/jev2.12238.
- F. Royo, C. Théry, J. M. Falcón-Pérez, R. Nieuwland and K. W. Witwer, Methods for Separation and Characterization of Extracellular Vesicles: Results of a Worldwide Survey Performed by the ISEV Rigor and Standardization Subcommittee, Cells, 2020, 9(9), 1955, DOI:10.3390/cells9091955.
- D. Kozhevnikova, V. Chernyshev and A. Yashchenok, Progress in Isolation and Molecular Profiling of Small Extracellular Vesicles via Bead-Assisted Platforms, Biosensors, 2023, 13(7), 688, DOI:10.3390/bios13070688.
- A. Meggiolaro, V. Moccia, P. Brun, M. Pierno, G. Mistura, V. Zappulli and D. Ferraro, Microfluidic Strategies for Extracellular Vesicle Isolation: Towards Clinical Applications, Biosensors, 2022, 13(1), 50, DOI:10.3390/bios13010050.
- M. T. Yaraki, A. Tukova and Y. Wang, Emerging SERS Biosensors for the Analysis of Cells and Extracellular Vesicles, Nanoscale, 2022, 14(41), 15242–15268, 10.1039/D2NR03005E.
- L. K. Chin, T. Son, J.-S. Hong, A.-Q. Liu, J. Skog, C. M. Castro, R. Weissleder, H. Lee and H. Im, Plasmonic Sensors for Extracellular Vesicle Analysis: From Scientific Development to Translational Research, ACS Nano, 2020, 14(11), 14528–14548, DOI:10.1021/acsnano.0c07581.
- J. Penders, A. Nagelkerke, E. M. Cunnane, S. V. Pedersen, I. J. Pence, R. C. Coombes and M. M. Stevens, Single Particle Automated Raman Trapping Analysis of Breast Cancer Cell-Derived Extracellular Vesicles as Cancer Biomarkers, ACS Nano, 2021, 15(11), 18192–18205, DOI:10.1021/acsnano.1c07075.
- I. Badillo-Ramírez, S. A. J. Janssen, G. Soufi, R. Slipets, K. Zór and A. Boisen, Label-Free SERS Assay Combined with Multivariate Spectral Data Analysis for Lamotrigine Quantification in Human Serum, Microchim. Acta, 2023, 190(12), 495, DOI:10.1007/s00604-023-06085-3.
- S. Gullace, V. Montes-García, V. Martín, D. Larios, V. Girelli Consolaro, F. Obelleiro, G. Calogero, S. Casalini and P. Samorì, Universal Fabrication of Highly Efficient Plasmonic Thin-Films for Label-Free SERS Detection, Small, 2021, 17(33), 2100755, DOI:10.1002/smll.202100755.
- H. Wu, G. Niu, W. Ren, L. Jiang, J. Zhao, Y. Quan, M. X. Ren, W. Yu, Y. Zhang, X. Cao, Y. Liu, R. An, L. Dai, Z. Wang, L. Zhao, Z. Xie and G. Zhao, Highly Sensitive Label-Free Detection of Analytes at Different Scales Using Uniform Graphene-Nanopyramids Hybrid SERS System, Sens. Actuators, B, 2022, 354, 131205, DOI:10.1016/j.snb.2021.131205.
- K. Zhu, Z. Wang, S. Zong, Y. Liu, K. Yang, N. Li, Z. Wang, L. Li, H. Tang and Y. Cui, Hydrophobic Plasmonic Nanoacorn Array for a Label-Free and Uniform SERS-Based Biomolecular Assay, ACS Appl. Mater. Interfaces, 2020, 12(26), 29917–29927, DOI:10.1021/acsami.0c03993.
- Q. Zhang, R. Ma, Y. Zhang, J. Zhao, Y. Wang and Z. Xu, Dual-Aptamer-Assisted Ratiometric SERS Biosensor for Ultrasensitive and Precise Identification of Breast Cancer Exosomes, ACS Sens., 2023, 8(2), 875–883, DOI:10.1021/acssensors.2c02587.
