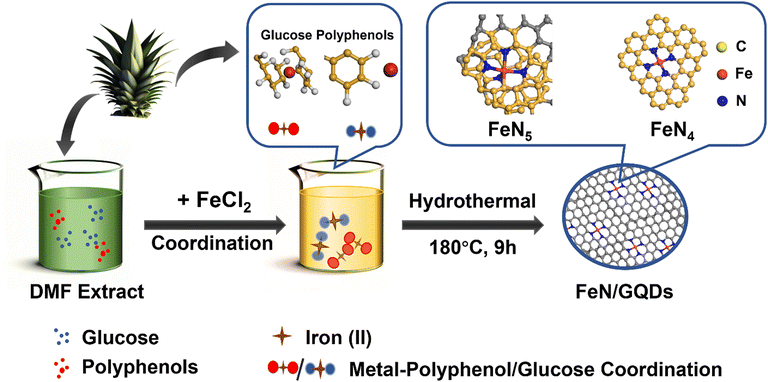
Green synthesis of iron-doped graphene quantum dots: an efficient nanozyme for glucose sensing†
Xinqi
Li‡
a,
Guanyou
Lin‡
a,
Lijun
Zhou
b,
Octavia
Prosser
a,
Mohammad H.
Malakooti
 *abc and
Miqin
Zhang
*abc and
Miqin
Zhang
 *ac
*ac
aDepartment of Materials Science and Engineering, University of Washington, Seattle, Washington 98195, USA. E-mail: mzhang@uw.edu
bDepartment of Mechanical Engineering, University of Washington, Seattle, WA 98195, USA. E-mail: malakoot@uw.edu
cInstitute for Nano-Engineered Systems, University of Washington, Seattle, WA 98195, USA
First published on 23rd March 2024
Abstract
Single-atom nanozymes with well-defined atomic structures and electronic coordination environments can effectively mimic the functions of natural enzymes. However, the costly and intricate preparation processes have hindered further exploration and application of these single-atom nanozymes. In this study, we presented a synthesis technique for creating Fe–N central single-atom doped graphene quantum dot (FeN/GQDs) nanozymes using a one-step solvothermal process, where individual iron atoms form strong bonds with graphene quantum dots through nitrogen coordination. Unlike previous studies, this method significantly simplifies the synthesis conditions for single-atom nanozymes, eliminating the need for high temperatures and employing environmentally friendly precursors derived from pineapple (ananas comosus) leaves. The resulting FeN/GQDs exhibited peroxidase-like catalytic activity and kinetics comparable to that of natural enzymes, efficiently converting H2O2 into hydroxyl radical species. Leveraging their excellent peroxide-like activity, FeN/GQDs nanozymes have been successfully applied to construct a colorimetric biosensor system characterized by remarkably high sensitivity for glucose detection. This achievement demonstrated a promising approach to designing single-atom nanozymes with both facile synthesis procedures and high catalytic activity, offering potential applications in wearable sensors and personalized health monitoring.
New conceptsThis study reveals a highly promising method for constructing single-atom nanozymes, particularly well-suited for applications in the biomedical field, such as glucose detection. Traditional noble metal nanozymes often present challenges related to biological toxicity or high costs in these applications. While Fe-based nanozymes exhibit lower biological toxicity, their preparation typically involves high-temperature conditions and intricate procedures. In this research, a novel approach was introduced by using plant extracts as precursors to coordinate single iron atoms, successfully achieving the synthesis of FeN/GQDs single-atom nanozymes. This method not only simplifies the synthesis process but also significantly reduces its complexity. The resulting FeN/GQDs single-atom nanozymes not only demonstrate outstanding peroxidase-like enzyme performance but also exhibit remarkably high sensitivity in glucose detection. This study offers new insights into designing single-atom nanozymes, advancing the synthesis of safe and cost-effective nanozymes. This promising strategy holds great potential for diverse applications in biosensing and various biomedical fields. |
1. Introduction
Natural enzymes are biocatalysts that accelerate reactions in living organisms and are essential to metabolic processes. However, their susceptibility to instability and inactivation under extreme conditions such as high heat and extreme pH limits their practical applications. In contrast, nanozymes represent an innovative category of nanomaterials that mimic the catalytic activity of natural enzymes while offering various advantages such as cost-effectiveness, ease of production, and enhanced stability.1–4 Since the first report on the peroxidase-like properties of Fe3O4 NPs by Yan et al. in 2007,5 nanozymes have gained significant attention as potential alternatives to natural enzymes across diverse biomedical fields, including bioimaging,6–8 disease diagnosis and treatment,9–12 and biosensing.11,13,14Among various nanozyme structures and compositions, single-atom nanozymes, a type of nanozyme that has only one metal atom as the active site for catalysis, have gained widespread attention due to their high catalytic activity, stability, and unique molecular structure.11,13,14 These single-atom nanozymes feature multiple isolated surface atoms in an unsaturated coordination environment, which maximizes atomic efficiency and active site density. This leads to highly efficient catalytic activity compared to bulk structures with multiple atoms bulk-in atom structures.15
Various single-atom nanozymes have been designed and synthesized using diverse elements, such as Au6,16–18 Pt,19,20 Ag,21,22 Co,23 and Mn,24 as catalytic centers, all showing promising results across a wide range of catalytic applications. However, certain challenges remain in their practical applications. For instance, many reported single-atom nanozymes rely on expensive noble metals such as Au and Pt as reaction sites.25,26 These noble metals are not biodegradable and may cause adverse effects as they accumulate in living organisms. Moreover, during the synthesis process, complex techniques are often employed to prevent metal overoxidation and cluster over-aggregation,27 which not only complicates production but also hinders large-scale manufacturing.26,28 Furthermore, the heavy-metal-based materials could release toxic ions, and raise their concerns about potential toxicity in biomedical applicationss.26–28 As a result, there is a growing interest in using more sustainable and environmentally friendly materials, such as Fe, to synthesize nanozymes.
Fe is a readily available and cost-effective element, making it a promising alternative to noble metals such as Au, Pt, and Cu for various catalytic systems.28,29 Furthermore, Fe-based nanozymes have demonstrated high stability and catalytic activity in aqueous solutions, making them well-suited for applications in biological systems.30–32 In recent years, the synthesis of single-atom nanozymes using Fe atoms has emerged as an promising area of research, with a growing body of research highlighting the diverse potential applications of Fe-based nanozymes. For instance, a high-temperature gas-migration strategy was utilized to create porphyrin-like single Fe sites on N-doped carbon nanomaterials, resulting in excellent peroxidase, oxidase, catalase enzyme-like, and Fenton-like activities.33 Alternatively, the use of Fe(phen)x as precursors, combined with a support-sacrificed strategy, has been employed to obtain single Fe atom nanozymes through pyrolysis and pickling under 600 °C.34 However, it is important to note the synthesis of Fe-based single-atom nanozymes often relies on metal–organic framework (MOF) materials as substrates for better anchoring of single-atom sites. This process is costly and involves high temperatures during MOF synthesis (Table 1),28,33–37 Additionally, the complex screening process further adds to the unsuitability of this method for large-scale production. Therefore, the development of a facile synthesis method for Fe-based single-atom nanozymes is necessary to fully harness their potential for widespread applications.
