Design and investigation of targeting agent orientation and density on nanoparticles for enhancing cellular uptake efficiency†
Weiwei
Fei
,
Xiuli
Wang
,
Jia
Guo
 and
Changchun
Wang
and
Changchun
Wang
 *
*
State Key Laboratory of Molecular Engineering of Polymers, Department of Macromolecular Science, and Laboratory of Advanced Materials, Fudan University, Shanghai 200433, P. R. China. E-mail: ccwang@fudan.edu.cn
First published on 3rd August 2023
Abstract
The design of targeting agent-conjugated systems is attracting much attention in cell targeted delivery and cancer therapy. However, quantitative study of the ligand density and binding efficiency is still limited due to the technical matters and tedious work involved. In this article, benzoboroxole-modified core–shell magnetic nanoparticles (MSP-AOPB NPs) as a drug carrier model were fabricated and transferrin (Tf) was immobilized on the nanoparticle surface in a site-oriented manner (Tf-MSP-AOPB NPs). The preparation conditions were investigated in detail to optimize the Tf binding efficiency. A suitable reaction temperature, time or initial feeding amount could significantly increase the Tf binding amount. The maximum Tf binding amount on the MSP-AOPB NPs was 184 mg g−1, and the targeting ligand density on the surface could be well controlled by simply adjusting the reaction conditions. In vitro studies demonstrated the excellent Tf-mediated targeting ability and enhanced cellular uptake efficacy by varying the ligand density. The optimal ligand binding amount for achieving the highest cellular uptake efficiency was 94 mg Tf/g, which corresponds to a ligand binding density of about 0.05 Tf/nm2, and the binding efficiency of conjugation was higher than 90%. Moreover, Tf-MSP-AOPB NPs prepared by a site-oriented conjugation strategy showed the best cell targeting ability, and their cellular uptake amount was 25 and 127 times higher than that of physical adsorption and EDC/NHS coupling reaction in HepG2 cells, respectively. This study provides a facile site-oriented bioconjugation technique for different kinds of antibodies, and a suitable ligand density can be easily attained to enhance the cellular uptake efficacy, which shows great significance for targeted delivery and cancer therapy.
1. Introduction
Nanoparticle-based drug delivery systems have attracted widespread attention in cancer diagnosis and therapy.1–4 Compared with traditional chemotherapeutic drugs, which usually suffer from quick clearance from the bloodstream and exhibit high cytotoxicity towards normal tissues, nanoparticle carriers possess many unique characteristics, such as tunable size, facile surface functionalization, high drug loading and good biocompatibility.5–7 To date, different kinds of nanoparticles have been employed in therapeutic studies, including liposomes,8,9 polymeric nanoparticles,10–12 silica nanoparticles,13–15 metal or metal oxide nanoparticles,16–18etc. Recently, much effort has been devoted to designing targeting nanoparticles to enhance the targeting efficacy and improve the accumulation of nanoparticles at the desired location of disease.19–21 A variety of bioconjugation methods have been developed to combine bioactive targeting ligands with nanoparticles. One method is physical adsorption of the targeting agents on the nanoparticles based on electrostatic interaction, hydrophobic interaction or van der Waals forces, etc.22,23 This kind of modification procedure is convenient, but the interactions between the targeting molecules and nanoparticles are not stable. Another method is realized by chemical conjugation of the targeting agents on the nanoparticles to obtain a stable structure. These strategies include amine-N-hydroxysuccinimide,24,25 amine-aldehyde,26 amine-epoxy27 and sulfhydryl-maleimide28,29 reactions due to the targeting agent (antibody) containing rich amine and sulfhydryl groups. However, it is usually randomly oriented and the receptor-binding region in the targeting ligand will be affected during the coupling process, causing partial loss of the targeting ability. Thus, a site-oriented conjugation method is urgently required to maintain the bioactivity of the targeting agents.Benzoboroxole-modified nanoparticles have been widely used in the field of glycoprotein enrichment30–32 and antibody purification33,34 with high selectivity. It is based on the formation of a five-membered cyclic ester after reacting with cis-diols in the sugar chains of glycoproteins, and the reaction conditions are mild. Some frequently used targeting agents such as transferrin (Tf) and antibodies are glycoproteins and have cis-diol moieties in the inactive structural domain.35 As a result, these targeting ligands can be easily immobilized on the benzoboroxole modified nanoparticles, and the receptor-binding region will not be interfered with during the conjugation procedure, achieving a site-oriented conjugation. In our previous studies, benzoboroxole-grafted polymer chain or polyacrylic acid nanohydrogels were fabricated for the modification of the targeting agent and protein loading, which exhibited enhanced cellular uptake and good therapeutic effect.36,37 Due to the targeting agents like antibodies being expensive, it is important to maximize the efficiency of the targeting agents. Meanwhile, binding efficiency should also be taken into consideration when preparing the targeting ligand-nanoparticle complex.
Moreover, in addition to the orientation of immobilization, the ligand density is also an important parameter in the targeting efficiency of ligands.38–40 Multivalent interactions exist in the association between nanoparticles and cells. Some reports have demonstrated that increasing the ligand density will enhance the cell binding and promote the cellular internalization. Besides, a high ligand density possibly results in a tight packing of the ligands due to steric interference and limits the mobility of the ligands to recognize receptors on the cell membrane.41 However, determining the cellular uptake efficacy by controlling the ligand density and binding efficiency through a site-oriented conjugation strategy is seldom reported. As a functional nanocarrier, magnetic nanoparticles can be usually used due to their various functionalities, such as magnetic resonance imaging (MRI),42 magnetic particle imaging (MPI)43 and magnetic hyperthermia,44etc. Meanwhile, they have the feature of easy separation from the reaction system, making it easy to determine the content and density of the modified moiety.
In this work, benzoboroxole-modified core–shell magnetic nanoparticles as a carrier model were fabricated for site-oriented conjugation of transferrin (Tf). Tf was chosen as the targeting agent, which could specifically bind with the transferrin receptor (TfR) on many cancer cells. The core–shell nanoparticles were firstly synthesized via reflux-precipitation polymerization to coat the poly(glycidyl methacrylate) (PGMA) layer on the magnetic nanoparticles, and then carboxyl groups were grafted on the core–shell nanoparticle surface by ring-opening reaction of the epoxy group with iminodiacetic acid (IDA). After that, the benzoboroxole moiety was fixed on the nanoparticles through an amidation reaction. Finally, Tf was conjugated on the nanoparticle surface in a site-oriented manner by reaction with the benzoboroxole group. Tf-modified nanoparticles with different ligand densities were employed to evaluate the cellular uptake efficacy.
2. Experimental section
2.1. Reagents and materials
Iron(III) chloride hexahydrate (FeCl3·6H2O), trisodium citrate dihydrate, anhydrous sodium acetate, 3-methacryloxypropyltrimethoxysilane (MPS), aqueous ammonia solution (NH3·H2O), glycidyl methacrylate (GMA), sodium phosphate monobasic (NaH2PO4), sodium phosphate dibasic (Na2HPO4), 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC·HCl) and N-hydroxysuccinimide (NHS) were purchased from Shanghai Aladdin Chemistry Co., Ltd. Sodium hydroxide (NaOH), ethylene glycol, anhydrous ethanol, sodium chloride (NaCl) and acetonitrile were purchased from Sinopharm Chemical Reagent Co., Ltd. Iminodiacetic acid (IDA) and 2,2′-azoisobutyronitrile (AIBN) were purchased from Macklin Biochemical Technology Co., Ltd. N,N′-Methylenebisacrylamide (MBA) was purchased from Fluka. 5-Amino-2-(hydroxymethyl)benzeneboronic acid hemiester hydrochloride (AOPB·HCl) was purchased from Alfa Aesar. Cyanine 7 amine was purchased from Xi’an Kaixin Biotechnology Co., Ltd. Holo-transferrin human (Tf) and bovine serum albumin (BSA) were purchased from Sigma-Aldrich Chemical Co., Ltd. Dulbecco's modified Eagle's medium (DMEM), penicillin–streptomycin solution (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 units penicillin and 10 mg streptomycin) and trypsin solution (with ethylene diamine tetraacetic acid, EDTA) were purchased from Thermo Fisher Scientific. Fetal bovine serum (FBS) was purchased from Biological Industries. PBS (1X) was purchased from Wisent Bioproducts. An enhanced BCA Protein Assay Kit was purchased from Beyotime Biotechnology Company. AIBN was recrystallized before use. Deionized water was used in all the experiments.