- S. Hosseini, P. Vazquez-Villegas, M. Rito-Palomares and S. O. Martinez-Chapa, Advantages Disadvantages and Modifications of Conventional ELISA, in SpringerBriefs in Applied Sciences and Technology, 2018, pp. 67–115. DOI:10.1007/978-981-10-6766-2_5.
- X. Xie, W. Yu, L. Wang, J. Yang, X. Tu, X. Liu, S. Liu, H. Zhou, R. Chi and Y. Huang, SERS-Based AI Diagnosis of Lung and Gastric Cancer via Exhaled Breath, Spectrochim. Acta, Part A, 2024, 314, 124181, DOI:10.1016/j.saa.2024.124181.
- H. Shin, H. Jeong, J. Park, S. Hong and Y. Choi, Correlation between Cancerous Exosomes and Protein Markers Based on Surface-Enhanced Raman Spectroscopy (SERS) and Principal Component Analysis (PCA), ACS Sens., 2018, 3(12), 2637–2643, DOI:10.1021/acssensors.8b01047.
- Q. Zhang, T. Ren, K. Cao and Z. Xu, Advances of Machine Learning-Assisted Small Extracellular Vesicles Detection Strategy, Biosens. Bioelectron., 2024, 251, 116076, DOI:10.1016/j.bios.2024.116076.
- Y. Ding, Y. Sun, C. Liu, Q.-Y. Jiang, F. Chen and Y. Cao, SERS-Based Biosensors Combined with Machine Learning for Medical Application, ChemistryOpen, 2023, 12(1), e202200192, DOI:10.1002/open.202200192.
- E. Gómez-de-Mariscal, M. Maška, A. Kotrbová, V. Pospíchalová, P. Matula and A. Muñoz-Barrutia, Deep-Learning-Based Segmentation of Small Extracellular Vesicles in Transmission Electron Microscopy Images, Sci. Rep., 2019, 9(1), 13211, DOI:10.1038/s41598-019-49431-3.
- Y.-F. Qin, X.-Y. Lu, Z. Shi, Q.-S. Huang, X. Wang, B. Ren and L. Cui, Deep Learning-Enabled Raman Spectroscopic Identification of Pathogen-Derived Extracellular Vesicles and the Biogenesis Process, Anal. Chem., 2022, 94(36), 12416–12426, DOI:10.1021/acs.analchem.2c02226.
- H. Shin, B. H. Choi, O. Shim, J. Kim, Y. Park, S. K. Cho, H. K. Kim and Y. Choi, Single Test-Based Diagnosis of Multiple Cancer Types Using Exosome-SERS-AI for Early Stage Cancers, Nat. Commun., 2023, 14(1), 1644, DOI:10.1038/s41467-023-37403-1.
- B. Shan, Y. Pu, Y. Chen, M. Liao and M. Li, Novel SERS Labels: Rational Design, Functional Integration and Biomedical Applications, Coord. Chem. Rev., 2018, 371, 11–37, DOI:10.1016/j.ccr.2018.05.007.
- T.-D. Li, R. Zhang, H. Chen, Z.-P. Huang, X. Ye, H. Wang, A.-M. Deng and J.-L. Kong, An Ultrasensitive Polydopamine Bi-Functionalized SERS Immunoassay for Exosome-Based Diagnosis and Classification of Pancreatic Cancer, Chem. Sci., 2018, 9(24), 5372–5382, 10.1039/C8SC01611A.
- M. Jalali, C. del Real Mata, L. Montermini, O. Jeanne, I. Hosseini, Z. Gu, C. Spinelli, Y. Lu, N. Tawil, M. C. Guiot, Z. He, S. Wachsmann-Hogiu, R. Zhou, K. Petrecca, W. W. Reisner, J. Rak and S. Mahshid, MoS2-Plasmonic Nanocavities for Raman Spectra of Single Extracellular Vesicles Reveal Molecular Progression in Glioblastoma, ACS Nano, 2023, 17(13), 12052–12071, DOI:10.1021/acsnano.2c09222.