| Single Fe nanozymes | Synthesis condition | Precursors | Application |
|---|---|---|---|
| a This work. | |||
| Fe–N–C Sazymes38 | Pyrolysis, 900 °C, N2 atmosphere | Glucose, DICY, and FeCl2·4H2O | H2O2 detection |
| FeBNC SACs12 | Pyrolysis | Glucose, dicyandiamide (DICY), boric acid, iron dichloride | Detection of acetylcholinesterase |
| Fe-SANs30 | Pyrolysis, 700 °C, N2 atmosphere | 1,10-Phenanthroline monohydrate (O-Phen), zinc acetate dihydrate (Zn(OAc)2·2H2O), phthalocyanine (FePc) | Drug detection |
| CNT/FeNC39 | Pyrolysis | CNTs, pyrrole, Fe(NO3)3, NaCl | Biosensing |
| Fe–Zn ZIFs/Fe–N/C36 | ZIFs pyrolysis | Fe(NO3)3·9H2O, Zn(NO3)2·6H2O, MIM | Detection of alkaline phosphatase |
| Fe–N–C SANs40 | Pyrolysis, 900 °C, N2 atmosphere | Ferric chloride, polyvinylpyrrolidone, potassium chloride, or sodium chloride | Selective determination of antioxidants |
| Fe–N–C Sazyme41 | Calcination, 350 °C + Pyrolysis, 800 °C | Pluronic F127, dopamine hydrochloride, (NH4)2Fe(SO4)2·6H2O, 1,3,5-trimethylbenzene, NH3·H2O | Antibacterial therapy |
| Porphyrin-like FeSAzyme42 | Pyrolysis, 800 °C | Fe(acac)3-ZIF-8@mSiO2 | Tumor therapy |
| Fe–N–C SAN43 | Pyrolysis, 900 °C, N2 atmosphere | Zn(NO3)2·6H2O, Fe(NO3)3·6H2O, methylimidazole | Biosensing |
| SA-Fe/NG44 | Pyrolysis, 800 °C | Urea, ferrous acetate, 1,10-phenanthroline monohydrate | Detection of Cr(VI) |
| Fe–N/C sazyme45 | Pyrolysis, 900 °C, N2 atmosphere | Fe(acac)3@ZIF-8 | Detection of malathion |
| Fe–N/S–C32 | 400 °C carbonized peanut shells, pyrolysis, 800 °C | Peanut shells, Fe(NO3)2·9H2O, CO(NH2)2 | Colorimetric detection of GSH and Hg2+ |
| FeSA-HNCS46 | Pyrolysis, 900 °C, N2 atmosphere | SiO2 nanospheres, TEOS, DA, Fe(acac)3 | Cell therapy |
| FeCu-DA/NC47 | Pyrolysis | Metal salts (Fe, Cu), PVP and nano-CaCO3 | Oxygen reduction |
| FeN/GQDsa | Solvothermal, 190 °C | Plant leaves, FeCl2 | Biosensing |
In this study, we introduced a liquid-phase chelation-assisted anchoring strategy for the preparation of single iron atom doped graphene dots (FeN/GQDs), simplifying the complex process required by the pyrolysis method. The synthesis of FeN/GQDs involved a facile solvothermal approach using chemicals extracted from ananas comosus leaves as precursors (Scheme 1). The single Fe atoms were anchored to graphene quantum dots through Fe–N coordinate bonds, serving as catalytic sites. Compared to traditional metal-doping methods involving high temperature and pressure, our process utilizes plant extracts, reducing environmental impact. Additionally, our solvothermal synthesis lowers energy consumption compared to the pyrolytic decomposition reactions previously reported studies.48,49 Notably, the solvothermal synthesis process used in this study was carried out at a significantly lower temperature of 180 °C, in contrast to the high temperature pyrolysis methods (>800 °C) with N2 atmosphere protection used in previous studies (Table 1). The extracts obtained from ananas comosus leaves served as carbon and nitrogen sources of graphene quantum dots and also contained plant polyphenols, which played a key role in fixing the Fe2+ ions.50,51 This natural source material may possess lower biotoxicity compared to synthetic chemicals.52 Dimethylformamide (DMF) served as the reaction solvent and helped prevent the oxidation of Fe2+ to Fe3+ during the reaction.53 The support from graphene quantum dot provided mechanical stability, while the nitrogen coordination structure acted as a protective layer, preventing oxidation and degradation of the Fe atoms. Furthermore, the confirmed biosafety of GQDs, combined with their synthesis from biomass precursors, underscores the potential for enhanced safety in biomedical applications.54–56 This unique combination of ingredients and synthesis conditions enabled the successful preparation of FeN/GQDs with enhanced stability and catalytic activity, making them promising candidates for various applications.
We also investigated the catalytic performance and potential application of FeN/GQDs. We found that FeN/GQDs exhibited remarkable peroxidase-like activity in acidic environments. This was demonstrated by their effective catalysis in various chromogenic systems using substrates such as 3,3′,5,5′-tetramethylbenzidine (TMB), o-phenylenediamine (OPD), and 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS). Furthermore, we assessed the performance of FeN/GQDs in colorimetric biosensing for glucose detection. Glucose plays a crucial role in the body's metabolic process, and maintaining physiological glucose level is essential for good health.57 Various methods have been explored for glucose detection, including colorimetry,58 fluorescence,59 surface-enhanced Raman scattering,60 chemiluminescence,61 and electrochemistry.62 Among these detection techniques, colorimetric approaches with nanozymes offer several advantages, including visual detectability, convenience, and cost-effectiveness, making them highly attractive options for glucose detection.63
In this study, we combined the colorimetric glucose detection system with glucose oxidase and FeN/GQDs. Through the innovative implementation of the chelation-assisted anchoring strategy and solvothermal synthesis, FeN/GQDs nanozymes exhibited not only exquisite Fe–N single-atom structures but also remarkable sensitivity to substrates and peroxidase-like activity. This approach emphasizes environmental sustainability, and significantly improves biocompatibility standards. Furthermore, the outstanding performance demonstrated in applications highlights the profound impact and promising future of this technology. As such, this study represents a significant step forward in adopting eco-conscious methodologies and lays a solid foundation for future developments in this field.
2. Materials and methods
Chemical reagents
All chemical reagents were of analytically pure grade and used without further purification. The following reagents were purchases from Sigma-Aldrich (St Louis, MO, USA): iron(II) chloride tetrahydrate (FeCl2·4H2O), 2,2-azinobis-3-ethylbenzthiazoline-6-sulphonate (ABTS, 98%), 3,3,5,5-tetramethylbenzidine (TMB, 99%), o-phenylenediamine (OPD, 98%), glucose oxidase (Gox), and horseradish peroxidase (HRP). The following reagents were purchased from Fisher Scientific (Walthem, MA, USA): hydrogen peroxide (H2O2, 30%), N,N-dimethylformamide (DMF, HPLC grade), acetic acid (HAc, 99.5%), sodium acetate (NaAc), and dimethyl sulfoxide (DMSO, HPLC grade). Ananas comosus leaves were sourced from pineapples purchased from Costco. Ethanol (anhydrate) was purchased from Decon Labs, Inc. (Montgomery County, PA, USA).Preparation of nanozymes
FeN/GQDs were synthesized using a green solvent thermal method, which involved ananas comosus leaves as the selected carbon and nitrogen sources. The leaves were washed and chopped into pieces prior to use. DMF was used as the solvent, serving multiple functions including supplementing nitrogen, reducing ferric ions (iron(III)), and protecting the ferrous ions (iron(II)) from oxidation.35To synthesize FeN/GQDs, a step-by-step process was followed. First, 2 g of chopped ananas comosus leaves were ultrasonicated in 20 mL of DMF to obtain an extract solution. Next, the extract solution was mixed with 1 g FeCl2 and sonicated again to obtain a homogeneous mixture. The resulting mixture was then transferred into a 50 mL PTFE liner and heated at 180 °C for a duration of 9 hours. After the solvothermal reaction, the final product underwent a purification process involving multiple rounds of centrifugation and dialysis. First, 6k rpm centrifuge for 10 min to initially remove the bulk products. Subsequently, 3.5 kDa dialysis was employed to eliminate residual small molecules. The final FeN/GQDs was isolated by collecting the supernatant following centrifugation at 14k rpm for 10 minutes. The resulting solution, containing nanoparticles with sizes of only 10–15 nm, was freeze-dried to obtain the FeN/GQD in powder form for further experiments. This method of synthesis ensures the production of FeN/GQDs with desirable properties, such as enhanced stability and catalytic activity.
For non-Fe–N doped graphene quantum dots (NGQDs), the synthesis method is similar to the one described above for FeN/GQDs, except with no FeCl2 added.