000 units penicillin and 10 mg streptomycin) and trypsin solution (with ethylene diamine tetraacetic acid, EDTA) were purchased from Thermo Fisher Scientific. Fetal bovine serum (FBS) was purchased from Biological Industries. PBS (1X) was purchased from Wisent Bioproducts. An enhanced BCA Protein Assay Kit was purchased from Beyotime Biotechnology Company. AIBN was recrystallized before use. Deionized water was used in all the experiments.
2.2. Synthesis of MSP-AOPB NPs
Firstly, magnetite supraparticles (MSPs) were prepared by a modified solvothermal reaction according to our previous work. Typically, 1.35 g of FeCl3·6H2O, 0.3 g of trisodium citrate dihydrate, 3.6 g of sodium acetate and 0.2 g of NaOH were dissolved in 35 mL of ethylene glycol. The mixture was stirred vigorously for 1 h at 50 °C and then transferred into a 50 mL Teflon-lined stainless-steel autoclave. The autoclave was maintained at 200 °C for 10 h. After cooling down to room temperature, the black product was washed with ethanol and water five times, respectively, and re-dispersed in ethanol for further use.Next, the MSP NPs were modified with 3-methacryloxypropyltrimethoxysilane (MPS) to form double bonds on the surface. 700 mg of MSP NPs, 2 mL of MPS, 3 mL of NH3·H2O and 20 mL deionized water were added into 80 mL of ethanol. The mixture was mechanically stirred at 60 °C for 24 h. The product was collected with a magnet and washed with ethanol three times and re-dispersed in water for subsequent use.
Then, the core/shell MSP-PGMA NPs were synthesized through a one-step reflux-precipitation polymerization of GMA, with MBA as the cross-linker and AIBN as the initiator. Briefly, 150 mg of MPS-modified MSP NPs, 270 μL of GMA, 270 mg of MBA and 12 mg of AIBN were added into 120 mL of acetonitrile. The mixture was sonicated for 15 min to form a homogeneous dispersion. Then, the mixture was heated to 95 °C and kept for 1 h. After the reaction, the obtained product was separated with a magnet and washed with ethanol five times.
Afterwards, MSP-IDA NPs were fabricated through the ring-opening reaction between the amino groups of IDA and epoxy groups on the surface of MSP-PGMA NPs. Briefly, 250 mg of MSP-PGMA NPs were dispersed in 100 mL of deionized water. 2 g of IDA and 1.2 g of NaOH were subsequently added into the suspension. The mixture was sonicated for 10 min and mechanically stirred at 70 °C for 12 h. The obtained product was washed with water five times.
MSP-AOPB NPs were prepared by grafting a benzoboroxole moiety onto the surface of the MSP-IDA NPs through an amidation reaction by the EDC/NHS method. Briefly, 100 mg of MSP-IDA NPs, 300 mg of EDC·HCl and 200 mg of NHS were dispersed in 50 mL of PBS (1X) to activate the carboxyl groups for 2 h at room temperature. Then, 40 mg of 5-amino-2-(hydroxymethyl)benzeneboronic acid hemiester hydrochloride was added into the mixture. The coupling reaction continued for 12 h at room temperature. The obtained product was washed with water three times and re-dispersed in water for further use.
Modification of Cy7 on the MSP-AOPB NPs was performed as follows. 25 mg of MSP-AOPB NPs, 75 mg of EDC·HCl and 50 mg of NHS were dispersed in 8 mL of PBS (1X) to activate for 2 h at room temperature. Then, 0.4 mg of amino Cy7 was added and the reaction continued for 12 h. The obtained product was washed with water three times and re-dispersed in water for further use.
2.3. Characterization
The morphologies of the nanoparticles were observed on a transmission electron microscope (TEM, Tecnai G2 20 TWIN, FEI) at an accelerating voltage of 200 kV. Samples dispersed at an appropriate concentration were cast onto a carbon-coated copper grid. Field emission scanning electron microscope (FESEM) images were taken on an Ultra 55 FESEM microscope (In-lens SE detector, Zeiss) at an accelerating voltage of 3 kV. The hydrodynamic diameter (Dh) and zeta potential were measured on a dynamic light scattering instrument (DLS, Zetasizer Nano ZS90, Malvern Instruments) using a He–Ne laser at a wavelength of 632.8 nm. X-ray diffraction (XRD) patterns were obtained using an X-ray diffraction meter (D8 Advance, Bruker) with Cu-Kα radiation at λ = 0.154 nm operating at 40 kV and 40 mA. Magnetic characterization was performed on a vibrating sample magnetometer (VSM, MPMS 3, Quantum Design) on a Model 6000 physical property measurement system at 300 K. Fourier transform infrared spectra (FT-IR) was recorded on a Nicolet 6700 (Thermofisher) spectrometer. Spectra were scanned over the range of 400-4000 cm−1. Thermogravimetric analysis (TGA) was conducted on a Pyris 1 instrument (Perkin Elmer) under a constant air flow of 40 mL min−1. The temperature increased from 100 °C to 800 °C at a rate of 20 °C min−1. Protein concentration was determined by measuring the absorbance at 562 nm according to the BCA assay using a multi-mode microplate reader (Cytation3, BioTek). Flow cytometry analysis was performed on a flow cytometer (Gallios, Beckman Coulter) using 638 nm excitation and 755 nm emission. Approximately 30,000 cells were measured for each sample.2.4. Conjugation of transferrin to MSP-AOPB NPs and quantitative analysis of transferrin binding amount
To obtain transferrin-conjugated MSP-AOPB NPs (Tf-MSP-AOPB NPs), 1 mg of MSP-AOPB NPs was dispersed in 1 mL of transferrin aqueous solution (100 μg mL−1) and the mixture was incubated at a certain temperature under mild shaking. Tf-MSP-AOPB NPs were collected from the mixture after separating them with a magnet. 200 μL of phosphate buffer solution (pH 7.4) was used as an eluent to remove the non-adsorbed transferrin. The concentrations of transferrin in the supernatant and eluent were determined by BCA assay. The Tf loading efficiency (TLE), Tf loading amount (TLA), Tf binding efficiency (TBE) and Tf binding amount (TBA) of Tf on the MSP-AOPB NPs were calculated according to the following equations:| TLE = (C0 − C1)/C0 × 100% |
| TBE = [(C0 − C1)V1 − C2V2]/(C0V1) × 100% |
| TLA = (C0 − C1)V1/M |
| TBA = [(C0 − C1)V1 − C2V2]/M |
To investigate the effects of the elution conditions on the Tf binding efficiency on the MSP-AOPB NPs, the NaCl concentration and phosphate concentration in the eluent were varied.
To optimize the reaction conditions for Tf binding on the MSP-AOPB NPs, the NaCl concentration in the reaction mixture, reaction temperature, reaction time, and initial Tf feeding amount were investigated.
