Open Access Article
Open Access ArticleTowards a classification strategy for complex nanostructures
V.
Castagnola
a,
J.
Cookman
a,
J. M.
de Araújo
ab,
E.
Polo
a,
Q.
Cai
a,
C. P.
Silveira
a,
Ž.
Krpetić
 ac,
Y.
Yan
a,
L.
Boselli
ac,
Y.
Yan
a,
L.
Boselli
 *a and
K. A.
Dawson
*a
*a and
K. A.
Dawson
*a
aCentre for BioNano Interactions, School of Chemistry, University College Dublin, Belfield, Dublin 4, Ireland. E-mail: kenneth.a.dawson@cbni.ucd.ie; luca.boselli@cbni.ucd.ie
bDepartamento de Física Teórica e Experimental, Universidade Federal do Rio Grande do Norte, 59078-970, Natal, RN, Brazil
cSchool of Environment and Life Sciences, University of Salford Manchester, Salford, M5 4WT, UK
First published on 13th March 2017
Abstract
The range of possible nanostructures is so large and continuously growing, that collating and unifying the knowledge connected to them, including their biological activity, is a major challenge. Here we discuss a concept that is based on the connection of microscopic features of the nanomaterials to their biological impacts. We also consider what would be necessary to identify the features that control their biological interactions, and make them resemble each other in a biological context.
1 Introduction
In the generations to come, mankind will be able to systematically make, characterise, and harness for use a great variety of nanoscale objects of different sizes, surfaces and shapes. The capacity to engineer at this scale will be of some importance for a very broad range of applications. Besides that, new approaches for engineering hard-materials ranging from orthopaedic implants to car tyres, and new processing scenarios, such as 3D printing and nanolithography, will lead to new varieties of shape or process-induced particles differing from those found in naturally occurring dusts. The future therefore involves many more forms of nanoparticles, all in contact with the living world, than those that have been discussed in recent years.We consider that the future will be marked by an increasing diversity of nanoparticle structures, including different surfaces, shapes, and topologies. While it has been striking just how little conventional short-term (acute) “toxicity” is associated with the vast majority of particles studied to date,1 clearly caution in the face of radically new scenarios is still wise. Furthermore, we fully expect, and our own experience suggests, that these novel forms will exhibit interesting biological properties, and that these will in many cases lead to important and useful medical applications.
To undertake investigations of such an extensive arena, future scientists will also consider not just the practical importance of the topic, but how it is positioned within the tapestry of conceptual and fundamental science. Succinctly put, is it important, and will it lead to new phenomena and ideas? We believe it will, partly based on our own experience, as well as the scientific context defined by the size.
Thus crucially, objects on the nanoscale engage with living systems in an entirely different manner to either small molecules, or large (micron sized) particles.2–5 Engineered particles being of some tens of nanometers are distinctive because they can engage with essentially all endogenous active biological processes, potentially leading to diverse complex biological outcomes.
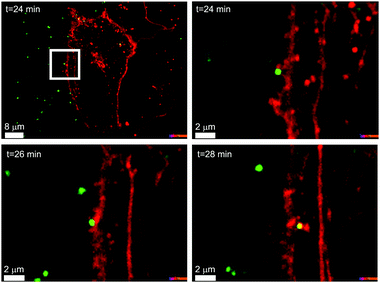
In Fig. 1 we can see an example of a common polystyrene nanoparticle (green) passing across the cell membrane (red), using the energy supplied by the cell, after biomolecules adsorbed on the particle surface have been recognised by specific cell receptors, rather than the non-specific diffusional processes by which the majority of small molecules enter cells.6–8 This example is one of many, and living organisms are usefully viewed as the most complex and effective nanoscale processing devices in existence. Whatever we, in our generation, are able to achieve, the capacity to engineer objects on the size scale of such biological phenomena clearly defines a unique new frontier that will grow and deepen across generations.
Now, our capacity to engineer NPs with different sizes, shapes and surfaces ensures that within cells, barriers and organs, a new universe of objects, some of them with surfaces and shapes never previously exposed to living organisms, will be brought into contact with the sophisticated biological processing machinery that can analyse them and respond to them using an extensive repertoire of pre-existing biological “pathways”, essentially biological processing subroutines. We do not know the boundaries and capacities of that endogenous processing machinery, when it confronts such novel scenarios.
The choices in making nanoparticles are so extensive and, unlike molecules, not governed by the rules and constraints of chemical bonding as to be almost limitless in composition and structure. This is rather akin to the situation with snowflakes: structures often grow by kinetic control, allowing for a range of shapes and variations that make it difficult to systematically name them, rather than simply showing pictures of them.
Thus, in practical terms, many new nanoscale objects are being reported in the literature or experienced in the living world without any form of categorisation, cross referencing to previous structures, or systematic naming strategy. There is no means to even check if such a structure has been identified and studied before. Therefore the basic desire would be that manufacturers of new nanoparticles could immediately identify if similar structures (according to the material, size, shape, and other physical features) have previously been made, if for no other reason than to avoid naming similar materials by different names.
If this process of systematisation could also support the progressive unveiling of the connection between the relevant (microscopic) control parameters of these nanomaterials and biological impacts, this would focus our energy on the drive for positive safe nanotechnology developments, and new health-promoting products. While fully recognising the magnitude of the challenge, these are compelling reasons now to address the whole question of categorisation, in a manner that intersects with the emerging understanding of biological interactions.
1.1 Categorisation and classification
We have stressed the idea that, at the nanoscale, biological interactions occur by active biological processes, intended to process naturally occurring biomolecules. However, current discussions about nanomaterial classification take place in a context where nanomaterials are under particular scrutiny for their potential to improve medical therapeutics9–11 and diagnostics,12,13 as well as their risk of exposure to ecosystems.14,15 Regulatory bodies, industry, infrastructures and institutions, aware of potential exposure risks, have maintained a watch on both risks and benefits of new nanotechnology. Therefore there is a need to understand, harness, and systematise the emerging scientific information that could impinge on these fields.Before beginning the discussion we should briefly consider the concepts of categorisation, identity, and similarity, as well as the fundamental purpose and drivers for the effort. We must clearly differentiate between a desire to (a) identify and parameterise nanoparticles (e.g., surface and shape) that enables them to be “searchable” in databases, and (b) organise and classify using key parameters that control, for example, important biological responses. The former is a typical challenge in any Internet-based search or match for similarity categorisation, possibly based on a few simple qualities. The meaning and complexity of this can be illustrated with the following analogy: motorcars might be identified as being in the same category quite readily from their images, but, for example, vacuum cleaners (given the novelty of design of several different recent versions) may not be, as they can look quite different. In the case of image-only-based searches for vacuum cleaners we would have to be satisfied to recognise those that are currently typical. On the other hand, if we genuinely seek to build in a deeper contextualisation of the data (for example, “items that use a vacuum to clean floors”) then an overall analysis of the physical form of the device may not be sufficient. For vacuum cleaners this is easy, because the manufacturers tell us clearly what the purpose is. Translating these considerations and concepts to nanoparticles is challenging because when a new nanoparticle type is made, the purpose and the context might not yet be clear.