To compare the properties of single iron atom nanozymes, a type of Fe3O4 nanozymes (Fe3O4 coated with polymer) was synthesized as a reference material. The detailed synthesis steps for these Fe3O4 can be found in the ESI.†
DLS measurements
Dynamic light scattering (DLS, Zetasizer Nano-ZS, Malvern Instruments, Worcestershire, UK) was utilized to determine the average particle size and zeta potential of FeN/GQDs in NaAc-Hac buffer. The polydispersity index (PDI) was collected as the indicator of the uniformity degree of nanoparticles.AFM imaging
Atomic force microscopy (AFM, Icon, Bruker, MA, USA) was employed to determine the height of FeN/GQDs. Images were acquired in tapping mode using a tip with a force constant of 0.3 N m−1 and a resonance frequency of 75 kHz. To ensure the dispersion of the sample, FeN/GQDs was diluted in DI-H2O to approximately 1 μg mL−1. The diluted solution was then dropped onto a freshly cleaved mica sheet and allowed to naturally dry in a clean and dry environment, facilitating the adsorption of the sample onto the mica surface.TEM imaging
Transmission electron microscopy (TEM, Ted Pella, Inc., Redding, CA, USA) was used to observe the morphological features of FeN/GQDs. High-resolution TEM (HR-TEM) images were acquired to provide a detailed view of FeN/GQDs, and element mapping was performed to confirm the presence of Fe, N, and C in samples.Ultraviolet-visible spectroscopy measurements
Ultraviolet-visible spectroscopy (UV-Vis, 8453 Diode Array UV-Vis Spectrophotometer, Agilent) was conducted to determine the optical properties of FeN/GQDs. The absorption spectra of FeN/GQDs in NaAc-Hac buffer were recorded and used to analyze the POD-like properties with the colorimetric method.Photoluminescence characterization
Photoluminescence (PL) spectroscopy (FL, LS-55 Luminescence Spectrophotometer, PerkinElmer) was performed to study the photoluminescence properties of FeN/GQDs. The PL spectra of FeN/GQDs were acquired to determine the photoluminescence emission characteristics of FeN/GQDs and to assess their potential as glucose detection nanozymes.FTIR characterizations
Fourier-transform infrared spectroscopy (FTIR, Nicolet 6700 spectrometer, Thermo Scientific Inc., Waltham, MA, USA) was used to determine the functional groups present in FeN/GQDs and to study their chemical structure. The spectra were obtained at 4 cm−1 resolution and the signal was averaged over 64 scans. The samples were pressed into a pellet with KBr for analysis.Raman spectroscopy
Raman spectroscopy (performed using an inVia™ confocal Raman microscope, RENISHAW) was employed to investigate the vibrational properties of FeN/GQDs and to elucidate their structural features. The excitation source selected for the measurements was a 514 nm laser, calibrated to ensure clear signal detection using silicon as the standard sample, with the signal located at 520 nm.X-ray photoelectron spectroscopy
X-ray photoelectron spectroscopy (XPS, AXIS Ultra DLD/Surface Science Instruments S-Probe, Kratos) was performed to assess the composition and electronic structure of FeN/GQDs. This instrument has a monochromatized Al Kα X-ray and a low-energy electron flood gun for charge neutralization. The X-ray spot size for these acquisitions was on the order of 700 μm × 300 μm. The electrostatic lens were used for data collection. The pressure in the analytical chamber during spectral acquisition was less than 5 × 10−9 Torr. The pass energy was 160 eV for survey spectra (composition) and 40 eV for the high-resolution spectra. The take-off angle (the angle between the sample normal and the input axis of the energy analyzer) was 0° and the sampling depth was ∼100 Å. The Kratos Vision2 software program was used to determine peak areas and to calculate the elemental compositions from peak areas. CasaXPS was used to peak-fit the high-resolution spectra. For the high-resolution spectra, a Shirley background was used, and all binding energies were referenced to the C ls C–C bonds at 285.0 eV. The XPS spectra of FeN/GQDs were recorded to determine the chemical compositions of the samples and to study their electronic structures.X-ray absorption spectra
X-ray absorption near edge structure (XANES) was employed to obtain the valence and coordination element information of FeN/GQDs. The X-ray absorption fine structure spectra (XAFS, Fe K-edge) were collected under the 7-BM beamline of the National Synchrotron Light Source II, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory.Peroxidase-like activity evaluation and kinetics analyze of nanozymes and HRP
The peroxidase (POD)-like activity was evaluated based on the recipes found elsewhere.64 The typical POD-like activity assays were performed in the presence of TMB (as the substrate) and H2O2 in buffer solution (0.1 M HAc-NaAc, pH 3.6). The absorbance of the blue-colored product (at 652 nm for TMB) was measured at a particular reaction time using a microplate reader spectrophotometer. Briefly, a certain quantity of FeN/GQDs or NGQDs was added into a buffer solution (1.0 mL) containing 10 μL TMB (0.4 mM) and H2O2 (100 μM) to assess the chromogenic reactions. Considering subsequent applications for blood glucose monitoring, the standard steady-state kinetic tests were conducted at 37 °C, which mimics the human body temperature. Then, the equal volume of GQDs-based nanozymes solution (10 μL of 1 mg mL−1) was allowed to react in the presence of TMB and H2O2 substrates. In the same way, the reaction kinetic assays of FeN/GQDs and NGQDs were recorded by adding the aliquots of 10 μL of H2O2 (10 mM, from 30% v/v stock solution) and variable amounts (0.5, 1, 2, 5, 7, 8, 10, 12.5, 15, 20, 50 μL) of TMB solution (in EtOH, 10 mg mL−1). Similarly, the POD-like kinetic assays were also performed by changing the H2O2 concentrations (0.5, 1, 2, 5, 10, 20, 30, 40, 50, 60, 70, 80, 100, 150, 200, 500 μL) of solutions while keeping the TMB (10 μL of 10 mg mL−1) and GQDs based catalyst (10 μL of 1 mg mL−1) conditions constant. The absorbance of each reaction was measured at various reaction time points during the process. The characteristic Michaelis–Menten constant (Km) and velocity (Vmax) were calculated by Lineweaver–Burk plot (eqn (1)) using the saturation curve in Origin Pro 2021b.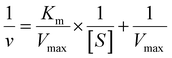 | (1) |
Free radical identification
The electron spin resonance (EPR) analyses of FeN/GQDs were performed by a Bruke EPR at room temperature. Briefly, 5,5-dimethyl-1-pyrroline N-oxide (DMPO) and FeN/GQDs were mixed in pH 3.6 buffer and the mixture was kept at 30 °C for 5 minutes. The reaction was initiated by adding 10 mM H2O2 solution, and the mixture was then incubated for 5 min before being transferred to a capillary tube. The final concentration of each component in the reaction mixture was 100 mM DMPO, 20 μg FeN/GQDs, and 1 mM H2O2. The EPR spectrum was acquired for detection of spin adducts using spin traps at the following settings: 1 G field modulation, 200 G scan range, and 20 mW microwave power. The spin-trap DMPO was used to verify the formation of hydroxyl radicals (OH˙) during the degradation of H2O2 in the presence of FeN/GQDs, NGQDs and HRP in 1.0 mL NaAc–HAc buffer (0.1 M, pH 3.6).Colorimetric detection of glucose
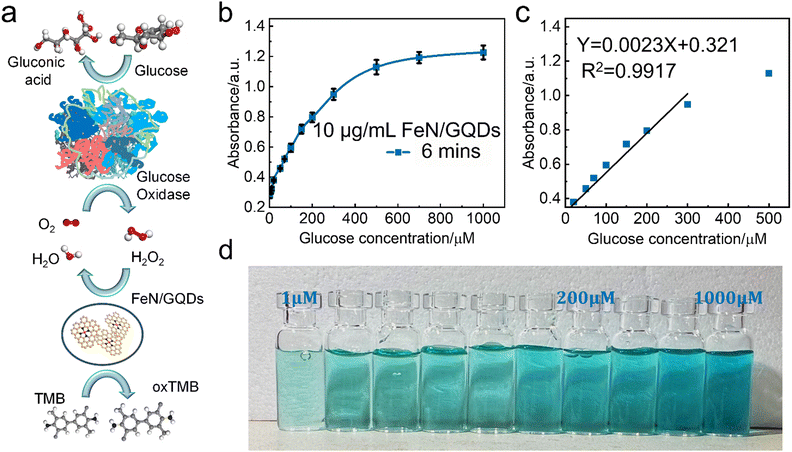