- Y. Li, X. Liu, J. Guo, Y. Zhang, J. Guo, X. Wu, B. Wang and X. Ma, Simultaneous Detection of Inflammatory Biomarkers by SERS Nanotag-Based Lateral Flow Assay with Portable Cloud Raman Spectrometer, Nanomaterials, 2021, 11(6), 1496, DOI:10.3390/nano11061496.
- J. U. Lee, W. H. Kim, H. S. Lee, K. H. Park and S. J. Sim, Quantitative and Specific Detection of Exosomal miRNAs for Accurate Diagnosis of Breast Cancer Using a Surface-Enhanced Raman Scattering Sensor Based on Plasmonic Head-Flocked Gold Nanopillars, Small, 2019, 15(17), 1804968, DOI:10.1002/smll.201804968.
- H. T. Ngo, H.-N. Wang, T. Burke, G. S. Ginsburg and T. Vo-Dinh, Multiplex Detection of Disease Biomarkers Using SERS Molecular Sentinel-on-Chip, Anal. Bioanal. Chem., 2014, 406(14), 3335–3344, DOI:10.1007/s00216-014-7648-4.
- H. He, Y. Zhang, S. Zhu, J. Ye and L. Lin, Resonant Strategy in Designing NIR-II SERS Nanotags: A Quantitative Study, J. Phys. Chem. C, 2022, 126(30), 12575–12581, DOI:10.1021/acs.jpcc.2c02512.
- G. McNay, D. Eustace, W. E. Smith, K. Faulds and D. Graham, Surface-Enhanced Raman Scattering (SERS) and Surface-Enhanced Resonance, Raman Scattering (SERRS): A Review of Applications, Appl. Spectrosc., 2011, 65(8), 825–837, DOI:10.1366/11-06365.
- L. Yang, J. Jia and S. Li, Advances in the Application of Exosomes Identification Using Surface-Enhanced Raman Spectroscopy for the Early Detection of Cancers, Front. Bioeng. Biotechnol., 2022, 9, 808933, DOI:10.3389/fbioe.2021.808933.
- X. Su, Y. Xie, X. Liu, M. Chen, C. Zheng, H. Zhong and M. Li, Absolute Quantification of Serum Exosomes in Patients with an SERS-Lateral Flow Strip Biosensor for Noninvasive Clinical Cancer Diagnosis, ACS Appl. Mater. Interfaces, 2023, 15(31), 37130–37142, DOI:10.1021/acsami.3c05039.
- X. Cao, Y. Sun, Y. Mao, M. Ran, Y. Liu, D. Lu and C. Bi, Rapid and Sensitive Detection of Dual Lung Cancer-Associated miRNA Biomarkers by a Novel SERS-LFA Strip Coupling with Catalytic Hairpin Assembly Signal Amplification, J. Mater. Chem. C, 2021, 9(10), 3661–3671, 10.1039/D0TC05737A.
- E. B. Bahadır and M. K. Sezgintürk, Lateral Flow Assays: Principles, Designs and Labels, TrAC, Trends Anal. Chem., 2016, 82, 286–306, DOI:10.1016/j.trac.2016.06.006.
- X. Su, X. Liu, Y. Xie, M. Chen, C. Zheng, H. Zhong and M. Li, Integrated SERS-Vertical Flow Biosensor Enabling Multiplexed Quantitative Profiling of Serological Exosomal Proteins in Patients for Accurate Breast Cancer Subtyping, ACS Nano, 2023, 17(4), 4077–4088, DOI:10.1021/acsnano.3c00449.
- K. Kim, L. Kashefi-Kheyrabadi, Y. Joung, K. Kim, H. Dang, S. G. Chavan, M.-H. Lee and J. Choo, Recent Advances in Sensitive Surface-Enhanced Raman Scattering-Based Lateral Flow Assay Platforms for Point-of-Care Diagnostics of Infectious Diseases, Sens. Actuators, B, 2021, 329, 129214, DOI:10.1016/j.snb.2020.129214.