On the other hand if we decide to add context to the nanoparticle concept of categorisation, by making a long list of properties, there is no reason to suppose this will lead to useful insights. Our perspective will be that we wish to capture key (microscopic) information from nanoparticles that could enable a connection to biological impacts. By analogy with the car, we do not wish to catalogue every bolt and plate used to manufacture it, nor simply to hope to recognize its purpose on the Internet from its “form”. Instead we wish to capture granular information (for example, it has four wheels, a steering wheel, pistons and cylinder) that is central to how it acts in meaningful environments. In our case this means that we are not seeking to describe it at a single atomic, molecular level, or by a list of “properties” but by semi-microscopic features that are likely to define its biological interactions. This allows us to “name” the object based on those features, and also to promote the meaningful link between its form and biological processes it induces, all within the same framework. For example, we will argue that features derived from particle shape, and particular biological recognition motifs presented at the surface have a similar status to the “wheels and pistons” of a car.
We note that quite different (non-microscopic) proposals have been published in an attempt to bring a generalised nomenclature system to nanostructures16,17 in addition to early classification systems.18–20 So far, classification of nanomaterials sometimes begins with their size definition, i.e. a nanomaterial is routinely defined as a material containing particles or constituents of nanoscale dimensions, in the size range from 1–100 nm.21–23 An early attempt to systematise the nomenclature of nanomaterials was based on a typographic string of minimalist hierarchical codes aimed at facilitating digital archiving and search strings for specific properties that address composition, size, shape and physicochemical properties.16 This schematic code includes the following five fields: chemical class, size and shape, core chemistry, ligand chemistry, and solubility. Efforts to address issues of toxicology based on physicochemical properties introduced the Minimal Information About Nanomaterials (MIAN) for physicochemical characteristics established by the Nanomaterial Registry.19,24 In parallel, progress has been made to further characterise nanomaterials for general purposes by organisations such as the Organisation for Economic Co-operation and Development (OECD), the International Organization for Standardization (ISO), the Food and Drug Administration (FDA), and the National Cancer Institute (NCI).10,17,25 These proposals are the best efforts so far in the midst of highly uncertain current knowledge of the real material control factors for biological outcomes. One cannot expect a single final broad and inclusive approach to categorisation to emerge for some time to come, so these are all useful contributions in an era when those discussions are being progressed. Thus, we propose to offer a “horizon” perspective on a quasi-microscopic conception of the arena, and show how that might grow over the coming decade.
Herein, we wish to illustrate a form of categorisation based on molecular and microscopic principles. Such considerations will link more directly to the biological interactions and biological processes, and the connections between them. We do not seek to minimise the long-term challenges in such a project, but stress that it does have the merit of providing a durable focus and purpose in understanding the biological impact of these materials, which could have many useful outcomes, besides the process of categorisation itself.
It is difficult to be very definitive, but in our discussion we have in mind particles that do not dissolve or degrade too rapidly so that their size, shape and surface coating (both manufactured and acquired from the environment) are relatively fixed for some time. Roughly speaking such particles carry their shape and key elements of their surface presentation into the cell where many are ultimately deposited in degradative organelles such as lysosomes or phagosomes.26 The soft outer surface may then be degraded within five hours, but other aspects of the particle form may be degraded only over weeks, months or years.27 Importantly, the initial interactions governed by molecular features at the interface between nanoparticles and biological systems strongly shape the biological properties of nanomaterials. For instance, in the first hours, the bio-distribution is determined, and immunological responses (e.g. inflammatory response) are initiated. Later on, as the whole particle degrades, it will be necessary to go much deeper into the molecular identities of the degradation products.
1.2 Current context of particle architecture
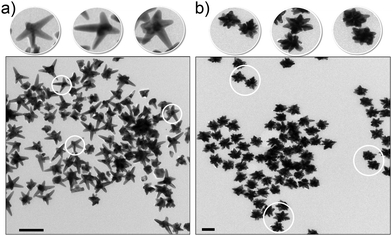
The way in which nanomaterials are made and reported is qualitatively different from chemicals. For instance, in synthesis, the promotion of one crystal plane growth and the slowing of another (using surface active agents for example) typically leads to non-spherical objects,28–35 and understanding how to control such parameters leads to increasingly rich structures. To clarify the magnitude of the problem, we remark that even fine-tuning the plasmon resonance of metallic gold nanoparticles can lead to creating a number of subtle variants of the particle shape that are often described with a large variety of arbitrary and increasingly exotic names. A few examples reported in the literature (Fig. 2) are nanodisco balls,36 describing the complex nanoarchitectures obtained by the self-assembly of spherical polymeric nanoparticles and nanocrystals, branched nanoparticles37,38 are often reported as nanourchins,39 nanoflowers,40–42 daisy shaped43 or star shaped29,44 and asymmetric silver nanoparticles are reported as nanocarrot structures.45 Sometimes the shape morphology given by specific surface modification is described as a nano-fruit texture, for example, nano-raspberries and nano-cranberries.46 The need to name these in this interim fashion is entirely unavoidable. However, these examples represent only a tiny part of the structures that scientists are currently, or soon will, fabricate using only metals.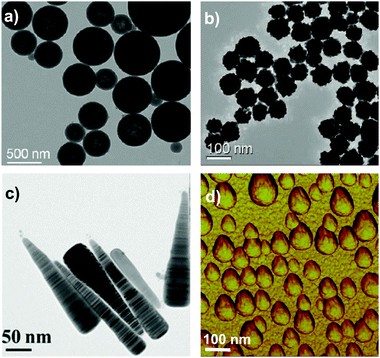 | ||
| Fig. 2 Examples of exotic shapes of NPs in the literature: (a) polymeric nanodisco balls (TEM micrograph),36 (b) gold nanoflowers (TEM micrograph),42 (c) silver nanocarrots (TEM micrograph),45 (d) polymeric nanoraspberries (phase AFM image).46 Reprinted with permission from American Chemical Society. | ||
Using different materials, there is a huge variety of potential structures and compositions, and the choice is limited only by the imagination. Moreover, their composition may not be uniform, and for many purposes different core materials have to be considered. The variety of materials used in these structures has implications for the biological impact. Some may leach free ions (that are themselves toxic) and have multiple biological actions.47,48 Examples include some quantum dots, and lead-based perovskites (film photovoltaic devices/solar cells).49,50 Leeching is the simplest form of biological processing and (for example liver) accumulation may lead to degradation, and the formation of secondary metabolites, all of which would need to be accommodated in a full (biologically relevant) molecular description of the nanomaterial.
These will all have to be addressed in due course, but as a first step we wish to illustrate how a form of biologically relevant categorisation could emerge based on key microscopic control parameters that are currently emerging. These are parameters that link directly to molecular or microscopic structural properties, and are currently believed to have a degree of universality in their impact on the biology, irrespective of the nature of the material. We are not yet sure we know all of them, but it is clear that the list includes size, shape, and surface presentation of the nanomaterial, and that these fix certain key control parameters affecting the biology.