A colorimetric glucose detection system was developed for biosensing application studies, employing a combination of glucose oxidase (GOx), FeN/GQDs, and TMB. In the detection process, GOx catalyzes the oxidation of glucose to glucose-acid, generating H2O2 as a byproduct. The presence of FeN/GQDs enhances the conversion of H2O2 into hydroxyl radicals. These radicals subsequently react with TMB, resulting in a distinct color change from colorless to blue (Scheme 1). The detection procedure consisted of the following steps: (1) incubation of a mixture containing 10 μL of GOx (10 mg mL−1) and 100 μL of glucose buffer solutions (pH 7.0) with varying concentrations of glucose (1, 5, 10, 20, 50, 100, 200, 500, 1000 μM) at 37 °C for 30 minutes. (2) Addition of 10 μL of TMB (30 mM), 10 μL of FeN/GQDs dispersion (1 mg mL−1), and 0.79 mL of 0.1 M HAc-NaAc buffer (pH 3.6) to the glucose mixture solution. (3) Incubation of the mixture solution at 37 °C for 6 minutes, followed by measurement of absorbance using a UV-vis spectrophotometer. The color changes in response to different glucose concentrations (1, 2, 5, 10, 20, 50, 100, 200, 500, 1000 μM) were monitored by analyzing the absorption spectra, specifically the intensity at 652 nm, and through visual observation without requiring specialized equipment. To ensure accuracy, the measurements were repeated three times.Statistical analysis
All experiments were conducted in triplicate and the results are presented as mean values ± standard error of the mean (DLS, TEM, AFM). The statistical differences in catalytic performance data were evaluated using a two-sided Student's t-test. The observed differences with p < 0.05 were considered statistically significant. For the analysis of the linear region of FeN/GQDs catalytic performance, a 99% confidence interval was utilized to assess the statistical significance of the observed correlations.3. Results and discussion
Synthesis and physicochemical properties of FeN/GQDs nanozymes
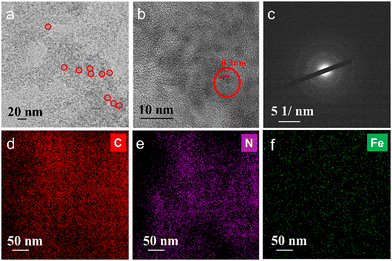
The synthesis of Fe–N doped graphene quantum dots (FeN/GQDs) involved a solvothermal process utilizing Ananas comosus (pineapple) leaves extract and FeCl2 as precursors, as depicted in Scheme 1. Initially, the leaves were cut into pieces, immersed in DMF, and then subjected to high intensity probe sonication. The resulting extract obtained from the leaves served as a precursor providing carbon source, nitrogen source, and metal protection agents.53 Glucose and other polyphenols species are reported as the main components from Ananas comosus leaves, which not only serve as carbon sources for graphene quantum dots formation but also can effectively chelate Fe2+ ions by the strong interaction between the oxygen-containing and phenolic groups with metal ions to prevent aggregation.65 Iron(II) chloride was added to the extract and subjected to 30 minutes of water bath sonication to disperse ferrous ions. Subsequently, centrifugation was carried out to eliminate large insoluble particles. The resulting solution was then transferred to a Teflon reactor and subjected to a temperature of 180 °C for 9 hours, ultimately yielding FeN/GQDs. The as-prepared FeN/GQDs nanozyme was purified with a combination of centrifuge and dialysis.In order to assess the colloidal properties of the purified nanozymes, we measured the hydrated size and zeta potential changes over time in PBS buffer solution using dynamics light scattering (DLS). Fig. S1a (ESI†) shows that FeN/GQD exhibited a uniform size of approximately 15.6 (± 2.3) nm in hydrated state, with a polydispersity index (PDI) of 0.122 (±0.05), indicating a relative narrow size distribution. The zeta potential of FeN/GQDs, as shown in Fig. S1b (ESI†), was measured to be 39.4 (±5.7) mV, which exceeds the stable value typically observed in colloidal systems.66 Furthermore, transmission electron microscopy (TEM) was utilized to examine the morphology and structure of these GQD-based nanozymes. The TEM image in Fig. 1a reveals that FeN/GQDs were spherical in shape and well dispersed, with an average size of 13.6 (±2.7) nm. The high-resolution TEM image confirmed the excellent crystallinity of FeN/GQDs, revealing a well-organized fringe pattern (Fig. 1b). The lattice interval of FeN/GQDs was determined to be 0.3 nm, indicating the D-spacing of the interlayer of the [100] facet of sp2 graphitic carbon pattern.67 The measured dimensions of FeN/GQDs are in good agreement with AFM measurements. The AFM phase plot demonstrated a diameter of 12.6 (±3.6) nm (Fig. S2a, ESI†). Furthermore, the AFM topography graph (Fig. S2b, ESI†) disclosed an average FeN/GQDs height of 0.75 (±0.18) nm. Given the interlayer spacing between two graphene layers of approximately 0.3 nm, the FeN/GQDs were estimated to consist of 2–3 graphene layers.
The selected area electron diffraction (SAED) pattern shown in Fig. 1c displayed ring-like features, indicating that the predominant structure of FeN/GQDs corresponds to the graphene structure.67 This SAED pattern further confirmed the presence of well-organized graphene-like arrangements in the FeN/GQDs, supporting their possible single-atom structure and crystalline nature. The combination of high-resolution TEM and SAED analysis provided strong evidence of the graphene-based composition and structural integrity of the FeN/GQDs. Furthermore, TEM-X-ray energy-dispersive spectrometer (EDS) mapping (Fig. S3, ESI†) was conducted to validate the uniform distribution of elements in FeN/GQDs including C (Fig. 1d), N (Fig. 1e), and Fe (Fig. 1f).
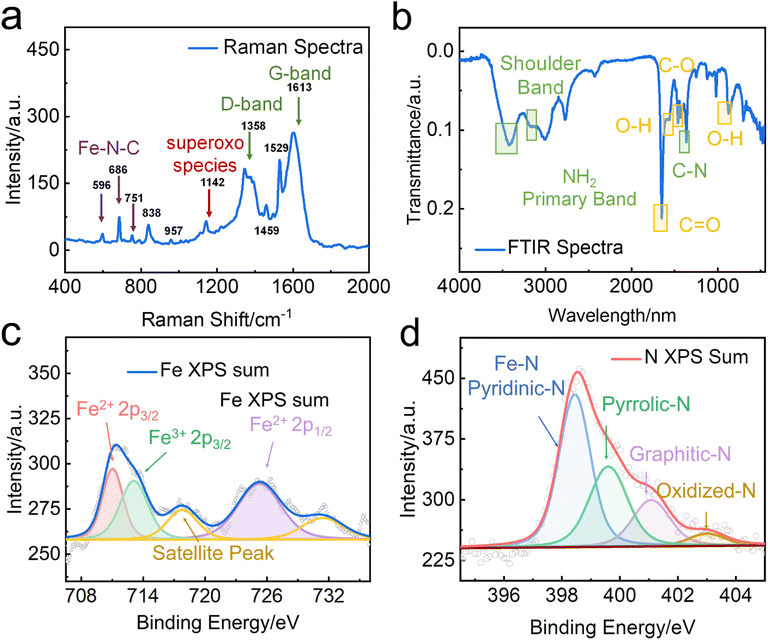
The high degree of crystallinity of FeN/GQDs was confirmed through Raman spectrum analysis. The characteristic Raman peaks of carbon were observed in the 1000–2000 cm−1 region. The band at 1640 cm−1 and 1380 cm−1 corresponded to the D-band and G-band, respectively. The G-band indicates the presence of sp2 graphitic carbon networks, while the D-band indicates sp3 amorphous carbon.68 As shown in Fig. 2a, the intensity ratio of the D-band to the G-band was found to be 0.68, indicating that the GQDs possessed a high level of crystallinity. Additionally, the Raman spectrum revealed peaks at 596 cm−1, 686 cm−1, and 751 cm−1, which can be attributed to the C/N displacement around the Fe center. Of particular significance is the peak at 596 cm−1, which can be assigned to the in-plane Fe–N stretching vibration of Fe–N4 moieties. This peak provided solid evidence for the existence of Fe–N structures within FeN/GQDs,69–71 further supporting the successful synthesis of single-atom Fe–N doped graphene quantum dots.
For exploring potential applications in biosensing and other purposes, the optical properties of FeN/GQDs were thoroughly investigated using UV-vis (Fig. S4, ESI†) and fluorescence spectroscopy (Fig. S5, ESI†). The UV-vis absorption spectrum of FeN/GQDs displayed a characteristic absorption peak at around 370 nm, which can be attributed to the Π–Π* transition of the graphitic sp2 domain.72 Additionally, two distinctive peaks were observed in the near-infrared (NIR) region, specifically at around 670 nm (NIR-I) and 850 nm (NIR-II). These peaks indicated the presence of a large conjugated system with extensive delocalized Π electrons in the stacked layer structure of FeN/GQDs.73 Importantly, no significant absorption peaks were found in the visible light region that could potentially overlap with the peaks corresponding to commonly used substrates for biosensing applications. This is a favorable characteristic for biosensing, as it reduces potential interference and background signals.
The fluorescence spectroscopy analysis of FeN/GQDs revealed a maximum near-infrared fluorescence emission at 750 nm when excited at 600 nm, resulting in a substantial Stokes shift of 150 nm (Fig. S5, ESI†). This pronounced Stokes shift could be attributed not only to the stacked aromatic carbon structures originating from the polyphenol precursors, as previously reported, but also to the Fe–N doping. The incorporation of nitrogen into the graphene quantum dots can significantly alter the band structure of the nanoparticles, further contributing to the observed large Stokes shift. These optical properties, including the absence of significant visible light absorption and the prominent near-infrared fluorescence emission with a substantial Stokes shift, make FeN/GQDs highly attractive for biosensing applications, particularly in the context of fluorescence-based detection and imaging in the near-infrared region.