2.5. Cell culture
HepG2 cells and HEK 293T cells were cultured in high-glucose Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 1% penicillin–streptomycin solution, and maintained in a humidified atmosphere containing 5% CO2 at 37 °C.2.6. Cellular uptake of Tf-MSP-AOPB NPs measured by flow cytometry
HepG2 cells were seeded in 6-well plates at a density of 3 × 105 cells per well. After 24 h of incubation, the culture medium was discarded and replaced by Tf-MSP-AOPB NPs in the serum-free culture medium (10 μg mL−1, 1 mL). After another 2 h incubation, the cells were rinsed with PBS twice and digested with 0.5 mL of trypsin for 2 min. The cell suspension was collected and centrifuged. Then, the cells were washed with 500 μL of PBS and centrifuged. Afterwards, the cells were resuspended in 400 μL of PBS for flow cytometry analysis. To detect the fluorescence intensity of Cy7, the excitation filter was 638 nm and the emission filter was 755 nm.2.7. Cell viability study
HEK 293T cells or HepG2 cells were seeded in 96-well plates at a density of 5000 cells per well. After 24 h incubation, the medium was replaced by different concentrations of MSP-AOPB NPs in the culture medium. The cells continued to be cultured for 24 h. Then, the medium was removed and serum-free medium containing 10% CCK-8 was added. After incubation for 1 h, the absorbance was measured at 450 nm through a microplate reader and cell viability was calculated.2.8. Statistical analysis
All results were presented as mean ± standard deviation (SD). All data were obtained from three independent experiments unless otherwise mentioned. The statistical analysis between groups was performed by a two-tailed Student's t-test, and the statistical differences were considered significant for *p < 0.05 and **p < 0.01.3. Results and discussion
3.1. Preparation and characterization of MSP-AOPB NPs
The fabrication procedure of the core–shell magnetic MSP-AOPB NPs and the transferrin-mediated targeting to cells are illustrated in Scheme 1. At first, magnetite supraparticles (MSPs) were prepared via a modified solvothermal method using trisodium citrate as the stabilizer in ethylene glycol. Then, abundant double bonds were formed on the surface through the hydrolysis reaction of 3-methacryloxypropyltrimethoxysilane (MPS). Next, a PGMA shell was coated on the MPS-modified MSP core via reflux-precipitation polymerization with GMA as the monomer and MBA as the cross-linker to obtain MSP-PGMA NPs. After that, an IDA moiety was grafted on the polymer shell according to the ring-opening reaction between the amino groups of IDA and epoxy groups on the PGMA shell to obtain carboxyl functionalized MSP-IDA NPs. Finally, an AOPB moiety could be immobilized on the surface through amidation reaction via the EDC/NHS coupling method to get MSP-AOPB NPs.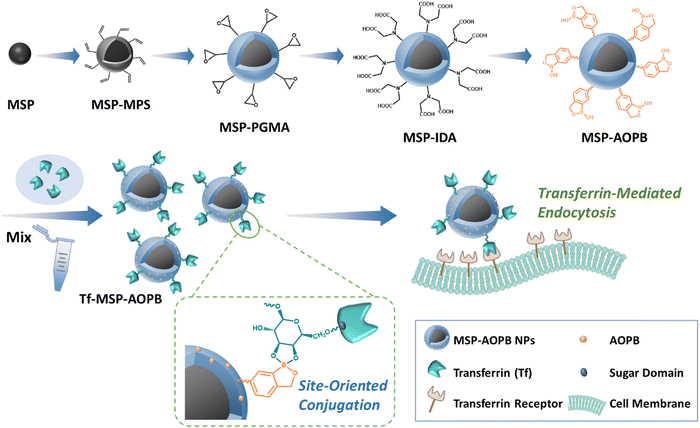 | ||
| Scheme 1 Illustration of the fabrication procedures of the MSP-AOPB nanoparticles and the interaction of the Tf-MSP-AOPB nanoparticles with cells by transferrin-mediated targeting. | ||
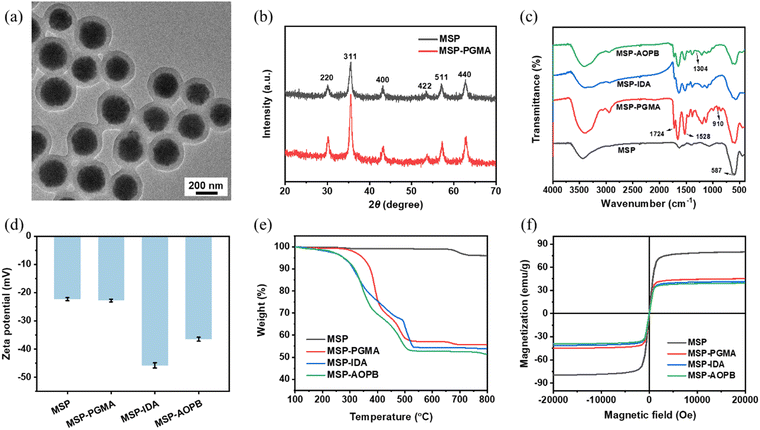
The MSP core particles shown in the transmission electron microscope (TEM) image had an average diameter of about 240 nm (Fig. S1, ESI†), and these supraparticles were composed of small-sized primary magnetite particles. After coating with a cross-linked PGMA shell via reflux-precipitation polymerization, the size of the MSP-PGMA NPs increased to 330 nm and the thickness of the PGMA shell was about 45 nm (Fig. 1a). The hydrodynamic diameters (Dh) of the MSP-PGMA NPs and MSP also confirmed the successful coating of a PGMA shell on MSPs (Fig. S3 and Table S1, ESI†). The diffraction peaks of the X-ray diffraction (XRD) patterns of the MSPs and MSP-PGMA NPs (Fig. 1b) matched well with the cubic structure of magnetite.
The functional groups on the nanoparticles were analyzed by FT-IR. As shown in Fig. 1c, the peaks at 587 cm−1 that appeared in all samples were attributed to the Fe–O stretching vibration in Fe3O4. The peaks at 1724 cm−1 for the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration of esters in GMA and 1528 cm−1 for N–H deformation vibration of amides in MBA demonstrated the formation of the PGMA shell. After the ring-opening reaction with IDA, the peak at 910 cm−1 for the C–O stretching vibration of epoxy groups disappeared in the MSP-IDA NPs. The appearance of a new peak at 1304 cm−1 for the B–O bond proved the effective immobilization of an AOPB moiety on the shell in the MSP-AOPB NPs.
O stretching vibration of esters in GMA and 1528 cm−1 for N–H deformation vibration of amides in MBA demonstrated the formation of the PGMA shell. After the ring-opening reaction with IDA, the peak at 910 cm−1 for the C–O stretching vibration of epoxy groups disappeared in the MSP-IDA NPs. The appearance of a new peak at 1304 cm−1 for the B–O bond proved the effective immobilization of an AOPB moiety on the shell in the MSP-AOPB NPs.
Besides, the results of zeta-potential and TGA also confirmed the successful fabrication of MSP-AOPB NPs. From Fig. 1d and Table S1 (ESI†), the zeta-potential values showed that MSP (−22.3 mV) and MSP-PGMA (−22.8 mV) had a negatively charged surface. After reacting with IDA, the zeta-potential value of the MSP-IDA nanoparticles became more negative (−45.8 mV). After grafting AOPB by amidation reaction, the zeta-potential of the AOPB-MSP NPs increased to −36.5 mV because some of the carboxyl groups were substituted by an AOPB moiety. Thermogravimetric analysis (TGA) was carried out to determine the composition of the nanoparticles. As shown in Fig. 1e, the weight loss of MSP (4 wt%) was attributed to the sodium citrate stabilizer, and the weight loss of MSP-PGMA (45 wt%) significantly increased due to the coating of the PGMA shell. After the subsequent modification of IDA and AOPB, the weight loss of the MSP-IDA and MSP-AOPB NPs increased further to 47 wt% and 49 wt%, respectively. The field emission scanning electron microscope (FESEM) image (Fig. S2, ESI†) indicated that the surface of the MSPs was rough and that of MSP-AOPB became much smoother due to the formation of a PGMA layer. The polydispersity indexes (PDIs) of the MSP-PGMA, MSP-IDA and MSP-AOPB NPs were 0.057, 0.015 and 0.045, respectively, which indicated that these nanoparticles were nearly uniform. These results demonstrated that the core–shell nanoparticles were well prepared and an AOPB moiety was successfully grafted onto the polymer layer.