2 Key microscopic control parameters for complex nanostructure parametrisation
We now have ample evidence that at least size, shape, and surface presentation impacts on the biological processes induced by nanoparticles. In understanding how to link properties to outcomes, one well-established approach is to form a reference library of objects in which those key parameters are evolved, and study the biological outcome. This is indeed one of the approaches we have taken. Thus, in Fig. 3a we show a partial catalogue of complex nanostructures (the “nanoparticle library”) developed within our laboratory. While such structures can have an almost limitless possibility of combinations, even for the limited set between size, shape and surface modifications, as summarised in Fig. 3b, we are attempting to distil the actual collection into a smaller set that highlights structural changes believed to induce qualitatively new (from spherical) biological processes, thereby illustrating the concept of key control parameters. The names of the particles (found in the caption) are related to their geometrical appearance whilst observing them by Transmission Electron Microscopy (TEM) and we remind the reader that all of these descriptions are somewhat idealised, because not all particles in a typical sample are identical. The majority of the general discussion is therefore centred on the typical, average, or “idealised” nanoparticle in the sample. This concept of diverse objects, and sample-containing “outliers” will be further discussed in the following section.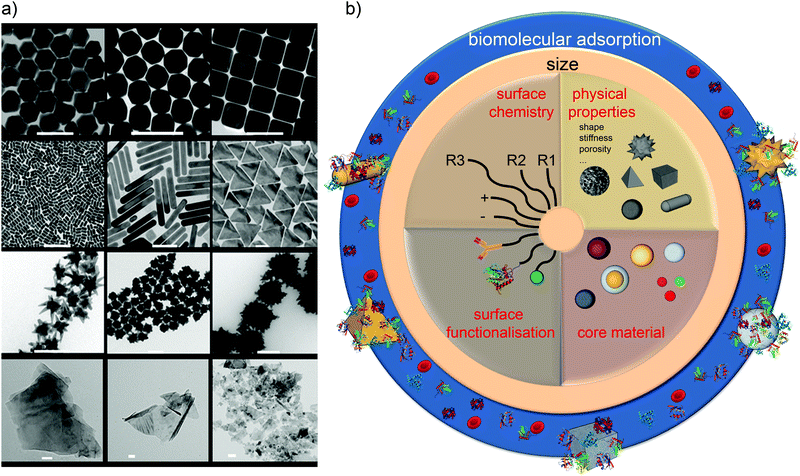 | ||
| Fig. 3 (a) Transmission Electron Microscopy (TEM) micrographs of Nanoparticles Shape Library developed within CBNI.51 From the top left: gold nanotrisoctahedra, faceted gold nanorods (top view), gold nanocubes, gold nanorods, gold nanoprisms, gold nanostars and gold urchin-like, few layer graphene flakes and commercial nanodiamonds. Scale bars are 100 nm. (b) The scheme summarises the multitude of possibilities in terms of the combination of size, core material, shape and other physical properties, surface chemistry, and functionalisation which will be further modified by the biological environment. | ||
2.1 Nanoparticle samples containing diverse objects with different individual properties
Before beginning, it is important to clarify a central message: individual nanoparticles interact with individual cells. The concept of an “average nanoparticle”, or indeed an “average cell” may be relevant if all nanoparticles interact with all cells in much the same way. However, all batches of nanomaterials made by current means, no matter how high their quality, are fundamentally samples containing huge numbers of somewhat different nanoparticles. Furthermore, some samples can contain a relatively small number of “exceptionally” shaped, or otherwise organised nanoparticles (Fig. 4). For their overall functional properties (whether in devices or consumer products) this presents no problems of any kind. On the contrary, tiny levels of “outliers” in, for example, surface, degree of crystallinity, or shape, are quite irrelevant, in much the same manner that small levels of chemical contaminants are irrelevant for the required function. The question here is not quite so simple. If a relatively small number of exceptional particles exist in a sample, with a highly specific shape or other notable properties that could lead to cellular damage then, because of the potential for damaged cells to multiply and amplify the effects (for example in tumours), then they could be significant. We note that there is at present no substantive reported case where this could raise concern. However, we noted that this article constitutes an attempt to clarify understanding and to support the framing of a long-term strategy. Small numbers of non-degradable, unclear nanoparticles could reside for long durations in novel biological contexts, and it would be wise to keep that in mind, albeit without undue emphasis, in the framing of a general strategy.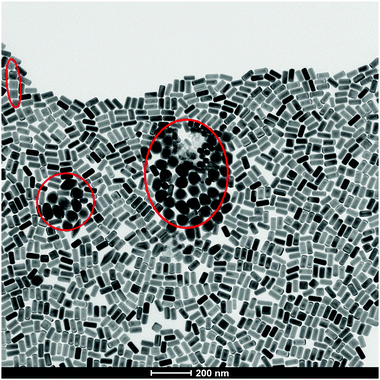 | ||
| Fig. 4 TEM micrograph of “exceptionally” shaped nanoparticles (red circles) in the same batch of seemingly monodisperse gold nanorods. | ||
It is also worth commenting that characterisation of complex nanostructures has presented significant challenges, but some progress is now being made. Most characterisation methods in dispersions (e.g. dynamic light scattering – DLS, and differential centrifugal sedimentation – DCS and others) involve measurements of relaxation time, from which one spatial dimension can be determined but have limitations to reveal complex features. The development of new techniques such as the Single Particle Scattering and Extinction technique (SPES)52,53 and Analytical Ultracentrifugation (AUC)54 especially with multi wavelength functionality (MLW-AUC)55 has broadened our capacity in analysing the complex shapes and size range of nanomaterials in dispersion. Combining these advanced techniques with more routine instrumentation (DCS and DLS) and high resolution imaging (High Resolution-Transmission Electron Microscopy, Scanning TEM) we can obtain a higher level of characterisation and shape recognition that will soon be indispensable.
Now, assuming that one has a set of particles in which key control parameters have been evolved, one must still be able to capture and quantify those control parameters on a scale that is meaningfully related to the biology. The degree to which current methods of characterisation, and those being developed, can link directly to key control parameters will be an important consideration for the future strategy in the development of nanoparticle characterisation technologies, and it is by no means clear that methods becoming available are sufficient to do so. To some degree one must accept that all of the developments supporting these efforts will be iterative. As we identify key control parameters, we will need to investigate the modes of characterisation necessary to capture these quantities. In the interim, we believe that some broad approaches, such as ones involving shape digitisation of images discussed below, are proving very fruitful in capturing the necessary information from which critical parameters can be identified.