The chemical composition of FeN/GQDs was characterized using Fourier-transform infrared spectroscopy (FTIR) and X-ray photoelectron spectroscopy (XPS). FTIR spectra of FeN/GQDs (Fig. 2b) indicated the presence of abundant oxygen and nitrogen-containing functional groups on the surface of the GQD-based nanoparticles. These functional groups included –COOH (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch around 1760–1690 cm−1, C–O stretch around 1320–1210 cm−1), –OH (O–H stretch around 3300–2500 cm−1, O–H bend from 1440–1395 and 950–910 cm−1), and –NH2 (N–H stretch around 3400–3250 cm−1, N–H bend around 1650–1580 cm−1, C–N stretch (aromatic amines) around 1335–1250 cm−1, N–H wag from 910–665 cm−1) groups,74,75 These functional groups contribute to the colloidal stability of FeN/GQDs. Peaks around 680, 520, and 440 cm−1 corresponding to Fe–O bonds were barely detected, indicating a limited content of oxidized iron in the sample.76
O stretch around 1760–1690 cm−1, C–O stretch around 1320–1210 cm−1), –OH (O–H stretch around 3300–2500 cm−1, O–H bend from 1440–1395 and 950–910 cm−1), and –NH2 (N–H stretch around 3400–3250 cm−1, N–H bend around 1650–1580 cm−1, C–N stretch (aromatic amines) around 1335–1250 cm−1, N–H wag from 910–665 cm−1) groups,74,75 These functional groups contribute to the colloidal stability of FeN/GQDs. Peaks around 680, 520, and 440 cm−1 corresponding to Fe–O bonds were barely detected, indicating a limited content of oxidized iron in the sample.76
XPS analysis was then performed to further investigate the surface elements and their binding states. The full survey of FeN/GQDs (Fig. S6a, ESI†) showed the presence of C(1s), O(1s), N(1s), and Fe(2p) peaks at 285, 531, 400, and 710 eV, respectively. High-resolution XPS spectra were used to determine the binding structure of C and N. The C 1s spectrum (Fig. S6b, ESI†) exhibited three main peaks at 285 eV, 287 eV, and 289 eV, which were assigned to sp2 C, O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, and C
O, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, respectively. The N 1s spectrum (Fig. 2c) exhibited four well-fitted peaks, indicating the coexistence of Pyridinic-N (Fe–N), graphitic-N, Pyrrolic-N, and Oxidized-N.77 Previous studies have suggested that the presence of pyridinic and pyrrolic nitrogen functionalities can facilitate the formation of coordination bonds with Fe, preventing the aggregation of single-atoms and the formation of larger particles.77 Furthermore, a strong metal–support (Fe–Nx structure) interaction with unsaturated coordination state of Fe plays a crucial role in enhancing catalytic performances by promoting efficient charge transfer processes.15,78
N, respectively. The N 1s spectrum (Fig. 2c) exhibited four well-fitted peaks, indicating the coexistence of Pyridinic-N (Fe–N), graphitic-N, Pyrrolic-N, and Oxidized-N.77 Previous studies have suggested that the presence of pyridinic and pyrrolic nitrogen functionalities can facilitate the formation of coordination bonds with Fe, preventing the aggregation of single-atoms and the formation of larger particles.77 Furthermore, a strong metal–support (Fe–Nx structure) interaction with unsaturated coordination state of Fe plays a crucial role in enhancing catalytic performances by promoting efficient charge transfer processes.15,78
To further validate the formation of the Fe–Nx structure, the Fe 2p spectra were acquired (Fig. 2d). The first doublet of Fe2+ 2p3/2 (711.2 eV) and Fe2+ 2p1/2 (725.3 eV) indicated the presence of Fe2+ in the FeN/GQDs, while the peaks located at 717.3 eV belonged to the satellite of Fe–Nx.79 The presence of Fe3+ 2p3/2 (713.8 eV) peak suggested the existence of different oxidation states, indicating a potentially unsaturated coordination environment around iron atoms.
Based on the above data, it can be concluded that Fe single-atoms were successfully doped into the graphene quantum dots through the Fe–Nx coordination structure. These findings provided further confirmation of the structure of FeN/GQDs, highlighting the strong metal–support interaction that creates a single-atom reaction site with potential for efficient catalysis.
Atomic structure of FeN/GQDs nanozymes
The coordination environment and chemical state of Fe species in FeN/GQDs were further investigated using X-ray absorption near edge structure (XANES) and extended X-ray absorption fine structure (EXAFS) spectroscopy at the Fe K edge. Based on the XANES spectra (Fig. 3a), the near edge absorption energy of FeN/GQDs was found to be situated between the Fe foil and Fe2O3, indicating the presence of positively charged Fe single-atoms in FeN/GQDs.12,31,34 Moreover, the EXAFS curve of FeN/GQDs exhibited a primary peak at about 1.5 Å, which was assigned to the Fe–N scattering paths.12,34 Notably, the Fe–Fe peak at 2.04 Å was not detected in the spectrum (Fig. 3b). According to the EXAFS fitting parameters (Table S1, ESI†), an average Fe–N coordination number of 4.6 was determined. Consequently, based on the XANES and EXAFS analysis, the presence of single Fe atom can be confirmed.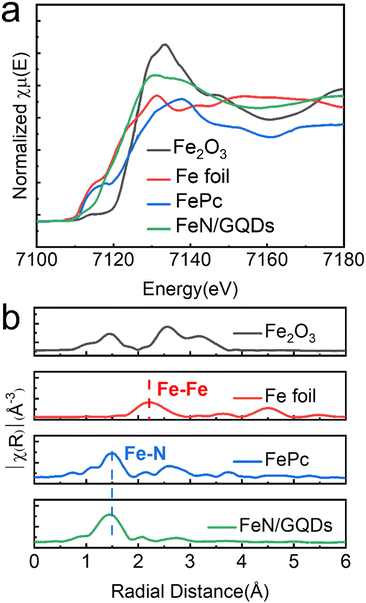 | ||
| Fig. 3 Atomic structural analysis of FeN/GQDs using (a) X-ray absorption near edge structure (XANES) and (b) extended X-ray absorption fine structure (EXAFS). | ||
Peroxidase-like activity and kinetics analysis
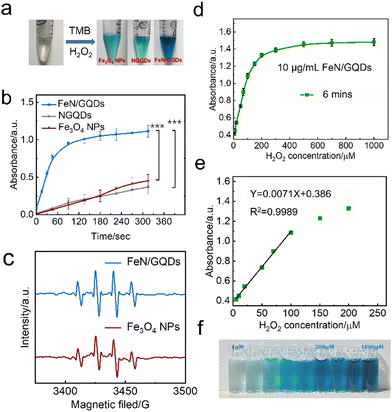
Peroxidase (POD) is a natural enzyme that plays a critical role in various biological processes by catalyzing the decomposition of hydrogen peroxide into water and reactive oxygen species (ROS).80 In our study, we evaluated the POD-like activity of Fe–N/GQDs nanozymes using a commonly used chromogenic substrate TMB (3,3,5,5-tetramethylbenzidine). TMB can undergo a catalytic reaction with OH˙ free radicals generated by the nanozymes, leading to the formation of a blue-colored TMB product (oxTMB) with a characteristic absorbance peak around 652 nm, as shown in eqn (2):| TMB + H2O2 → oxTMB + H2O | (2) |
To understand the effect of the Fe–N coordination structure on the peroxidase-like activity, the catalytic efficiency of two other types of nanozymes were compared. Specifically, NGQDs, which are graphene quantum dots without FeN single-atom structure, and Fe3O4 nanoparticles, representing bulk-like iron-based nanozymes, were used for comparison. The color change of the solution and the absorbance at 652 nm were monitored over time (Fig. 4a and b). The results clearly demonstrated that FeN/GQDs exhibited superior catalytic efficiency compared to both NGQDs and Fe3O4 nanoparticles, as evidenced by the solution's color change and the absorbance at 652 nm. These findings strongly suggested that the Fe–N single-atom structure plays a crucial role in enhancing the reaction efficiency, making FeN/GQDs highly efficient nanozymes for POD-like activity.
Since the reaction efficiency mainly depends on the superoxide radical produced by the catalysis, the generation of the ROS such as OH˙ and OOH˙ in the FeN/GQDs catalytic reaction system was confirmed using the EPR spin-trap technique of DMPO conjunction (Fig. 4c). As shown in Fig. 4c, after the addition of H2O2, EPR spectra exhibited a characteristic 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 signal pattern of OH˙ in the sample containing both H2O2 and FeN/GQDs. This signal pattern is similar to the one observed in Fe3O4 nanoparticles, which have been reported to possess the POD-like properties.35 The presence of the characteristic EPR signal pattern indicated the production of OH˙ radicals during the catalytic reaction of FeN/GQDs with H2O2. This observation provided further evidence of the POD-like activity of FeN/GQDs and their capability to generate reactive oxidative species, making them suitable candidates for various catalytic applications.
1 signal pattern of OH˙ in the sample containing both H2O2 and FeN/GQDs. This signal pattern is similar to the one observed in Fe3O4 nanoparticles, which have been reported to possess the POD-like properties.35 The presence of the characteristic EPR signal pattern indicated the production of OH˙ radicals during the catalytic reaction of FeN/GQDs with H2O2. This observation provided further evidence of the POD-like activity of FeN/GQDs and their capability to generate reactive oxidative species, making them suitable candidates for various catalytic applications.
To determine the optimal reaction condition, the effects of pH and temperature were also investigated separately. The results show that the optimal pH value for the catalytic reaction of FeN/GQDs falls between pH 2 and 10, with the highest efficiency observed at pH 3.6 (Fig. S8a, ESI†). Similarly, the best temperature range for the reaction is found to be between 17 and 60 °C, with the highest efficiency observed at 30 °C (Fig. S8b, ESI†).