- X. Fu, J. Wen, J. Li, H. Lin, Y. Liu, X. Zhuang, C. Tian and L. Chen, Highly Sensitive Detection of Prostate Cancer Specific PCA3 Mimic DNA Using SERS-Based Competitive Lateral Flow Assay, Nanoscale, 2019, 11(33), 15530–15536, 10.1039/c9nr04864b.
- W. C. Mak, V. Beni and A. Turner, Lateral-Flow Technology: From Visual to Instrumental, TrAC, Trends Anal. Chem., 2016, 79, 297–305, DOI:10.1016/j.trac.2015.10.017.
- K. J. Land, D. I. Boeras, X.-S. Chen, A. R. Ramsay and R. W. Peeling, REASSURED Diagnostics to Inform Disease Control Strategies, Strengthen Health Systems and Improve Patient Outcomes, Nat. Microbiol., 2019, 4(1), 46–54, DOI:10.1038/s41564-018-0295-3.
- R. Xiao, L. Lu, Z. Rong, C. Wang, Y. Peng, F. Wang, J. Wang, M. Sun, J. Dong, D. Wang, L. Wang, N. Sun and S. Wang, Portable and Multiplexed Lateral Flow Immunoassay Reader Based on SERS for Highly Sensitive Point-of-Care Testing, Biosens. Bioelectron., 2020, 168, 112524, DOI:10.1016/j.bios.2020.112524.
- V. Tran, B. Walkenfort, M. König, M. Salehi and S. Schlücker, Rapid Quantitative, and Ultrasensitive Point-of-Care Testing: A Portable SERS Reader for Lateral Flow Assays in Clinical Chemistry, Angew. Chem., Int. Ed., 2019, 58(2), 442–446, DOI:10.1002/anie.201810917.
- L. Y. Yeo, H.-C. Chang, P. P. Y. Chan and J. R. Friend, Microfluidic Devices for Bioapplications, Small, 2011, 7(1), 12–48, DOI:10.1002/smll.201000946.
- L. Wang, M.-M. Pan, L. Xu, X. Yu and S. Zheng, Recent Advances of Emerging Microfluidic Chips for Exosome Mediated Cancer Diagnosis, Smart Mater. Med., 2021, 2, 158–171, DOI:10.1016/j.smaim.2021.06.001.
- Z. Han, X. Peng, Y. Yang, J. Yi, D. Zhao, Q. Bao, S. Long, S.-X. Yu, X.-X. Xu, B. Liu, Y.-J. Liu, Y. Shen and L. Qiao, Integrated Microfluidic-SERS for Exosome Biomarker Profiling and Osteosarcoma Diagnosis, Biosens. Bioelectron., 2022, 217, 114709, DOI:10.1016/j.bios.2022.114709.
- W. Zhang, L. Wang, D. Li, D. H. Campbell, B. J. Walsh, N. H. Packer, Q. Dong, E. Wang and Y. Wang, Phenotypic Profiling of Pancreatic Ductal Adenocarcinoma Plasma-Derived Small Extracellular Vesicles for Cancer Diagnosis and Cancer Stage Prediction: A Proof-of-Concept Study, Anal. Methods, 2022, 14(23), 2255–2265, 10.1039/D2AY00536K.
- W. Lee, A. Nanou, L. Rikkert, F. A. W. Coumans, C. Otto, L. W. M. M. Terstappen and H. L. Offerhaus, Label-Free Prostate Cancer Detection by Characterization of Extracellular Vesicles Using Raman Spectroscopy, Anal. Chem., 2018, 90(19), 11290–11296, DOI:10.1021/acs.analchem.8b01831.
- E.-K. Vetsika, P. Sharma, I. Samaras, A. Markou, V. Georgoulias, T. L. Whiteside and A. Kotsakis, Small Extracellular Vesicles in Pre-Therapy Plasma Predict Clinical Outcome in Non-Small-Cell Lung Cancer Patients, Cancers, 2021, 13(9), 2041, DOI:10.3390/cancers13092041.