2.2 Capture of key control parameters: shape digitisation
Unlike for simple spherical nanomaterials, there are no obvious means to describe the particle form or shape. For nanoparticles with well-known geometrical shape (rods, cubes) the classification and naming can appear straightforward, but on the other hand, more complex anisotropic or branched nanostructures require special geometrical descriptors. Cubic or spherical nanoparticles can be easily identified and classified based on classic geometry while the so-called “nanostars” represent a variety of complex shapes with several similarities (Fig. 5).One approach is therefore to capture the structures in digital form so that the forms can be analysed, stored and processed, and key control parameters progressively identified from them. Mathematical quantities derived from their shape can be computed directly, and hypotheses of “search driven” efforts made to relate those to biological impacts. This is not a simple question of characterisation, but of sufficiently resolved information to ensure that key parameters are encompassed. The simplest and most generally applicable method of determining shape is electron microscopy, applied to sufficient numbers of particles to form a statistical basis.56 In the case of complex nanoparticles and nanoparticle assemblies it is possible to capture the 3D model of particles from TEM-based tomography using image reconstruction processes.57 Systematic construction of an extensive collection of wire-frame models of complex nanoparticles can then be used to yield a database of descriptors for each particle kind using algorithms to process the individual samples in a high throughput fashion.58–60
2.3 The importance of the nanoparticle surface to biological impacts
The nature of the “bare” nanoparticle surface can be extraordinarily complex, and certainly it is this “bare” surface that will be important in understanding long-term effects when coatings have been degraded off the surface within biological subsystems such as phagosomes and lysosomes. However, we are now essentially certain that the early stages of biological response for materials depends on what is presented on the surface2,61,62in situ rather than only the original bare surface. Molecules, especially biomolecules derived from the biological milieu, such as blood and the lining of the lungs, are adsorbed onto the surface (see scheme in Fig. 6a). If this does not occur, then, of course, efforts to study particle interactions with cells will lead to spurious and damaging interactions due to the high energy surface disrupting the cell.2,63 Thus, for cell level studies, the simplest role of the corona coating is to ensure sensible exposure conditions for cells. Recent innovations now allow the recognition motifs presented at the surface of nanoparticles to be fully mapped out using immuno-reporters and other simple and inexpensive devices.64 This means that a rather complete description of the in situ surface may be given.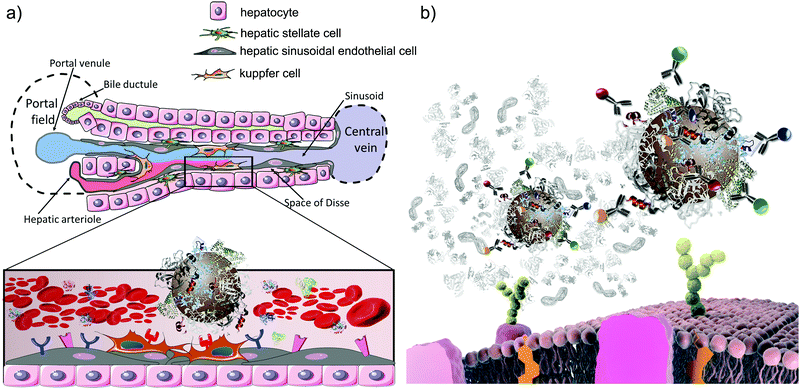 | ||
| Fig. 6 (a) Schematic representation of the liver architecture and zoomed-in view of the interaction between the nanoparticle–corona complex and cell receptors in the liver sinusoid; Kupffer cells express a multitude of receptors able to selectively recognise specific epitopes on the nanoparticle surface. (b) Schematic representation of the in situ immuno quantum dot epitope mapping approach.92 The identified epitopes’ presentation can be then linked to specific receptor-mediated uptake pathways (adapted from Monopoli et al.).93 | ||
It is essential to stress that simply nanoparticle adsorbed molecules may not themselves induce a specific biological response, but for molecules with specific biological functions such as receptor recognition motifs, such functions can be preserved and amplified when presented on the nanoparticle surface. Also, other value chain modifications of the nanoparticle could be of importance either by being the source of some new biological outcomes or by modifying the adsorbed coronas. Thus, if biologically active molecules from the environment adsorb onto the surface, this could be important. A simple example is the case of lipopolysaccharide (LPS) derived from bacterial debris and widely present in the environment. This is a biologically active molecule that, when presented on the nanoparticle surface could lead to a high level of biological activity.65 Succinctly put, when nano-objects are presented to living organisms, the sum of all (particularly bio-active) molecules presented on the outer surface, whether from the value chain or from body entry by different routes, is a key factor to understanding early biological impacts.61,66–72
Most experience so far suggests that the majority of biologically active molecules (apart from LPS and other microbial components) on the surface of nanoparticles in vivo are derived after exposure to the endogenous biomolecules of the organism itself, rather than derived from the broader environment. In any case, the methods to screen for surface presentation of molecules are becoming so inexpensive and simple that it is practical to screen for a huge variety of outcomes, and identify if there are any other (biologically important) surface anomalies that are derived from the broader environment.
In summary, we are now essentially certain that (besides size) the shape and in situ presentation of biomolecules at the surface of nanoparticles are key control parameters for biological impacts. Furthermore, methods to study these are now available, or emerging, and it is thus feasible to show how a categorisation approach could be built on this kind of input. We stress that this does not exclude many other known (for example degradation or dissolution) effects or as yet unknown parameters. It merely begins the process of organising the data around what we are most confident in.
3 Biological pathways as the fundamental biological targets
The enormous variety of possibilities for nanoparticle properties and the difficulty in condensing those into clearly defined key control parameters are matched by the complexity of biological processes that could be affected by nanoparticles.One broad approach, based on our own experience and accumulated knowledge from different disciplines, suggests that part of the effort could be focused on collecting and systematically rationalising the activation of the currently known biological pathways by nanomaterials, and connecting them to specific features of the nanoparticles.7,73–79 Such pathway maps already comprise a high level of widely available accumulated knowledge on the potential biological targets of the system. Briefly, these pathways constitute a basic grouping of the ‘subroutines’ of a cell (or cell assembly) that are required to make the cell function, elements of which can be up-regulated, or down-regulated in the context of an external perturbation.
While far from a complete description of all biological events, they constitute a well-organised body of information that has developed over time about the connected biological processes that make cells function. Thus, knowledge of the impact of nanomaterials on these pathways will suggest many of the largest and most significant biological outcomes.
If one finds that different particles activate predominantly similar biological actions, or groups of pathways, then it will be natural to ask if those similarities in activation of pathways are based on a common material property, possibly as yet unidentified. If so, this will progressively unveil the detailed features of nanomaterials that represent “commonalities” between particles. This idea is closely related to the capacity to “read-across” mentioned earlier, between the known impacts of one set of materials to the likely outcomes of another, and how to apply this knowledge in a systematic way. Such an approach to systematising the biological impact is far from complete, but fits rather well in seeking to understand the microscopic connection between biological processes and key control parameters.