Furthermore, the relationship between the reaction efficiency and the dosage of chemical reagents was also surveyed. The results show that the catalytic efficiency is directly correlated with the dosage of H2O2 and TMB (Fig. S8c and d, ESI†). By optimizing the pH, temperature, and reagent dosage, the catalytic performance of FeN/GQDs can be enhanced, providing valuable insights for their practical application in various catalytic processes and biosensing assays.
To gain further insights into the POD-like activity of FeN/GQDs, the steady state kinetics were analysed by varying the concentration of H2O2 or TMB (Fig. S9a and c, ESI†). The Lineweaver–Burk or double reciprocal plots were used to estimate the Michaelis constant (Km) and maximal reaction velocity (Vmax) (Fig. S9b and d, ESI†). Km is a measure of the binding capacity between an enzyme and its substrates, and a lower value indicates a stronger affinity between the enzyme and substrates. The Km values of FeN/GQDs were found to be 0.52 for TMB and 0.49 for H2O2, respectively. When compared to the natural enzyme horseradish peroxidase (HRP), FeN/GQDs exhibited significantly improved catalytic performance. The Km value for H2O2 was 7.5 times lower, and the Vmax for H2O2 was 10 times higher than those of HRP, indicating enhanced substrate affinity and catalytic ability towards H2O2 (Table S2, ESI†).
To further elucidate the catalytic performance of FeN/GQDs, a comprehensive comparison of reaction constants was conducted with other Fe-based nanozymes, including oxide-based, single-atom-based, and composite nanozymes (Table S2, ESI†). FeN/GQDs displayed much lower Km values and higher Vmax compared to oxide-based and composite nanozymes, suggesting that the single-atom structure provides superior POD-like mimicry capability. When compared with other reported single-atom nanozymes, FeN/GQDs synthesized through the solvothermal method exhibited comparable performance, highlighting the effectiveness and feasibility of the solvothermal approach in obtaining high-performing nanozymes for various applications.
It is important to highlight that FeN/GQDs demonstrate universal catalytic activity as a nanozyme for various chromogenic substrates in the POD-like reactions. Besides TMB, other chromogenic substrates such as ABTS and OPD (Fig. S10, ESI†) can also undergo oxidation by FeN/GQDs, leading to distinct color changes (green and orange) in the catalytic system. This versatility in catalytic capacity underscores the potential of FeN/GQDs as a powerful tool for diverse chromogenic substrate-based assays.
Moreover, the oxidase-like, catalase-like, and superoxide dismutase-like activities of FeN/GQDs nanozymes were also measured (Fig. S11–S13, ESI†). The results demonstrated higher catalytic efficiency exhibited by FeN/GQDs, providing further evidence that the FeN structure plays a dominant role in enhancing the catalytic reactions. These properties expand the application scope of FeN/GQDs as a potential candidate for colorimetric detection in various fields.
Glucose detection through the catalytic activity of FeN/GQDs
Based on the observed POD-like enzyme catalytic performance of FeN/GQDs, the linear relationship between H2O2 concentration of substrate and ROS production is first analyzed to establish 99% confidence intervals. As shown in Fig. 4d, the absorbance at 652 nm gradually reached saturation as the H2O2 concentration increased, indicating the catalytic activity of FeN/GQDs in the presence of H2O2. Based on the concentration–response plot, a linear relationship could be achieved in the H2O2 concentration range of 5–100 μM (Fig. 4e). Fig. 4f illustrates the color change of the H2O2/TMB/FeN-GQDs solution with different H2O2 concentrations. As the concentration of H2O2 solution increased, the color changed to a deeper blue color in the detection system. The color change diminished when the concentration of H2O2 reached 200 μM, suggesting that the catalytic capacity of FeN/GQDs reached saturation, which was consistent with the result shown in Fig. 4d. Based on the analysis, the detection limit (DL) of H2O2 was calculated to be 0.78 μM (DL = 3.3σ/S). This result implies that the detection limit of glucose could be lower than 0.78 μM since H2O2 was a downstream product of glucose catalysis.FeN/GQDs, with their promising POD-like activity, were utilized to construct a colorimetric glucose sensing system. The system comprised the combination of glucose oxidase (GOx) and the colorimetric substrate of TMB. The detection process relies on cascade reactions shown in Fig. 5a. In step 1, glucose undergoes oxidation by the GOx enzyme, resulting the production of H2O2. In step 2, the FeN/GQDs nanozyme catalyzes the conversion of H2O2 to hydroxyl radicals (OH˙). These hydroxyl radicals then initiate the oxidation of the colorless TMB, leading to the formation of a blue-colored product, oxTMB.
To quantify the amount of produced H2O2 and, consequently, the amount of glucose present, the changes in intensity of the UV-vis absorption peak at 652 nm of oxTMB were monitored. By measuring the color change of the solution at the specific wavelength, the concentration of glucose can be determined accurately and quantitatively.
To further investigate the glucose detection capability of FeN/GQDs, we employed UV-Vis to measure absorbance changes of glucose solutions at 652 nm with glucose concentrations ranging from 1 μM to 1000 μM (Fig. 5a). The glucose solution first reacted with the glucose oxidase (GOx). After being placed in a 36 °C-water bath for 30 minutes, the reacted glucose solution was added into the TMB/FeN/GQDs solution. The glucose/GOx/TMB/FeN/GQDs solution showed a quick response within 6 min upon mixing (Fig. 5c). The mixed solution's absorbance increased as the glucose concentration increased due to the production of H2O2 from the reaction between glucose and Gox. The absorbance exhibited a linear relationship with the glucose concentration between 1 μM and 300 μM (Fig. 5b). The detection limit was evaluated to be 0.36 μM with a relative standard deviation of 0.83%. These results demonstrate the high sensitivity and accuracy of FeN/GQDs as a nanozyme for colorimetric glucose detection. Additionally, Table S3 (ESI†) underscores the outstanding performance of FeN/GQDs in glucose detection when compared to various nanozymes and detection platforms. FeN/GQDs exhibited shorter detection time, lower detection limits, and a wider detection range, demonstrating their advantage in these critical aspects of glucose detection.
Stability and reusability of FeN/GQDs nanozymes
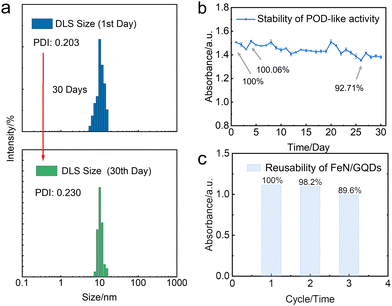
In this study, we further investigated the stability and reusability of FeN/GQDs nanozymes, which are critical indicators for their practical applications. The nanozymes were stored at room temperature for 30 days to assess their colloidal stability and catalytic activity. The analysis of particle size distribution and polydispersity index (PDI) by dynamic light scattering (DLS) revealed that the nanozymes exhibited exceptional colloidal stability even after the 30-day storage period (Fig. 6a). This remarkable stability can be attributed to the presence of amino groups on the surface of the nanoparticles, resulting in a positive zeta potential exceeding 30 mV, which ensures their long-term colloidal stability.66Furthermore, the TMB experiment demonstrated that the catalytic activity of the nanozymes remained largely unaffected, further confirming their remarkable stability (Fig. 6b). These results indicate the structural integrity and functional reliability of FeN/GQDs nanozymes for an extended duration.
To evaluate the reusability of the nanozymes, a recycling experiment was conducted. The post-TMB reaction nanozymes were collected and washed through an ultrafiltration membrane. Subsequently, the washed nanozymes were subjected to a TMB assay to assess their catalytic activity. Notably, the catalytic performance of the washed nanozymes remained almost unchanged, indicating their remarkable reusability (Fig. 6c). This suggests that FeN/GQDs nanozymes can be efficiently recovered and employed for subsequent catalytic reactions without significant loss in their catalytic performance.
Overall, the results obtained from the stability and reusability experiments validate the long-term stability and functionality of FeN/GQDs nanozymes, positioning them as promising candidates for a wide range of catalytic applications.
4. Conclusions
This study has successfully synthesized single Fe atom nanozymes using solvothermal process. This approach offers a simple, cost-effective, and environmentally friendly method for their preparation. The characterization of the nanozymes using XANES, TEM, and XPS confirmed the presence of single Fe atoms in the nanozymes, validating their unique structure and composition.The EPR spectroscopy analysis further demonstrated the POD-like activity of the Fe atom nanozymes, as evidenced by a clear POD-like signal. The glucose detection assay revealed that the nanozymes exhibited excellent sensitivity and selectivity for glucose detection with a detection limit of 0.36 μM and a wide linear range up to 300 mM, which can be attributed to the single Fe atom structure providing numerous active sites for efficient glucose oxidation. This colorimetric glucose sensing system, based on FeN/GQDs as nanozymes, provides a practical and efficient approach for glucose detection, which holds significant potential for various applications in biomedical research, diagnostics, and other fields requiring glucose quantification. Furthermore, the FeN/GQDs nanozymes demonstrated exceptional stability and reusability, maintaining their catalytic activity and structural integrity even after prolonged storage and repeated use. These remarkable properties make them highly promising for various practical applications.