The magnetic properties of the nanoparticles were investigated by vibrating sample magnetometer (VSM). As shown in Fig. 1f, there was no hysteresis observed in the curves and all samples had almost zero remanence and coercivity, which suggested that these samples were superparamagnetic. The saturation magnetization (Ms) values of the MSP, MSP-PGMA, MSP-IDA and MSP-AOPB NPs were 79.9, 45.1, 41.3 and 39.4 emu g−1, respectively. Although the Ms value decreased during the polymer coating and functional modification, the final obtained MSP-AOPB NPs still possessed good magnetic responsiveness and could be quickly separated from the solution within 30 s under an external magnetic field.
3.2. Study of the elution conditions for determining the Tf binding amount on the MSP-AOPB NPs
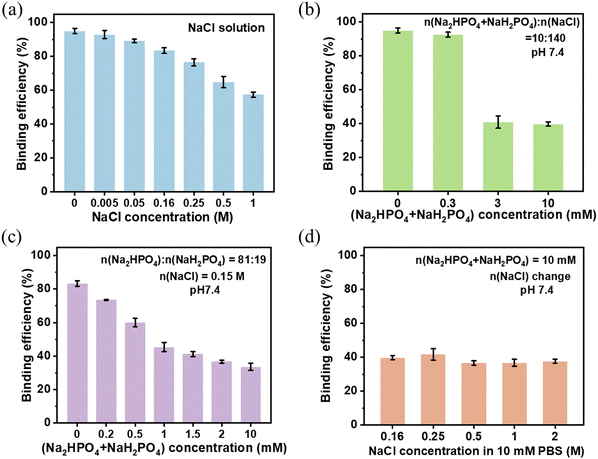
Transferrin (Tf) is a widely used targeting ligand and can selectively bind with the transferrin receptor (TfR) on the cancer cell membrane, suggesting that Tf is a good choice in the application of cell targeted delivery and disease treatment.45–47 Human Tf (hTf, 79 kDa) is a kind of monomeric glycoprotein and the carbohydrate moieties approximately represent 6% of its mass.48 Based on the cis-diols structure in the sugar chain of glycoproteins, Tf can be covalently conjugated on the benzoboroxole modified nanoparticles in a site-oriented manner by the formation of a five-membered cyclic ester. Nevertheless, part of the Tf being physically adsorbed on the nanoparticles during the reaction is unavoidable. When these nanoparticles enter a physiological environment, they will be affected by salts, leading to the detachment of Tf from the surface and thus a decrease of the cell targeting efficacy.In order to investigate the influence of salts on the Tf binding capacity, various eluents containing different concentrations of sodium chloride and phosphate were used to determine the Tf binding efficiency. After simply mixing the Tf solution (200 μg mL−1) and 1 mg MSP-AOPB NPs at the mass ratio of Tf![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NPs = 0.1
NPs = 0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, different concentrations of NaCl solution (0–1 M) were used to elute the weakly adsorbed Tf. As shown in Fig. 2a, the Tf binding efficiency decreased from 95% to 57% when the NaCl concentration increased from 0 M to 1 M. The physically adsorbed Tf via electrostatic interaction could be released from the surface. After using different dilutions of PBS (n(Na2HPO4 + NaH2PO4):n(NaCl) = 10
1, different concentrations of NaCl solution (0–1 M) were used to elute the weakly adsorbed Tf. As shown in Fig. 2a, the Tf binding efficiency decreased from 95% to 57% when the NaCl concentration increased from 0 M to 1 M. The physically adsorbed Tf via electrostatic interaction could be released from the surface. After using different dilutions of PBS (n(Na2HPO4 + NaH2PO4):n(NaCl) = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 140, pH 7.4) as an eluent, the Tf binding efficiency dramatically decreased from 93% to 40% when the PBS concentration increased from 0.3 mM to 3 mM (Fig. 2b), which meant that the phosphate caused a significant decline of the Tf binding efficiency. Thus, the influence of phosphate was studied further. To keep pH 7.4, different concentrations of phosphate buffer solution (n(Na2HPO4):n(NaH2PO4) = 81
140, pH 7.4) as an eluent, the Tf binding efficiency dramatically decreased from 93% to 40% when the PBS concentration increased from 0.3 mM to 3 mM (Fig. 2b), which meant that the phosphate caused a significant decline of the Tf binding efficiency. Thus, the influence of phosphate was studied further. To keep pH 7.4, different concentrations of phosphate buffer solution (n(Na2HPO4):n(NaH2PO4) = 81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19) containing 0.15 M NaCl were used as eluents. The Tf binding efficiency decreased when the total phosphate concentration (n(Na2HPO4) + n(NaH2PO4)) increased from 0 mM to 10 mM (Fig. 2c). The possible reason was that small phosphate ions might compete with large-sized Tf molecules and bind with benzoboroxole, leading to the release of Tf and decrease of the binding efficiency. Besides, when increasing the NaCl concentration in 10 mM phosphate buffer solution from 0.16 M to 2 M in the eluent, there was almost no change in the Tf binding efficiency (Fig. 2d). From the above results, it could be inferred that phosphate would affect the binding process between Tf and MSP-AOPB NPs, and cause the decrease of the Tf binding efficiency to a greater extent when compared with sodium chloride. Considering the physiological environment, we employed 10 mM Na2HPO4/NaH2PO4 buffer solution (pH 7.4) containing 0.15 M NaCl as the eluent in the following experiments.
19) containing 0.15 M NaCl were used as eluents. The Tf binding efficiency decreased when the total phosphate concentration (n(Na2HPO4) + n(NaH2PO4)) increased from 0 mM to 10 mM (Fig. 2c). The possible reason was that small phosphate ions might compete with large-sized Tf molecules and bind with benzoboroxole, leading to the release of Tf and decrease of the binding efficiency. Besides, when increasing the NaCl concentration in 10 mM phosphate buffer solution from 0.16 M to 2 M in the eluent, there was almost no change in the Tf binding efficiency (Fig. 2d). From the above results, it could be inferred that phosphate would affect the binding process between Tf and MSP-AOPB NPs, and cause the decrease of the Tf binding efficiency to a greater extent when compared with sodium chloride. Considering the physiological environment, we employed 10 mM Na2HPO4/NaH2PO4 buffer solution (pH 7.4) containing 0.15 M NaCl as the eluent in the following experiments.
3.3. Optimizing the reaction conditions for Tf binding on MSP-AOPB NPs
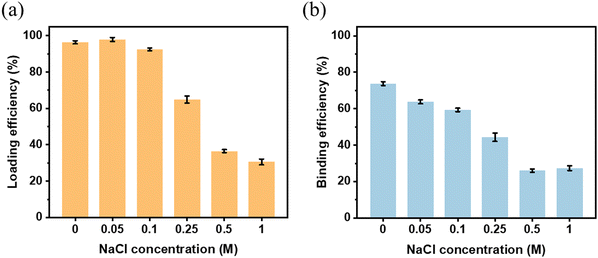
Given that the reaction conditions have a great impact on the Tf binding capacity on the MSP-AOPB NPs, it is absolutely necessary to optimize the reaction conditions to obtain the best conditions for Tf binding. Some important parameters such as ionic strength, reaction temperature, reaction time and initial feeding amount were investigated. The Tf loading efficiency and binding efficiency were calculated by determining the Tf concentration change according to a Bicinchoninic Acid (BCA) Assay. The Tf loading amount (TLA) was defined as the total Tf amount on the MSP-AOPB NPs after reaction, and the Tf binding amount (TBA) was defined as the remaining Tf amount on the MSP-AOPB NPs after treatment with a specific eluent. The Tf loading efficiency (TLE) was defined as the percent of Tf loading amount compared with the initial Tf feeding amount, and the Tf binding efficiency (TBE) was defined as the percent of Tf binding amount compared with the initial Tf feeding amount.As shown in Fig. 3, the NaCl concentration (ionic strength) of the reaction medium was varied between 0 M and 1 M, and it greatly influenced the Tf loading and binding behavior on the MSP-AOPB NPs. When the NaCl concentration increased from 0 M to 0.1 M, the Tf loading efficiency (TLE) exhibited almost no change (>90%) and the binding efficiency (TBE) decreased from 74% to 59%. Further increasing the NaCl concentration from 0.1 M to 1 M, the TLE and TBE dropped to 30% and 27%, respectively. The addition of NaCl in the reaction medium would prohibit the attachment of Tf onto the MSP-AOPB NP surface due to weakening of electrostatic interaction, causing a loss of opportunity for covalent conjugation between Tf and AOPB-modified nanoparticles.