3.1 Hypothesis driven exploration of nanoparticle-biological impacts
For some nanoparticle key parameters (shape and surface presentation) we can readily hypothesise the involvement of specific pathways, and even potentially suggest higher level in vivo impacts. One example is the connection of surface presentation of recognition motifs of nanoparticles in situ to specific biological pathways. To illustrate this, we focus on biological pathways of practical importance and show how surface presentation can be used to organise information about them. There is nothing special about our choice of example, and the same process can be carried out for many others.Thus, we will consider the issue of liver accumulation, which is important in understanding the toxicity of nanomaterials, both for general safety, and for medical applications.80–82 The role of the liver is to monitor the bloodstream for the presence of undesirable foreign materials, which it may then remove and degrade. Evidently, if it removes more foreign material than it is designed to accommodate, or if that foreign material degrades to produce new toxins, this could lead to liver damage. For medical applications, it is usually desirable to evade the liver, as one is targeting other locations. Thus determining and categorising the likely outcome of nanomaterials on the liver is an important task undertaken in any safety assessment, for all products, whether consumer or medicinal. Extensive knowledge of the liver architecture (Fig. 6a) suggests that the nature of the cells, and thereby the outer membrane receptors, leads to the accumulation of biomolecules and various debris that should be cleared. In practice, for example, there are less than twenty key receptors believed to dominate the process of receptor mediated liver accumulation. Thus, by matching the molecules presented at the surface of nanoparticles in situ (Fig. 6b) with the receptors (and associated pathways) on the surface of cells of the liver, we can suggest how those particles could interact with the liver, and via which mechanisms and processes. To illustrate, in a recent study we have shown that many forms of amorphous silica in human serum and plasma present recognition motifs for both low-density lipoprotein (LDL) and immunoglobulin G (IgG), suggesting that they will be recognised by liver using a mechanism designed to handle cholesterol and infections.64
Moreover, the molecular structures and the relative abundance of these surface biomolecules have been obtained through mass spectrometry and various proteomics, lipidomics, and glycomics, amongst other approaches,83–90 and the results of a more complete mapping exercise are now becoming available.91,92 These mapping methods now give information at the molecular level, establishing if all of the potential recognition motifs are actually expressed on the nanoparticle surface in an accessible manner to receptors. This now leaves us with a clear set of hypotheses that links the nature of the molecules on the surface to the impact on cells. It is significant that, by working at this level of description it now becomes meaningful to ask how “similar” (at these early timescales) nanoparticles are on the basis of their surface presentation of recognition motifs (“epitope maps”). This therefore provides an avenue to categorise them, at least in relation to such things as bio-distributions, early immunological processing, etc.
3.2 Databasing based on microscopic principles of interaction
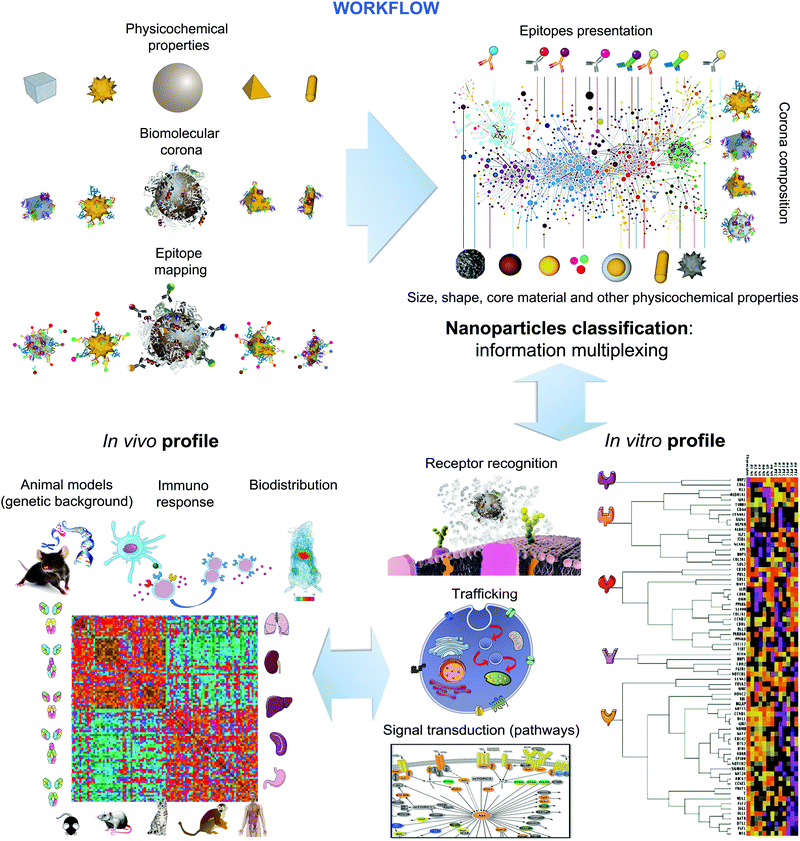
The use of microscopic and mesoscopic structural information and molecular (recognition) principles as the basis for linking properties to biological outcomes makes possible new ways of collecting and organising the information. Furthermore, the methods are available (for example epitope mapping and shape digitisation) to approach the set of tasks in a logical and systematic manner. While the approach does require significant effort, some of the methods are also relatively inexpensive, and in vivo is a final specific step, rather than the basis of the great body of knowledge. The workflow depicted in Fig. 7, can then be used to capture systematic knowledge, gathered in different laboratories, potentially ultimately providing the basis to predict the nature of the early stages of biological interaction on living organisms.This then constitutes a clear strategy and successful approach to both categorise and to read-across, for early stage impacts based on the nature of the surface of nanomaterials. Obviously such a full categorisation will also have to include any other major overarching effects, one of which we believe to be shape, as well as any specific properties of interest for a specific material.
4. Conclusions
There is little doubt that a review such as this, written a decade from now, when so many of the issues that are currently uncertain will have been resolved will be much more complete. However, such an idealised academic perspective does not make useful connection to the real world in which we live. Key questions are being posed now, and decisions being made based on current discussions. Rather, our aim here has been instead to make concrete “horizon” proposals that can be acted on now, with the hope of being more mature within a decade.We stress it is too early to decide whether the approach expounded here, in which microscopic information is linked to biological outcomes, will become most central. For example, at the time of writing, more “informatics” related approaches that a posteriori harvest information that is accumulated without any driving force from microscopic science are favoured by some scientists. And there are many other alternatives also. The relative merits and difficulties of all of these options will only be clarified over time, and it is premature to impose a single direction on our thinking.
However, if one wishes to retain the option of linking microscopic and structural nanomaterial principles to biology, then information collected over the coming decade must respect some guiding vision, of categorisation or systematisation, or it will be difficult to integrate later into any meaningful overall picture. Currently we have high levels of confidence that size, shape and surface presentation in situ will determine much of the early stages of biological impacts. At later stages of biological interaction we believe (but still there is much to be done to prove) that the degradation products produced in intracellular degradative organelles will be important.
It need hardly be stressed, after a decade of such discussions, that one should characterise nanomaterials (for example particle surface properties) by every approach available, and record as many properties as possible. Still, for truly microscopic understanding it will be necessary to identify the key (microscopic) control parameters, and this may require detail that does not flow directly from current characterisation methods. While we believe we are beginning to understand these issues, it makes sense now to capture all of the information that could encompass the control parameters that could be used to check those hypotheses, and may ultimately be useful in framing a systematisation of the field.
Thus, it makes sense now to have initiatives to fully capture (for example digitise) the shapes (and their distributions) of particles, for this will allow the progressive uncovering of those (control) structural features that lead to different outcomes, at a time when we do not know all the features that lead to significant new outcomes. Similarly, the knowledge of the biological recognition fragments (via epitope mapping and other methods) presented at the surface will also allow for a direct link to immediate biological outcomes, including immune response, biodistribution, and others.
Also, increased focus on the manner in which the materials are processed and degraded (leading to specific molecular details of metabolites) within cells will also be valuable as one seeks to understand longer time scales of biological impacts.