In summary, this study presents a novel approach for synthesizing single-atom nanozymes with excellent properties. Our findings provided strong evidence of the single-atom Fe–N coordination structure, supporting the feasibility of utilizing FeN/GQDs as highly efficient nanozymes for various catalytic applications. This research could have a significant impact on the advancement of nanomaterials for biosensing and medical applications.
Author contributions
Conceptualization/validation: Xinqi Li, Guanyou Lin, Miqin Zhang, Mohammad H. Malakooti; methodology: Xinqi Li, Lijun Zhou; formal analysis/data curation: Xinqi Li, Octavia Prosser; investigation: Xinqi Li, Lijun Zhou, Miqin Zhang; writing—original draft preparation: Xinqi Li.; writing—review and editing: Xinqi Li, Guanyou Lin, Lijun Zhou, Miqin Zhang, Mohammad H. Malakooti; supervision: Miqin Zhang, Mohammad H. Malakooti. Funding acquisition: Mohammad H. Malakooti, Miqin Zhang. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work was supported by Chronic Disease Pilot Grant from Novo Nordisk through the University of Washington Population Health Initiative and Professor Zhang Kyocera professor endowment. We acknowledge the use of the equipment on NP characterization from the Nanoengineering and Science Institute and Molecular Engineering and Science Institute supported by NSF (NNCI-2025489, grant NNCI-1542101), UW Department of Chemical Engineering (grant NIH S10 OD030224-01A1).References
- S. Cai and R. J. F. I. C. Yang, Front. Chem., 2020, 8, 565940–565957 CrossRef CAS PubMed.
- J. He, P. Liu, R. Ran, W. Wang, W. Zhou and Z. Shao, J. Mater. Chem. A, 2022, 10, 6835–6871 RSC.
- H. Zhang, X. F. Lu, Z.-P. Wu and X. W. D. Lou, ACS Cent. Sci., 2020, 6, 1288–1301 CrossRef CAS PubMed.
- H. Wei and E. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 RSC.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang and S. Perrett, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- C. He, D. Liu and W. Lin, ACS Nano, 2015, 9, 991–1003 CrossRef CAS PubMed.
- F. Cao, L. Zhang, Y. You, L. Zheng, J. Ren and X. Qu, Angew. Chemie, 2020, 132, 5146–5153 CrossRef.
- Z. Wang and D. C. Baulcombe, Nat. Commun., 2020, 11, 1221–1230 CrossRef CAS PubMed.
- X. Zhang, X. Chen and Y. Zhao, Nano-Micro Lett., 2022, 14, 95–122 CrossRef CAS PubMed.
- X. Zhang, S. Wang, G. Cheng, P. Yu and J. Chang, Engineering, 2022, 13, 18–30 CrossRef CAS.
- S. Huang, B. Wang, X. Zhang, F. Lu, Z. Wang, S. Tian, D. Li, J. Yang, F. Cao and L. Cheng, Biomaterials, 2020, 238, 119829–119843 CrossRef CAS PubMed.
- L. Jiao, J. Wu, H. Zhong, Y. Zhang, W. Xu, Y. Wu, Y. Chen, H. Yan, Q. Zhang and W. Gu, ACS Catal., 2020, 10, 6422–6429 CrossRef CAS.
- S. Ahmadi, N. Rabiee, M. Bagherzadeh, F. Elmi, Y. Fatahi, F. Farjadian, N. Baheiraei, B. Nasseri, M. Rabiee and N. T. Dastjerd, Nano Today, 2020, 34, 100914–100935 CrossRef CAS PubMed.
- Z. Pu, I. S. Amiinu, R. Cheng, P. Wang, C. Zhang, S. Mu, W. Zhao, F. Su, G. Zhang and S. Liao, Nano-Micro Lett., 2020, 12, 1–29 CrossRef PubMed.
- J. Liu, M. Jiao, L. Lu, H. M. Barkholtz, Y. Li, Y. Wang, L. Jiang, Z. Wu, D.-J. Liu and L. Zhuang, Nat. Commun., 2017, 8, 15938–15947 CrossRef CAS PubMed.
- Y. Chao, L. Xu, C. Liang, L. Feng, J. Xu, Z. Dong, L. Tian, X. Yi, K. Yang and Z. Liu, Nat. Biomed. Eng., 2018, 2, 611–621 CrossRef CAS PubMed.
- Y. Kang, C. Li, H. Shi, A. Zhang, C. Huang, C. Zhou and N. Jia, Chin. J. Chem., 2023, 41, 3189–3196 CrossRef CAS.
- L. Zuo, H. King, M. A. Hossain, F. Farhana, M. M. Kist, R. L. Stratton, J. Chen and H. Shen, Chem. Biomed. Imaging, 2023, 1, 760–766 CrossRef CAS PubMed.
- S. Liu, H. Xu, D. Liu, H. Yu, F. Zhang, P. Zhang, R. Zhang and W. Liu, J. Am. Chem. Soc., 2021, 143, 15243–15249 CrossRef CAS PubMed.
- R. Yan, S. Sun, J. Yang, W. Long, J. Wang, X. Mu, Q. Li, W. Hao, S. Zhang and H. Liu, ACS Nano, 2019, 13, 11552–11560 CrossRef CAS PubMed.
- D. Wang, B. Zhang, H. Ding, D. Liu, J. Xiang, X. J. Gao, X. Chen, Z. Li, L. Yang and H. Duan, Nano Today, 2021, 40, 101243–101254 CrossRef CAS PubMed.
- S. Dell'Elce, F. Liscio, A. Kovtun, S. Allegri, O. M. Roscioni, C. Albonetti, G. De Luca, H. W. Amenitsch, N. Demitri and L. Giorgini, Nanoscale, 2018, 10, 23018–23026 RSC.
- H. Wang, Y. Wang, L. Lu, Q. Ma, R. Feng, S. Xu, T. D. James and L. Wang, Adv. Funct. Mater., 2022, 32, 2200331–2200340 CrossRef CAS.
- Y. Zhu, W. Wang, J. Cheng, Y. Qu, Y. Dai, M. Liu, J. Yu, C. Wang, H. Wang and S. Wang, Angew. Chemie, 2021, 133, 9566–9574 CrossRef.
- J. Chen, X. Liu, G. Zheng, W. Feng, P. Wang, J. Gao, J. Liu, M. Wang and Q. Wang, Small, 2023, 19, 2205924–2205943 CrossRef CAS PubMed.
- X. Liu, X. Liang, J. Yu, K. Xu, J.-W. Shen, W. Duan and J. Zeng, TrAC, Trends Anal. Chem., 2023, 117386–117393 CrossRef CAS.
- M. Rai, A. P. Ingle, S. Birla, A. Yadav and C. A. D. Santos, Crit. Rev. Microbiol., 2016, 42, 696–719 CAS.
- Q. Liu, A. Zhang, R. Wang, Q. Zhang and D. Cui, Nano-Micro Lett., 2021, 13, 1–53 CrossRef PubMed.
- L. Shen, D. Ye, H. Zhao and J. Zhang, Anal. Chem., 2020, 93, 1221–1231 CrossRef PubMed.
- W. Wu, L. Huang, X. Zhu, J. Chen, D. Chao, M. Li, S. Wu and S. Dong, Chem. Sci., 2022, 13, 4566–4572 RSC.
- S. Ji, B. Jiang, H. Hao, Y. Chen, J. Dong, Y. Mao, Z. Zhang, R. Gao, W. Chen and R. Zhang, Nat. Catal., 2021, 4, 407–417 CrossRef CAS.
- S. Wang, Z. Wang, Z. Li, X. Zhang, H. Zhang, T. Zhang, X. Meng, F. Sheng and Y. Hou, Sci. Adv., 2022, 8, eabn3883 CrossRef CAS PubMed.
- C. Zhao, C. Xiong, X. Liu, M. Qiao, Z. Li, T. Yuan, J. Wang, Y. Qu, X. Wang and F. Zhou, Chem. Commun., 2019, 55, 2285–2288 RSC.
- M. Chen, H. Zhou, X. Liu, T. Yuan, W. Wang, C. Zhao, Y. Zhao, F. Zhou, X. Wang and Z. Xue, Small, 2020, 16, 2002343–2002349 CrossRef CAS PubMed.
- C. Zhao, C. Xiong, X. Liu, M. Qiao, Z. Li, T. Yuan, J. Wang, Y. Qu, X. Wang and F. Zhou, Chem. Commun., 2019, 55, 2285–2288 RSC.
- Q. Chen, S. Li, Y. Liu, X. Zhang, Y. Tang, H. Chai and Y. Huang, Sens. Actuators, B, 2020, 305, 127511–127520 CrossRef CAS.