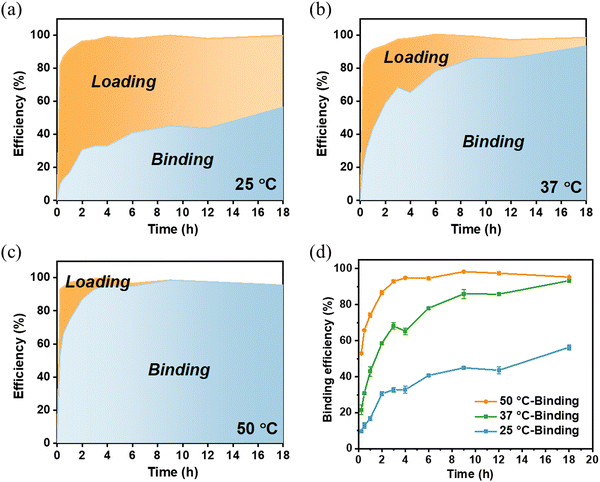
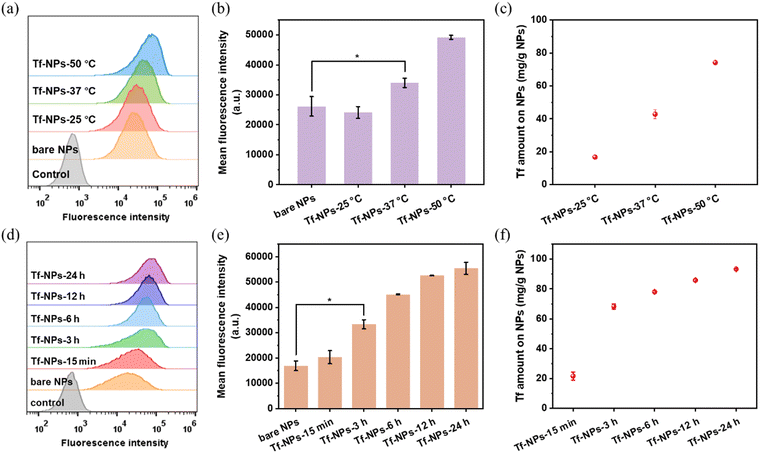
The effects of reaction temperature and time were also evaluated to determine the Tf loading efficiency and binding efficiency on MSP-AOPB NPs. We mixed the Tf solution with MSP-AOPB NPs at different temperatures (25 °C, 37 °C and 50 °C) for different incubation times (0–18 h). As shown in Fig. 4a–c and Fig. S4 (ESI†), Tf could rapidly adsorb on the MSP-AOPB NPs within 15 min at these temperatures with a loading efficiency larger than 80%. After reacting for 3 h, the Tf binding efficiency at the temperature of 25 °C, 37 °C and 50 °C was 33%, 68% and 93%, respectively (Fig. 4d). Prolonging the incubation time to 18 h, the Tf binding efficiency could reach 95% at the temperature of 37 °C and 50 °C. These results indicated that a higher reaction temperature and longer incubation time would increase the binding efficiency of Tf on the nanoparticle surface and promote the covalent coupling between Tf molecules and MSP-AOPB NPs.
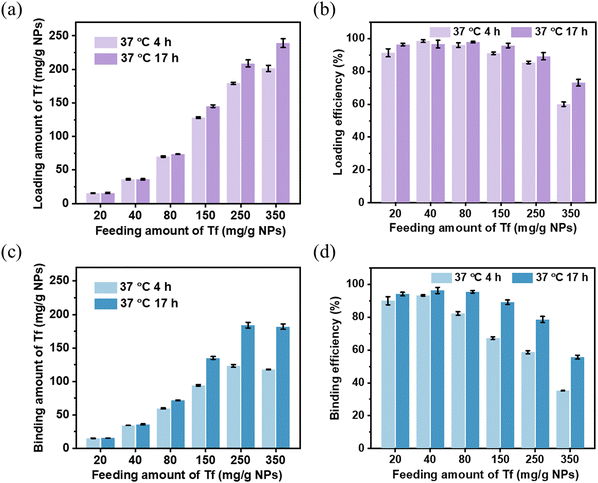
The effect of Tf initial feeding amount on the binding capacity was also studied. As presented in Fig. 5a and c, the Tf loading and binding capacity of MSP-AOPB NPs increased when the Tf feeding amount was in the range from 20 to 350 mg g−1 NPs, and it could attain the maximum loading capacity of 239 mg g−1 NPs. The effect of reaction time on the Tf binding properties showed no obvious difference at low feeding amounts (20–80 mg g−1 NPs) and significantly changed at high feeding amounts (150–350 mg g−1 NPs). Increasing the reaction time could encourage the formation of a five-membered cyclic ester and enhance the Tf binding capacity on MSP-AOPB NPs at high feeding amounts. The Tf loading and binding efficiency decreased when increasing the Tf initial feeding amount (Fig. 5b and d). However, the Tf binding property of the MSP-AOPB NPs still remained at a satisfactory level with high binding efficiency (90%) and high binding amount (135 mg g−1 NPs) at the feeding amount of 150 mg g−1 NPs. Besides, the reaction temperature had no obvious impact on the Tf loading efficiency, but could greatly influence the Tf binding efficiency at different feeding amounts from 15 to 120 mg g−1 NPs, indicating that a higher reaction temperature could contribute to a higher binding efficiency (Fig. S5, ESI†).
From the above results, the Tf binding efficiency on the MSP-AOPB NPs could be optimized after changing the reaction conditions. Meanwhile, the Tf ligand density could also be well controlled by simply adjusting the reaction parameters, such as reaction temperature, reaction time and initial feeding amount.