The progressive digitisation of such information now would allow us to build a searchable and addressable database that will identify if similar forms (based on computable automated metrics) have previously been synthesised, and thereby attach attendant information, and new information derived, for example, from the study of other pathways as that becomes available. This database would thus index those material forms also with what we believe to be the key metrics of biological responses, and provide a list of expected biological pathway response that could arise, allowing the user to confirm these, or critically annotate that database.
While for some (non-degrading) materials we expect the material, size, shape, and (intra-organelle) surface to continue to dominate the subsequent story, the tools that we have to link all of these ideas together are currently more limited. For materials that evolve significantly within organisms (dissolve, degrade etc.) we can at least be sure that the (molecular) classification of degradative pathways will be important. No doubt, for chronic long-term exposure there is lack of tools for credible mechanistic studies. Still we are confident that the basic framework outlined here will evolve smoothly into new forms that will allow us to deal with longer-term issues, and have begun work on that topic.
We argued at the beginning that the nanoscale in contact with living processes is distinctive, because biology was (uniquely) built to process on the nanoscale. Thus, we consider that, whatever the trends and fashions of science, knowledge and understanding of the engineered nanoscale in contact with biology is of durable value, and this is particularly true of understanding based on microscopic principles. Thus, unlike many incremental scientific enterprises, the systematisation and microscopic understanding of engineered nanoscale objects interacting with living organisms, while it has been commenced by this generation of scientists, belongs to all generations, and will attract the durable attention and interest of scientists across generations. Investment, if wisely crafted, will not be wasted in such a scientific arena.
Acknowledgements
We gratefully acknowledge John Rumble (CODATA) for helpful discussions. The authors acknowledge Mattia Bramini, Laurence Fitzpatrick, Luciana M. Herda and Diana Hudecz for parts of Fig. 1–3. Nanoparticles shown in Fig. 3–5 were synthesised in the framework of EU FP7 FutureNanoNeeds project.V. C. acknowledges the support of funding from the European Union Seventh Framework Programme under grant agreement no. 604391 Graphene Flagship and Irish Research Council under the Government of Ireland Postdoctoral Fellowship scheme (GOIPD/2016/128). J. C. and Ž. K. greatly acknowledge the financial support of the EU FP7 FutureNanoNeeds project (NMP/2013/1.3-3) under the grant agreement no. 604602. J. M. A. acknowledges CNPq – Conselho Nacional de Desenvolvimento Científico e Tecnológico, of the Ministry of Science, Technology and Innovation of Brazil. E. P. acknowledges Science Foundation Ireland (SFI) Principal Investigator Award 298 (agreement no. 12/IA/1422). Q. C. acknowledges Chinese Scholarship Council (agreement no. 201408300003), and Irish Research Council under the Government of Ireland Postgraduate Scholarship scheme (GOIPG/2014/874). C. P. S. acknowledges the financial support of CNPq – Conselho Nacional de Desenvolvimento Científico e Tecnológico, of the Ministry of Science, Technology and Innovation of Brazil (agreement no. 205146/2014-7). L. B. acknowledges the financial support of the EU H2020 Nanofacturing projects (grant agreement no. 646364).
References
- K. A. Dawson, Science for Environment Policy – European Commission, 2015, 3–5 Search PubMed
.
- A. Lesniak, F. Fenaroli, M. P. Monopoli, C. Åberg, K. A. Dawson and A. Salvati, ACS Nano, 2012, 6, 5845–5857 CrossRef CAS PubMed
.
- A. Verma, O. Uzun, Y. Hu, Y. Hu, H.-S. Han, N. Watson, S. Chen, D. J. Irvine and F. Stellacci, Nat. Mater., 2008, 7, 588–595 CrossRef CAS PubMed
.
- M. Ferrari, Nat. Nanotechnol., 2008, 3, 131–132 CrossRef CAS PubMed
.
- L. Shang, K. Nienhaus and G. U. Nienhaus, J. Nanobiotechnol., 2014, 12, 1 CrossRef PubMed
.
- M. Bramini, D. Ye, A. Hallerbach, M. Nic Raghnaill, A. Salvati, C. Åberg and K. A. Dawson, ACS Nano, 2014, 8, 4304–4312 CrossRef CAS PubMed
.
- P. Sandin, L. W. Fitzpatrick, J. C. Simpson and K. A. Dawson, ACS Nano, 2012, 6, 1513–1521 CrossRef CAS PubMed
.
- C. Åberg, J. A. Varela, L. W. Fitzpatrick and K. A. Dawson, Sci. Rep., 2016, 6, 34457 CrossRef PubMed
.
- M. Ferrari, Nat. Rev. Cancer, 2005, 5, 161–171 CrossRef CAS PubMed
.
- M. L. Etheridge, S. A. Campbell, A. G. Erdman, C. L. Haynes, S. M. Wolf and J. McCullough, Nanomed.: Nanotechnol., Biol. Med., 2013, 9, 1–14 CrossRef CAS PubMed
.
- T. Tanaka, P. Decuzzi, M. Cristofanilli, J. H. Sakamoto, E. Tasciotti, F. M. Robertson and M. Ferrari, Biomed. Microdevices, 2009, 11, 49–63 CrossRef CAS PubMed
.
- I. Brigger, C. Dubernet and P. Couvreur, Adv. Drug Delivery Rev., 2002, 54, 631–651 CrossRef CAS PubMed
.
- M. M.-C. Cheng, G. Cuda, Y. L. Bunimovich, M. Gaspari, J. R. Heath, H. D. Hill, C. A. Mirkin, A. J. Nijdam, R. Terracciano and T. Thundat, Curr. Opin. Chem. Biol., 2006, 10, 11–19 CrossRef CAS PubMed
.
- C. Watson, J. Ge, J. Cohen, G. Pyrgiotakis, B. P. Engelward and P. Demokritou, ACS Nano, 2014, 8, 2118–2133 CrossRef CAS PubMed
.
- S. L. Harper, J. L. Carriere, J. M. Miller, J. E. Hutchison, B. L. Maddux and R. L. Tanguay, ACS Nano, 2011, 5, 4688–4697 CrossRef CAS PubMed
.
- D. J. Gentleman and W. C. Chan, Small, 2009, 5, 426–431 CrossRef CAS PubMed
.
- ISO/TR 14786:2014(E) 2014.
- I. Lynch, C. Weiss and E. Valsami-Jones, Nano Today, 2014, 9, 266–270 CrossRef CAS
.
- K. C. Mills, D. Murry, K. A. Guzan and M. L. Ostraat, J. Nanopart. Res., 2014, 16, 1–9 CrossRef
.
-
M. Hull and D. Bowman, Nanotechnology Environmental Health and Safety: Risks, Regulation, and Management, William Andrew, 2014 Search PubMed
.
- S. Foss Hansen, B. H. Larsen, S. I. Olsen and A. Baun, Nanotoxicology, 2007, 1, 243–250 CrossRef
.
- A. Dowling, R. Clift, N. Grobert, D. Hutton, R. Oliver, O. O’neill, J. Pethica, N. Pidgeon, J. Porritt and J. Ryan, London: The Royal Society & The Royal Academy of Engineering Report, 2004, 61, e64 Search PubMed
.
- M. Auffan, J. Rose, J.-Y. Bottero, G. V. Lowry, J.-P. Jolivet and M. R. Wiesner, Nat. Nanotechnol., 2009, 4, 634–641 CrossRef CAS PubMed
.
- A. V. Eletskii, A. O. Erkimbaev, G. A. Kobzev, M. S. Trachtengerts and V. Y. Zitserman, Data Sci. J., 2012, 11, 126–139 CrossRef
.
- NIEHS and NCL/NCI Announce Partnership to Study Nanotechnology Safety.
- F. Bertoli, G. L. Davies, M. P. Monopoli, M. Moloney, Y. K. Gun'ko, A. Salvati and K. A. Dawson, Small, 2014, 10, 3307–3315 CrossRef CAS PubMed
.
- F. Bertoli, D. Garry, M. P. Monopoli, A. Salvati and K. A. Dawson, ACS Nano, 2016, 10, 10471–10479 CrossRef CAS PubMed
.
- T. K. Sau and C. J. Murphy, J. Am. Chem. Soc., 2004, 126, 8648–8649 CrossRef CAS PubMed
.
- H. Yuan, C. G. Khoury, H. Hwang, C. M. Wilson, G. A. Grant and T. Vo-Dinh, Nanotechnology, 2012, 23, 075102 CrossRef PubMed
.
- X. Ye, L. Jin, H. Caglayan, J. Chen, G. Xing, C. Zheng, V. Doan-Nguyen, Y. Kang, N. Engheta and C. R. Kagan, ACS Nano, 2012, 6, 2804–2817 CrossRef CAS PubMed
.
- L. Manna, E. C. Scher and A. P. Alivisatos, J. Am. Chem. Soc., 2000, 122, 12700–12706 CrossRef CAS
.
- Y. Sun and Y. Xia, Science, 2002, 298, 2176–2179 CrossRef CAS PubMed
.
- T. S. Ahmadi, Z. L. Wang, T. C. Green, A. Henglein and M. A. El-Sayed, Science, 1996, 272, 1924 CrossRef CAS PubMed
.
- N. R. Jana, L. Gearheart and C. J. Murphy, Adv. Mater., 2001, 13, 1389 CrossRef CAS
.
- S. Chen, Z. L. Wang, J. Ballato, S. H. Foulger and D. L. Carroll, J. Am. Chem. Soc., 2003, 125, 16186–16187 CrossRef CAS PubMed
.
- P. Chhour, N. Gallo, R. Cheheltani, D. Williams, A. Al-Zaki, T. Paik, J. L. Nichol, Z. Tian, P. C. Naha and W. R. Witschey, ACS Nano, 2014, 8, 9143–9153 CrossRef CAS PubMed
.
- G. H. Jeong, Y. W. Lee, M. Kim and S. W. Han, J. Colloid Interface Sci., 2009, 329, 97–102 CrossRef CAS PubMed
.
- Y. Lee and T. G. Park, Langmuir, 2011, 27, 2965–2971 CrossRef CAS PubMed
.
- J. Li, J. Wu, X. Zhang, Y. Liu, D. Zhou, H. Sun, H. Zhang and B. Yang, J. Phys. Chem. C, 2011, 115, 3630–3637 CAS
.
- Y. Luo, X. Ji, J. Zhuang and W. Yang, Colloids Surf., A, 2014, 463, 28–36 CrossRef CAS
.
- L. Zhao, X. Ji, X. Sun, J. Li, W. Yang and X. Peng, J. Phys. Chem. C, 2009, 113, 16645–16651 CAS
.
- J. Xie, Q. Zhang, J. Y. Lee and D. I. Wang, ACS Nano, 2008, 2, 2473–2480 CrossRef CAS PubMed
.
- S. Reculusa, C. Mingotaud, E. Bourgeat-Lami, E. Duguet and S. Ravaine, Nano Lett., 2004, 4, 1677–1682 CrossRef CAS
.
- C. G. Khoury and T. Vo-Dinh, J. Phys. Chem. C, 2008, 112, 18849–18859 CAS
.
- H. Liang, D. Rossouw, H. Zhao, S. K. Cushing, H. Shi, A. Korinek, H. Xu, F. Rosei, W. Wang and N. Wu, J. Am. Chem. Soc., 2013, 135, 9616–9619 CrossRef CAS PubMed
.
- J. Ford, S. R. Marder and S. Yang, Chem. Mater., 2009, 21, 476–483 CrossRef CAS
.
- C. Levard, E. M. Hotze, G. V. Lowry and G. E. Brown Jr, Environ. Sci. Technol., 2012, 46, 6900–6914 CrossRef CAS PubMed
.
- T. Xia, M. Kovochich, M. Liong, L. Mädler, B. Gilbert, H. Shi, J. I. Yeh, J. I. Zink and A. E. Nel, ACS Nano, 2008, 2, 2121–2134 CrossRef CAS PubMed
.
- W. Zhang, M. Saliba, S. D. Stranks, Y. Sun, X. Shi, U. Wiesner and H. J. Snaith, Nano Lett., 2013, 13, 4505–4510 CrossRef CAS PubMed
.
- M. A. Green, A. Ho-Baillie and H. J. Snaith, Nat. Photonics, 2014, 8, 506–514 CrossRef CAS
.
- EU FP7 FutureNanoNeeds: a framework to respond to the regulatory needs of future nanomaterials and markets.
- M. Potenza, Z. Krpetic, T. Sanvito, Q. Cai, M. Monopoli, J. M. de Araújo, C. Cella, L. Boselli, V. Castagnola and P. Milani, Nanoscale, 2017, 9, 2778–2784 RSC
.
- M. Potenza, T. Sanvito, S. Argentiere, C. Cella, B. Paroli, C. Lenardi and P. Milani, Sci. Rep., 2015, 5, 18228 CrossRef CAS PubMed
.
- R. P. Carney, J. Y. Kim, H. Qian, R. Jin, H. Mehenni, F. Stellacci and O. M. Bakr, Nat. Commun., 2011, 2, 335 CrossRef PubMed
.
- J. Walter, K. Löhr, E. Karabudak, W. Reis, J. Mikhael, W. Peukert, W. Wohlleben and H. Cölfen, ACS Nano, 2014, 8, 8871–8886 CrossRef CAS PubMed
.
- B. D. Levin, E. Padgett, C.-C. Chen, M. Scott, R. Xu, W. Theis, Y. Jiang, Y. Yang, C. Ophus and H. Zhang, Sci. Data, 2016, 3, 160041 CrossRef PubMed
.
- S. Bals, B. Goris, L. M. Liz-Marzán and G. Van Tendeloo, Angew. Chem., Int. Ed., 2014, 53, 10600–10610 CrossRef CAS PubMed
.
- R. Osada, T. Funkhouser, B. Chazelle and D. Dobkin, ACM Trans. Graphics, 2002, 21, 807–832 CrossRef
.
- J. W. Tangelder and R. C. Veltkamp, Multimedia Tools Appl., 2008, 39, 441–471 CrossRef
.
- A. S. Keys, C. R. Iacovella and S. C. Glotzer, J. Comput. Phys., 2011, 230, 6438–6463 CrossRef CAS
.
- A. Salvati, A. S. Pitek, M. P. Monopoli, K. Prapainop, F. B. Bombelli, D. R. Hristov, P. M. Kelly, C. Åberg, E. Mahon and K. A. Dawson, Nat. Nanotechnol., 2013, 8, 137–143 CrossRef CAS PubMed
.
- D. Walczyk, F. B. Bombelli, M. P. Monopoli, I. Lynch and K. A. Dawson, J. Am. Chem. Soc., 2010, 132, 5761–5768 CrossRef CAS PubMed
.
- F. Wang, L. Yu, M. P. Monopoli, P. Sandin, E. Mahon, A. Salvati and K. A. Dawson, Nanomed.: Nanotechnol., Biol. Med., 2013, 9, 1159–1168 CrossRef CAS PubMed
.
- S. Lara, F. Alnasser, E. Polo, D. Garry, M. C. Lo Giudice, D. R. Hristov, L. Rocks, A. Salvati, Y. Yan and K. A. Dawson, ACS Nano, 2017, 11, 1884–1893 CrossRef CAS PubMed
.
- M. A. Dobrovolskaia and S. E. McNeil, Nat. Nanotechnol., 2007, 2, 469–478 CrossRef CAS PubMed
.
- A. Albanese, C. D. Walkey, J. B. Olsen, H. Guo, A. Emili and W. C. Chan, ACS Nano, 2014, 8, 5515–5526 CrossRef CAS PubMed
.
- J. A. Kim, A. Salvati, C. Åberg and K. A. Dawson, Nanoscale, 2014, 6, 14180–14184 RSC
.
- I. Lynch, A. Ahluwalia, D. Boraschi, H. J. Byrne, B. Fadeel, P. Gehr, A. C. Gutleb, M. Kendall and M. G. Papadopoulos, BioNanoMaterials, 2013, 14, 195–216 CrossRef
.
- E. Brun and C. Sicard–Roselli, Cancer Nanotechnol., 2014, 5, 1–13 CrossRef PubMed
.
- K. Prapainop, D. P. Witter and P. Wentworth Jr, J. Am. Chem. Soc., 2012, 134, 4100–4103 CrossRef CAS PubMed
.
- Y. Yan, K. T. Gause, M. M. Kamphuis, C.-S. Ang, N. M. O’Brien-Simpson, J. C. Lenzo, E. C. Reynolds, E. C. Nice and F. Caruso, ACS Nano, 2013, 7, 10960–10970 CrossRef CAS PubMed
.
- Z. J. Deng, M. Liang, M. Monteiro, I. Toth and R. F. Minchin, Nat. Nanotechnol., 2011, 6, 39–44 CrossRef CAS PubMed
.
- T.-G. Iversen, T. Skotland and K. Sandvig, Nano Today, 2011, 6, 176–185 CrossRef CAS
.
- O. Harush-Frenkel, E. Rozentur, S. Benita and Y. Altschuler, Biomacromolecules, 2008, 9, 435–443 CrossRef CAS PubMed
.
- J. Gilleron, W. Querbes, A. Zeigerer, A. Borodovsky, G. Marsico, U. Schubert, K. Manygoats, S. Seifert, C. Andree, M. Stoter, H. Epstein-Barash, L. Zhang, V. Koteliansky, K. Fitzgerald, E. Fava, M. Bickle, Y. Kalaidzidis, A. Akinc, M. Maier and M. Zerial, Nat. Biotechnol., 2013, 31, 638–646 CrossRef CAS PubMed
.
- K. L. Douglas, C. A. Piccirillo and M. Tabrizian, Eur. J. Pharm. Biopharm., 2008, 68, 676–687 CrossRef CAS PubMed
.
- M. S. Cartiera, K. M. Johnson, V. Rajendran, M. J. Caplan and W. M. Saltzman, Biomaterials, 2009, 30, 2790–2798 CrossRef CAS PubMed
.
- S. K. Lai, K. Hida, C. Chen and J. Hanes, J. Controlled Release, 2008, 125, 107–111 CrossRef CAS PubMed
.
- S. K. Lai, K. Hida, S. T. Man, C. Chen, C. Machamer, T. A. Schroer and J. Hanes, Biomaterials, 2007, 28, 2876–2884 CrossRef CAS PubMed
.
- G. Sonavane, K. Tomoda and K. Makino, Colloids Surf., B, 2008, 66, 274–280 CrossRef CAS PubMed
.
- E. Sadauskas, G. Danscher, M. Stoltenberg, U. Vogel, A. Larsen and H. Wallin, Nanomed.: Nanotechnol., Biol. Med., 2009, 5, 162–169 CrossRef CAS PubMed
.
- E. Sadauskas, H. Wallin, M. Stoltenberg, U. Vogel, P. Doering, A. Larsen and G. Danscher, Part. Fibre Toxicol., 2007, 4, 1 CrossRef PubMed
.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz, R. Hecht, F. Schlenk, D. Fischer, K. Kiouptsi and C. Reinhardt, Nat. Nanotechnol., 2013, 8, 772–781 CrossRef CAS PubMed
.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. Baldelli Bombelli and K. A. Dawson, J. Am. Chem. Soc., 2011, 133, 2525–2534 CrossRef CAS PubMed
.
- C. D. Walkey, J. B. Olsen, F. Song, R. Liu, H. Guo, D. W. H. Olsen, Y. Cohen, A. Emili and W. C. Chan, ACS Nano, 2014, 8, 2439–2455 CrossRef CAS PubMed
.
- M. Lundqvist, J. Stigler, G. Elia, I. Lynch, T. Cedervall and K. A. Dawson, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14265–14270 CrossRef CAS PubMed
.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggård, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050–2055 CrossRef CAS PubMed
.
- E. Casals, T. Pfaller, A. Duschl, G. J. Oostingh and V. Puntes, ACS Nano, 2010, 4, 3623–3632 CrossRef CAS PubMed
.
- S. Wan, P. M. Kelly, E. Mahon, H. Stöckmann, P. M. Rudd, F. Caruso, K. A. Dawson, Y. Yan and M. P. Monopoli, ACS Nano, 2015, 9, 2157–2166 CrossRef CAS PubMed
.
- A. L. Barrán-Berdón, D. Pozzi, G. Caracciolo, A. L. Capriotti, G. Caruso, C. Cavaliere, A. Riccioli, S. Palchetti and A. Laganá, Langmuir, 2013, 29, 6485–6494 CrossRef PubMed
.
- P. M. Kelly, C. Åberg, E. Polo, A. O'Connell, J. Cookman, J. Fallon, Ž. Krpetić and K. A. Dawson, Nat. Nanotechnol., 2015, 10, 472–479 CrossRef CAS PubMed
.
- M. C. Lo Giudice, L. M. Herda, E. Polo and K. A. Dawson, Nat. Commun., 2016, 7, 13475 CrossRef CAS PubMed
.
- M. P. Monopoli, C. Aberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2017 |