- S. Rodríguez Zorrilla, M. Pérez-Sayans, S. Fais, M. Logozzi, M. Gallas Torreira and A. García García, A Pilot Clinical Study on the Prognostic Relevance of Plasmatic Exosomes Levels in Oral Squamous Cell Carcinoma Patients, Cancers, 2019, 11(3), 429, DOI:10.3390/cancers11030429.
- J. Langer, D. Jimenez de Aberasturi, J. Aizpurua, R. A. Alvarez-Puebla, B. Auguié, J. J. Baumberg, G. C. Bazan, S. E. J. Bell, A. Boisen, A. G. Brolo, J. Choo, D. Cialla-May, V. Deckert, L. Fabris, K. Faulds, F. J. García de Abajo, R. Goodacre, D. Graham, A. J. Haes, C. L. Haynes, C. Huck, T. Itoh, M. Käll, J. Kneipp, N. A. Kotov, H. Kuang, E. C. Le Ru, H. K. Lee, J.-F. Li, X. Y. Ling, S. A. Maier, T. Mayerhöfer, M. Moskovits, K. Murakoshi, J.-M. Nam, S. Nie, Y. Ozaki, I. Pastoriza-Santos, J. Perez-Juste, J. Popp, A. Pucci, S. Reich, B. Ren, G. C. Schatz, T. Shegai, S. Schlücker, L.-L. Tay, K. G. Thomas, Z.-Q. Tian, R. P. Van Duyne, T. Vo-Dinh, Y. Wang, K. A. Willets, C. Xu, H. Xu, Y. Xu, Y. S. Yamamoto, B. Zhao and L. M. Liz-Marzán, Present and Future of Surface-Enhanced Raman Scattering, ACS Nano, 2020, 14(1), 28–117, DOI:10.1021/acsnano.9b04224.
- X. Zhang, T. Takeuchi, A. Takeda, H. Mochizuki and Y. Nagai, Comparison of Serum and Plasma as a Source of Blood Extracellular Vesicles: Increased Levels of Platelet-Derived Particles in Serum Extracellular Vesicle Fractions Alter Content Profiles from Plasma Extracellular Vesicle Fractions, PLoS One, 2022, 17(6), e0270634, DOI:10.1371/journal.pone.0270634.
- Y.-S. Chen, Y.-D. Ma, C. Chen, S.-C. Shiesh and G.-B. Lee, An Integrated Microfluidic System for On-Chip Enrichment and Quantification of Circulating Extracellular Vesicles from Whole Blood, Lab Chip, 2019, 19(19), 3305–3315, 10.1039/C9LC00624A.
- X. Gao, P. Zheng, S. Kasani, S. Wu, F. Yang, S. Lewis, S. Nayeem, E. B. Engler-Chiurazzi, J. G. Wigginton, J. W. Simpkins and N. Wu, Paper-Based Surface-Enhanced Raman Scattering Lateral Flow Strip for Detection of Neuron-Specific Enolase in Blood Plasma, Anal. Chem., 2017, 89(18), 10104–10110, DOI:10.1021/acs.analchem.7b03015.
- M. Konoshenko, G. Sagaradze, E. Orlova, T. Shtam, K. Proskura, R. Kamyshinsky, N. Yunusova, A. Alexandrova, A. Efimenko and S. Tamkovich, Total Blood Exosomes in Breast Cancer: Potential Role in Crucial Steps of Tumorigenesis, Int. J. Mol. Sci., 2020, 21(19), 7341, DOI:10.3390/ijms21197341.
- N. Karimi, R. Dalirfardouei, T. Dias, J. Lötvall and C. Lässer, Tetraspanins Distinguish Separate Extracellular Vesicle Subpopulations in Human Serum and Plasma - Contributions of Platelet Extracellular Vesicles in Plasma Samples, J. Extracell. Vesicles, 2022, 11(5), e12213, DOI:10.1002/jev2.12213.
| This journal is © The Royal Society of Chemistry 2025 |