3.4. Study on the cellular uptake efficiency of Tf-mediated NPs by site-oriented conjugation
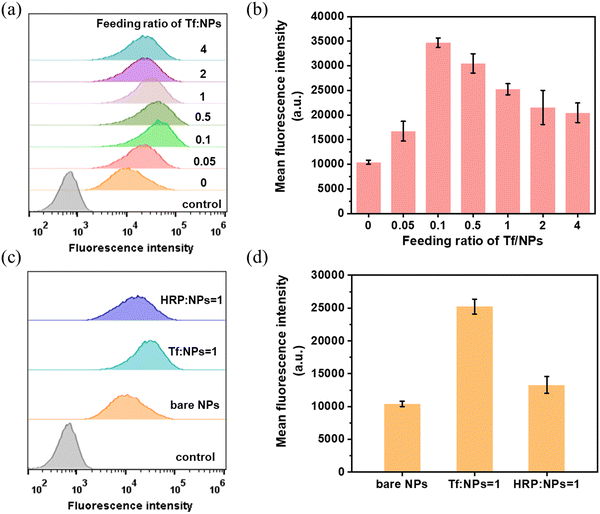
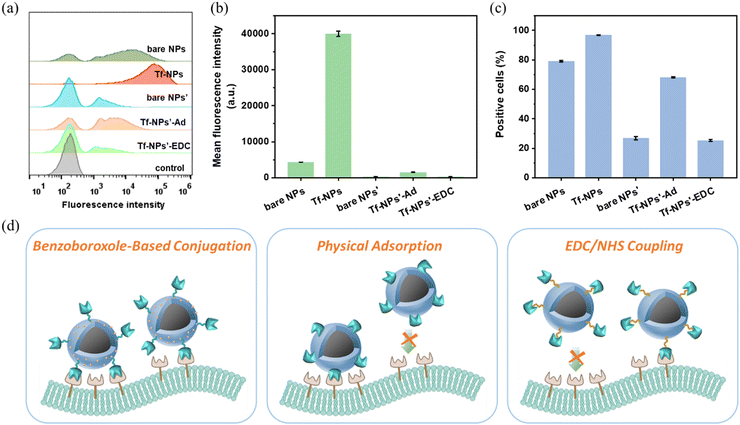
The Tf receptor is found to be overexpressed in many cancer cells compared with normal cells. To investigate the cellular uptake efficacy of Tf-MSP-AOPB NPs, HepG2 cells were used as a cell model due to the overexpression of TfR on the cell membrane. MSP-AOPB NPs were labeled with Cy7 (fluorescent dye). Different amounts of Tf were conjugated on the nanoparticle surface to obtain Tf-MSP-AOPB NPs, followed by incubation with HepG2 cells. Flow cytometry analysis was employed to measure the cell targeting ability by detecting the fluorescence intensity.Tf-MSP-AOPB NPs were prepared by mixing Tf solution and MSP-AOPB NPs at different reaction temperatures and reaction times. The initial Tf feeding amount in the coupling process was 100 mg g−1 NPs. Tf-MSP-AOPB NPs prepared here were abbreviated as Tf-NPs. MSP-AOPB NPs without Tf modification served as a control and were abbreviated as bare NPs. As shown in Fig. 6c, the Tf binding amount on the Tf-NPs at a reaction temperature of 25 °C, 37 °C and 50 °C was 16, 43 and 74 mg g−1 NPs, respectively. The targeting ability of the Tf-NPs prepared at 37 °C and 50 °C increased by 30% and 88% compared with the bare NPs, respectively, while that of Tf-NPs prepared at 25 °C showed no significant statistical difference (Fig. 6a and b), which means that too low a density of Tf on the nanoparticles will greatly reduce the targeting property. When the Tf-NPs prepared at different reaction time points (15 min, 3 h, 6 h, 12 h, 24 h) were incubated with HepG2 cells, a significant difference in cell uptake amount could be observed when the Tf-NPs were prepared for longer reaction time (≥3 h) compared with the bare NPs (Fig. 6d–f). Tf binding amount on Tf-NPs-15 min was 22 mg g−1 NPs, and the cell uptake of Tf-NPs-15 min had no significant difference with bare NPs. After increasing the reaction time, the Tf binding amount on Tf-NPs-3 h was 68 mg g−1 NPs. The cellular uptake efficacy of Tf-NPs-3 h increased by 97% compared with bare NPs. Moreover, the cell targeting ability of Tf-NPs-24 h could increase by 228% with the Tf binding amount of 93 mg g−1 NPs on the surface, which corresponds to a ligand binding density of about 0.05 Tf/nm2. Tf ligand density could be well controlled by simply adjusting the reaction temperature and time. Low Tf ligand density could not provide enough opportunities for nanoparticles to bind with TfR on cell membranes, and the interactions between nanoparticles and cells were weak without enough Tf modification. Higher Tf ligand density obtained by increasing the reaction temperature and time could effectively promote the interactions between nanoparticles and cell membranes, and thus enhance the cellular uptake of nanoparticles.
When we further incubated HepG2 cells with Tf-NPs prepared by changing the initial feeding mass ratio of Tf/NPs, there was an increase in the cellular uptake first, and then a decrease when the feeding ratio of Tf/NPs was larger than 0.1 (Fig. 7a and b). It is very important information that the ligand density has an optimized level. The optimal ligand density for achieving the highest cellular uptake amount was 0.05 Tf/nm2 (94 mg Tf/g NPs) by using the feeding ratio of Tf/NPs = 0.1, and the binding efficiency of the conjugation process was larger than 90%. Besides, the cellular uptake amounts of nanoparticles also depended on the nanoparticle dosage and incubation time. This indicated that increasing the nanoparticle dosage (Fig. S6, ESI†) or increasing the incubation time (Fig. S7, ESI†) could significantly enhance the cellular uptake. In order to further investigate the targeting ability of Tf, we conjugated another kind of glycoprotein HRP on the MSP-AOPB NPs. As presented in Fig. 7c and d, Tf conjugated nanoparticles exhibited the highest cellular uptake amount when compared with bare NPs and HRP-MSP-AOPB NPs. Due to the abnormal expression level of glycoproteins on cancer cell membranes, the bare NPs which were modified with benzoboroxole groups on the surface could interact with the glycoproteins on HepG2 cells, and this would cause an increased mean fluorescence intensity of bare NPs. However, Tf-conjugated nanoparticles still showed an improved targeting property based on receptor-mediated endocytosis.
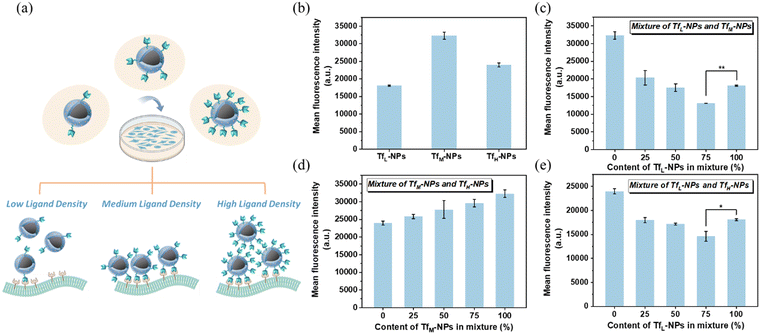
In order to investigate the effect of ligand density on the cellular uptake efficacy, we prepared three kinds of Tf-MSP-AOPB NPs with low, medium and high Tf ligand density (abbreviated as TfL-NPs (low Tf density), TfM-NPs (medium Tf density) and TfH-NPs (high Tf density)). TfL-NPs, TfM-NPs and TfH-NPs were obtained by using the feeding ratio of Tf/NPs = 0.05, 0.1 and 1. The Tf binding amount on TfL-NPs, TfM-NPs and TfH-NPs was 46, 94 and 184 mg g−1 NPs, respectively. Then, we incubated these nanoparticles with HepG2 cells and measured the cellular uptake by flow cytometry. As presented in Fig. 8b, TfM-NPs showed the best cellular uptake efficacy. For TfL-NPs, low ligand density would decrease the opportunity for nanoparticles to recognize cells and weaken the interactions, leading to a lower cellular uptake. In addition, it also demonstrated that excessively high Tf ligand density could suppress the cellular uptake of nanoparticles. One possible reason is that high Tf ligand density will inhibit the Tf from being recognized by TfR on cells due to steric interference. Another possible reason should be that the nanoparticles conjugated with high ligand density would bind with more receptors on the cell membranes, causing reduced chances for other free nanoparticles to interact with available receptors, and thus decreasing the cellular uptake of nanoparticles (Fig. 8a).
To further study the effect of homogeneity of ligand density on the cellular uptake efficacy, mixtures of two kinds of nanoparticles at different proportions were incubated with HepG2 cells. For the mixed system, the data indicated that the addition of TfL-NPs to TfM-NPs or TfH-NPs to form a mixture could lead to a decrease in cellular uptake compared with TfM-NPs or TfH-NPs only (Fig. 8c and e), which was consistent with the previous result that higher ligand density could improve cellular uptake efficacy. Besides, 75% of TfL-NPs added in the mixture could even cause less cellular uptake amount compared with TfL-NPs only, the possible reason should be that the addition of nanoparticles with high ligand density could consume more available receptors and thus inhibited the interactions between the nanoparticles with low ligand density and cells. This was also a possible reason that the mixture of TfM-NPs with TfH-NPs showed an increase in cellular uptake when decreasing the content of TfH-NPs in the mixture (Fig. 8d). The above results showed that cellular uptake efficacy was dependent on ligand density and the homogeneity of ligand densities, which were of prime importance in cell targeting delivery. Meanwhile, the experimental results showed that the MSP-AOPB NPs had no cytotoxicity against normal cells (HEK 293T cells), and could not influence the cell viability of HepG2 cells (Fig. S8, ESI†).
3.5. Comparison of cellular uptake efficacy for different conjugation methods
Based on the AOPB moiety on nanoparticles, Tf could be conjugated on the nanocarrier surface in a site-oriented manner through the coupling reaction between cis-diols in the sugar chain of Tf and benzoboroxoles on MSP-AOPB NPs, which meant that the function of Tf to recognize receptors on cell membranes could be maintained in the fabrication procedure. In order to inspect the advantage of the benzoboroxole-based coupling method, we also prepared another two kinds of Tf-NPs by physical adsorption or the traditional EDC/NHS method to modify Tf on MSP-IDA NPs, which were abbreviated as Tf-NPs’-Ad and Tf-NPs’-EDC, respectively. Nanoparticles prepared by the benzoboroxole-based method were abbreviated as Tf-NPs. As shown in Fig. 9a and b, Tf-NPs showed the highest cellular uptake amount among these nanoparticles. The mean fluorescence intensity of the Tf-NPs detected in cells was 25 times and 127 times higher than that of Tf-NPs’-Ad and Tf-NPs’-EDC in cells, respectively. The highest percentage of cells with fluorescence detected was observed for Tf-NPs (97% positive cells), indicating that almost all the cells had uptake of nanoparticles (Fig. 9c). Just as we know, the evaluation of targeting-agent orientation is usually associated with the cellular uptake activity,49–51 and oriented immobilization of targeting agents can effectively improve the targeting properties of the nanocarriers. Herein, we prepared several samples through different immobilization methods to discuss the orientation of the targeting ligands. For Tf-NPs, Tf was directionally conjugated on the nanoparticles through the cis-diols in the sugar chain, which is irrelevant to the receptor-binding reaction, and other regions of Tf will not be interfered with. Thus, it can create more opportunities for Tf to bind with receptors on the cell membranes and the receptor-binding region is less affected (Fig. 9d). Meanwhile, for Tf immobilized on nanoparticles by physical adsorption (Tf-NPs’-Ad), the Tf was randomly adsorbed on the surface through the positive regions by electrostatic interaction. And for Tf immobilized on nanoparticles by the EDC/NHS coupling method (Tf-NPs’-EDC), the Tf was randomly conjugated to the carboxyl modified nanoparticles through the amino groups, which were distributed on the whole protein surface by amidation reaction. Thus, the receptor-binding region of Tf would be largely influenced, and it could lead to a lower cellular uptake (Fig. 9d). The results shown in Fig. 9b and c demonstrate that Tf immobilized nanoparticles by the benzoboroxole method exhibited the best cellular uptake performance. This confirmed the advantage of the benzoboroxole-based coupling method in the oriented immobilization of the targeting ligand on the nanoparticles.4. Conclusions
In summary, we have established a benzoboroxole-modified core–shell magnetic nanoparticle platform for facile site-oriented immobilization of a targeting ligand (transferrin, Tf), which was based on the covalent reaction between cis-diols in the sugar chain of Tf and benzoboroxoles on the MSP-AOPB NPs. The maximum Tf binding amount on the MSP-AOPB NPs was 184 mg g−1, and the ligand density on the nanoparticle surface could be easily controlled by simply adjusting the reaction conditions. In vitro studies revealed the excellent Tf-mediated targeting ability of the magnetic nanoparticles and enhanced cellular uptake efficacy by optimizing the ligand density. The experimental results demonstrated the advantage of site-oriented conjugation with the benzoboroxole-based nanoparticle system compared with physical adsorption or the traditional EDC coupling reaction, and their cellular uptake amount detected in HepG2 cells was 25 and 127 times higher than that of physical adsorption and the EDC/NHS coupling reaction. This approach can be expected to be applicable in the site-oriented immobilization of antibodies via the sugar moiety on the constant region (Fc) without affecting the targeting ability of the antigen-binding region (Fab). Considering the ligand density-dependent cellular uptake and the high cost of targeting agents, this article provides a feasible technique to optimize the ligand density in an effective and economic way, which can facilitate the targeted delivery research of nanoparticles in vivo. Therefore, this work shows great potential in surface functionalization with a targeting agent in nanomedicine design and drug delivery.Author contributions
C. W. and W. F. conceived the concept. W. F. and X. W. conducted the experimental material preparation and characterization. W. F. and C. W. drafted the manuscript. C. W. and J. G. supervised the project. All the authors have given approval to the final version of the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
We acknowledge the financial support by the National Natural Science Foundation of China (Grant No. 52131308, 51721002, 51873040).References
- A. L. Papa, N. Korin, M. Kanapathipillai, A. Mammoto, T. Mammoto, A. Jiang, R. Mannix, O. Uzun, C. Johnson, D. Bhatta, G. Cuneo and D. E. Ingber, Biomaterials, 2017, 139, 187–194 CrossRef CAS PubMed
.
- F. Soetaert, P. Korangath, D. Serantes, S. Fiering and R. Ivkov, Adv. Drug Delivery Rev., 2020, 163–164, 65–83 CrossRef CAS PubMed
.
- Y. Islam, A. G. Leach, J. Smith, S. Pluchino, C. R. Coxon, M. Sivakumaran, J. Downing, A. A. Fatokun, M. Teixidò and T. Ehtezazi, Adv. Sci., 2021, 8, 2002085 CrossRef CAS PubMed
.
- J. Zhang, Y. Lin, Z. Lin, Q. Wei, J. Qian, R. Ruan, X. Jiang, L. Hou, J. Song, J. Ding and H. Yang, Adv. Sci., 2022, 9, 1–27 Search PubMed
.
- S. Lv, Y. Wu, K. Cai, H. He, Y. Li, M. Lan, X. Chen, J. Cheng and L. Yin, J. Am. Chem. Soc., 2018, 140, 1235–1238 CrossRef CAS PubMed
.
- Y. Wang, Y. Wang, Y. Hu, W. J. Yang and L. Wang, Polym. Chem., 2021, 12, 4159–4166 RSC
.
- Y. Geng, J. Xiang, S. Shao, J. Tang and Y. Shen, J. Controlled Release, 2022, 342, 122–133 CrossRef CAS PubMed
.
- L. Yang, Y. Zhang, Y. Zhang, Y. Xu, Y. Li, Z. Xie, H. Wang, Y. Lin, Q. Lin, T. Gong, X. Sun, Z. Zhang and L. Zhang, ACS Nano, 2022, 16, 9799–9809 CrossRef CAS PubMed
.
- M. Wu, Q. Wang, S. Chen, Z. Zhou, J. Li, H. Sun, J. Liu, G. Wang, F. Zhou and M. Sun, J. Controlled Release, 2022, 350, 1–10 CrossRef CAS PubMed
.
- S. Xu, L. Wang and Z. Liu, Angew. Chem., Int. Ed., 2021, 60, 3858–3869 CrossRef CAS PubMed
.
- X. Ling, W. Han, X. Jiang, X. Chen, M. Rodriguez, P. Zhu, T. Wu and W. Lin, Biomaterials, 2021, 270, 120690 CrossRef CAS PubMed
.
- J. Chen, J. Yang and J. Ding, J. Mater. Chem. B, 2022, 10, 7173–7182 RSC
.
- M. Manzano and M. Vallet-Regí, Adv. Funct. Mater., 2020, 30, 3–5 CrossRef
.
- Y. Tan, J. Feng, Y. Xiao and C. Bao, J. Mater. Chem. B, 2022, 10, 4840–4855 RSC
.
- J. Zhang, X. Su, L. Weng, K. Tang, Y. Miao, Z. Teng and L. Wang, J. Colloid Interface Sci., 2023, 633, 102–112 CrossRef CAS PubMed
.
- Y. Weng, S. Guan, L. Wang, H. Lu, X. Meng, G. I. N. Waterhouse and S. Zhou, Small, 2020, 16, 1–10 Search PubMed
.
- Q. Mu, G. Lin, M. Jeon, H. Wang, F. C. Chang, R. A. Revia, J. Yu and M. Zhang, Mater. Today, 2021, 50, 149–169 CrossRef CAS PubMed
.
- X. Zhao, J. Zhang, B. Chen, X. Ding, N. Zhao and F. J. Xu, Small Methods, 2023, 2201595, 1–12 Search PubMed
.
- F. Gao, B. Yu, H. Cong and Y. Shen, J. Mater. Chem. B, 2022, 10, 6896–6921 RSC
.
- Z. Qiao, K. Zhang, J. Liu, D. Cheng, B. Yu, N. Zhao and F. J. Xu, Nat. Commun., 2022, 13, 1–16 Search PubMed
.
- M. Hou, Y. Wei, Z. Zhao, W. Han, R. Zhou, Y. Zhou, Y. Zheng and L. Yin, Adv. Mater., 2022, 34, 1–14 Search PubMed
.
- X. Wang, M. Wang, R. Lei, S. F. Zhu, Y. Zhao and C. Chen, ACS Nano, 2017, 11, 4606–4616 CrossRef CAS PubMed
.
- D. Lou, L. Ji, L. Fan, Y. Ji, N. Gu and Y. Zhang, Langmuir, 2019, 35, 4860–4867 CrossRef CAS PubMed
.
- B. Saha, P. Songe, T. H. Evers and M. W. J. Prins, Analyst, 2017, 142, 4247–4256 RSC
.
- R. Duwa, A. Banstola, F. Emami, J. H. Jeong, S. Lee and S. Yook, J. Drug Delivery Sci. Technol., 2020, 60, 101928 CrossRef CAS
.
- D. Caballero, J. Samitier, J. Bausells and A. Errachid, Small, 2009, 5, 1531–1534 CrossRef CAS PubMed
.
- M. Aydın, E. B. Aydın and M. K. Sezgintürk, Biosens. Bioelectron., 2018, 107, 1–9 CrossRef PubMed
.
- X. Suo, B. N. Eldridge, H. Zhang, C. Mao, Y. Min, Y. Sun, R. Singh and X. Ming, ACS Appl. Mater. Interfaces, 2018, 10, 33464–33473 CrossRef CAS PubMed
.
- H. Yang, Q. V. Le, G. Shim, Y. K. Oh and Y. K. Shin, Acta Pharm. Sin. B, 2020, 10, 2212–2226 CrossRef CAS PubMed
.
- Y. Zhang, W. Ma, D. Li, M. Yu, J. Guo and C. Wang, Small, 2014, 10, 1379–1386 CrossRef CAS PubMed
.
- H. Xiao, W. Chen, J. M. Smeekens and R. Wu, Nat. Commun., 2018, 9, 1692 CrossRef PubMed
.
- J. Luan, X. Zhu, L. Yu, Y. Li, X. He, L. Chen and Y. Zhang, Talanta, 2023, 251, 123772 CrossRef CAS PubMed
.
- S. A. S. L. Rosa, R. Dos Santos, M. R. Aires-Barros and A. M. Azevedo, Sep. Purif. Technol., 2016, 160, 43–50 CrossRef CAS
.
- R. dos Santos, S. A. S. L. Rosa, M. R. Aires-Barros, A. Tover and A. M. Azevedo, J. Chromatogr. A, 2014, 1355, 115–124 CrossRef CAS PubMed
.
- S. Wu, F. Wu and X. Chen, Adv. Mater., 2022, 34, 2109210 CrossRef CAS PubMed
.
- J. Wan, Y. Li, K. Jin, J. Guo, J. Xu and C. Wang, ACS Appl. Mater. Interfaces, 2020, 12, 23717–23725 CrossRef CAS PubMed
.
- J. Sun, Y. Li, X. Wang, W. Fei, J. Guo and C. Wang, ACS Appl. Mater. Interfaces, 2021, 13, 12888–12898 CrossRef CAS PubMed
.
- K. G. Reuter, J. L. Perry, D. Kim, J. C. Luft, R. Liu and J. M. DeSimone, Nano Lett., 2015, 15, 6371–6378 CrossRef CAS PubMed
.
- M. Li, J. Dong, F. Cheng, C. Li, H. Wang, T. Sun, W. He and Q. Wang, Mol. Pharm., 2021, 18, 1196–1207 CrossRef CAS PubMed
.
- C. Chen, Y. Zhou, C. Chen, S. Zhu and X. Yan, ACS Nano, 2022, 16, 6886–6897 CrossRef CAS PubMed
.
- D. R. Elias, A. Poloukhtine, V. Popik and A. Tsourkas, Nanomedicine, 2013, 9, 194–201 CrossRef CAS PubMed
.
- A. J. Grippin, B. Wummer, T. Wildes, K. Dyson, V. Trivedi, C. Yang, M. Sebastian, H. R. Mendez-Gomez, S. Padala, M. Grubb, M. Fillingim, A. Monsalve, E. J. Sayour, J. Dobson and D. A. Mitchell, ACS Nano, 2019, 13, 13884–13898 CrossRef CAS PubMed
.
- C. Lu, L. Han, J. Wang, J. Wan, G. Song and J. Rao, Chem. Soc. Rev., 2021, 50, 8102–8146 RSC
.
- Y. Park, A. A. Demessie, A. Luo, O. R. Taratula, A. S. Moses, P. Do, L. Campos, Y. Jahangiri, C. R. Wyatt, H. A. Albarqi, K. Farsad, O. D. Slayden and O. Taratula, Small, 2022, 18, 1–15 Search PubMed
.
- L. Messori and F. Kratz, Met.-Based Drugs, 1994, 1, 161–167 CrossRef CAS PubMed
.
- H. Kawabata, Free Radic. Biol. Med., 2019, 133, 46–54 CrossRef CAS PubMed
.
- B. Xue, D. DasGupta, M. Alam, M. S. Khan, S. Wang, A. Shamsi, A. Islam and M. I. Hassan, J. Cell. Biochem., 2022, 123, 1381–1393 CrossRef CAS PubMed
.
- A. M. N. Silva, T. Moniz, B. de Castro and M. Rangel, Coord. Chem. Rev., 2021, 449, 214186 CrossRef CAS
.
- M. K. Greene, D. A. Richards, J. C. F. Nogueira, K. Campbell, P. Smyth, M. Fernández, C. J. Scott and V. Chudasama, Chem. Sci., 2018, 9, 79–87 RSC
.
- W. Zhang, Q. A. Besford, A. J. Christofferson, P. Charchar, J. J. Richardson, A. Elbourne, K. Kempe, C. E. Hagemeyer, M. R. Field, C. F. McConville, I. Yarovsky and F. Caruso, Nano Lett., 2020, 20, 2660–2666 CrossRef CAS PubMed
.
- K. Alt, F. Carraro, E. Jap, M. Linares-Moreau, R. Riccò, M. Righetto, M. Bogar, H. Amenitsch, R. A. Hashad, C. Doonan, C. E. Hagemeyer and P. Falcaro, Adv. Mater., 2021, 34, 2106607 CrossRef PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb01375h |
| This journal is © The Royal Society of Chemistry 2023 |