- C. Peng, R. Pang, J. Li and E. Wang, Adv. Mater., 2023, 2211724–2211751 Search PubMed.
- L. Jiao, W. Xu, H. Yan, Y. Wu, C. Liu, D. Du, Y. Lin and C. Zhu, Anal. Chem., 2019, 91, 11994–11999 CrossRef CAS PubMed.
- N. Cheng, J. C. Li, D. Liu, Y. Lin and D. Du, Small, 2019, 15, 1901485–1901492 CrossRef CAS PubMed.
- L. Shen, M. A. Khan, X. Wu, J. Cai, T. Lu, T. Ning, Z. Liu, W. Lu, D. Ye and H. Zhao, Biosens. Bioelectron., 2022, 205, 114097–114106 CrossRef CAS PubMed.
- Y. Feng, J. Qin, Y. Zhou, Q. Yue and J. Wei, J. Colloid Interface Sci., 2022, 606, 826–836 CrossRef CAS PubMed.
- N. Feng, Q. Li, Q. Bai, S. Xu, J. Shi, B. Liu and J. Guo, J. Colloid Interface Sci., 2022, 618, 68–77 CrossRef CAS PubMed.
- X. Niu, Q. Shi, W. Zhu, D. Liu, H. Tian, S. Fu, N. Cheng, S. Li, J. N. Smith and D. Du, Biosens. Bioelectron., 2019, 142, 111495–111503 CrossRef CAS PubMed.
- Y. Mao, S. Gao, L. Yao, L. Wang, H. Qu, Y. Wu, Y. Chen and L. Zheng, J. Hazard. Mater., 2021, 408, 124898–124907 CrossRef CAS PubMed.
- J. Ge, L. Yang, Z. Li, Y. Wan, D. Mao, R. Deng, Q. Zhou, Y. Yang and W. Tan, J. Hazard. Mater., 2022, 436, 129199–129208 CrossRef CAS PubMed.
- R. Niu, Y. Liu, Y. Wang and H. Zhang, Chem. Commun., 2022, 58, 7924–7927 RSC.
- C. Du, Y. Gao, H. Chen, P. Li, S. Zhu, J. Wang, Q. He and W. Chen, J. Mater. Chem. A, 2020, 8, 16994–17001 RSC.
- J. A. Libra, K. S. Ro, C. Kammann, A. Funke, N. D. Berge, Y. Neubauer, M.-M. Titirici, C. Fühner, O. Bens and J. Kern, Biofuels, 2011, 2, 71–106 CrossRef CAS.
- N. D. Berge, L. Li, J. R. Flora and K. S. Ro, Waste Manage., 2015, 43, 203–217 CrossRef PubMed.
- C. Ma, S.-Y. Xiao, Z.-G. Li, W. Wang and L.-J. Du, J. Chromatogr. A, 2007, 1165, 39–44 CrossRef CAS PubMed.
- Y. Choi, S. Bae, B.-S. Kim and J. Ryu, J. Mater. Chem. A, 2021, 9, 13874–13882 RSC.
- W. Wang, X. Liu, X. Zheng, H. J. Jin and X. Li, Adv. Healthcare Mater., 2020, 9, 2001117–2001141 CrossRef CAS PubMed.
- T. Nagata and Y. Obora, RSC Adv., 2019, 5, 98–103 Search PubMed.
- C. Zhu, Z. Chen, S. Gao, B. L. Goh, I. B. Samsudin, K. W. Lwe, Y. Wu, C. Wu and X. Su, Prog. Nat. Sci.: Mater. Int., 2019, 29, 628–640 CrossRef CAS.
- O. Akhavan, E. Ghaderi and A. Akhavan, Biomaterials, 2012, 33, 8017–8025 CrossRef CAS PubMed.
- S. Zhang, X. Pei, Y. Xue, J. Xiong and J. Wang, Chin. Chem. Lett., 2020, 31, 1654–1659 CrossRef CAS.
- H. Teymourian, A. Barfidokht and J. Wang, Chem. Soc. Rev., 2020, 49, 7671–7709 RSC.
- X. Liu, D. Huang, C. Lai, L. Qin, G. Zeng, P. Xu, B. Li, H. Yi and M. Zhang, Small, 2019, 15, 1900133–1900160 CrossRef PubMed.
- M. Adeel, K. Asif, M. M. Rahman, S. Daniele, V. Canzonieri and F. Rizzolio, Adv. Funct. Mater., 2021, 31, 2106023–2106051 CrossRef CAS.
- K. Xu, R. Zhou, K. Takei and M. Hong, Adv. Sci., 2019, 6, 1900925–1900948 CrossRef PubMed.
- X. Gan, H. Zhao, R. Schirhagl and X. Quan, Microchim. Acta, 2019, 186, 1–10 CrossRef PubMed.
- R. G. Mahmudunnabi, F. Z. Farhana, N. Kashaninejad, S. H. Firoz, Y.-B. Shim and M. J. A. Shiddiky, The Analyst, 2020, 145, 4398–4420 RSC.
- W. Zhao, G. Zhang, Y. Du, S. Chen, Y. Fu, F. Xu, X. Xiao, W. Jiang and Q. Ji, J. Mater. Chem. B, 2021, 9, 4726–4734 RSC.
- K. Fan, J. Xi, L. Fan, P. Wang, C. Zhu, Y. Tang, X. Xu, M. Liang, B. Jiang, X. Yan and L. Gao, Nat. Commun., 2018, 9, 1440–1451 CrossRef PubMed.
- L. Zhao, Y. Zhang, L.-B. Huang, X.-Z. Liu, Q.-H. Zhang, C. He, Z.-Y. Wu, L.-J. Zhang, J. Wu and W. Yang, Nat. Commun., 2019, 10, 1278–1289 CrossRef PubMed.
- G. V. Lowry, R. J. Hill, S. Harper, A. F. Rawle, C. O. Hendren, F. Klaessig, U. Nobbmann, P. Sayre and J. Rumble, Environ. Sci.: Nano, 2016, 3, 953–965 RSC.
- J. Peng, W. Gao, B. K. Gupta, Z. Liu, R. Romero-Aburto, L. Ge, L. Song, L. B. Alemany, X. Zhan and G. Gao, Nano Lett., 2012, 12, 844–849 CrossRef CAS PubMed.
- E. Dervishi, Z. Ji, H. Htoon, M. Sykora and S. K. Doorn, Nanoscale, 2019, 11, 16571–16581 RSC.
- A. Desbois, M. Lutz and R. Banerjee, Biochemistry, 1979, 18, 1510–1518 CrossRef CAS PubMed.
- I. C.-Y. Chang, Y.-S. Sun, Y.-W. Yang, C.-H. Wang, S.-L. Cheng and W.-W. Hu, ACS Appl. Nano Mater., 2019, 3, 858–868 CrossRef.
- J. Wei, D. Xia, Y. Wei, X. Zhu, J. Li and L. Gan, ACS Catal., 2022, 12, 7811–7820 CrossRef CAS.
- D. Pan, J. Zhang, Z. Li and M. Wu, Adv. Mater., 2010, 22, 734–738 CrossRef CAS PubMed.
- L. Tang, R. Ji, X. Li, G. Bai, C. P. Liu, J. Hao, J. Lin, H. Jiang, K. S. Teng and Z. Yang, ACS Nano, 2014, 8, 6312–6320 CrossRef CAS PubMed.
- G. N. Balistreri, I. R. Campell, X. Li, J. Amorim, S. Zhang, E. Nance and E. Roumeli, RSC Appl. Polym., 2024, 2, 172–183 RSC.
- R. Riaz, M. Ali, H. Anwer, M. J. Ko and S. H. Jeong, J. Colloid Interface Sci., 2019, 557, 174–184 CrossRef CAS PubMed.
- H. Namduri and S. Nasrazadani, Corrosion Sci., 2008, 50, 2493–2497 CrossRef CAS.
- L. Lin, Q. Zhu and A.-W. Xu, J. Am. Chem. Soc., 2014, 136, 11027–11033 CrossRef CAS PubMed.
- L. Jiao, H. Yan, Y. Wu, W. Gu, C. Zhu, D. Du and Y. Lin, Angew. Chemie, 2020, 132, 2585–2596 CrossRef.
- P. Muhammad, S. Hanif, J. Li, A. Guller, F. U. Rehman, M. Ismail, D. Zhang, X. Yan, K. Fan and B. Shi, Nano Today, 2022, 45, 101530–101545 CrossRef CAS.
- B. Jiang, D. Duan, L. Gao, M. Zhou, K. Fan, Y. Tang, J. Xi, Y. Bi, Z. Tong and G. F. Gao, Nat. Protocols, 2018, 13, 1506–1520 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental section. Fig. S1–S13. See DOI: https://doi.org/10.1039/d4nh00024b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |