Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Classification of chemically modified red blood cells in microflow using machine learning video analysis†
R. K. Rajaram
Baskaran
 ,
A.
Link
,
A.
Link
 ,
B.
Porr
,
B.
Porr
 and
T.
Franke
and
T.
Franke
 *
*
Division of Biomedical Engineering, School of Engineering, University of Glasgow, Oakfield Avenue, Glasgow G12 8LT, UK. E-mail: thomas.franke@glasgow.ac.uk
First published on 4th December 2023
Abstract
We classify native and chemically modified red blood cells with an AI based video classifier. Using TensorFlow video analysis enables us to capture not only the morphology of the cell but also the trajectories of motion of individual red blood cells and their dynamics. We chemically modify cells in three different ways to model different pathological conditions and obtain classification accuracies for all three classification tasks of more than 90% between native and modified cells. Unlike standard cytometers that are based on immunophenotyping our microfluidic cytometer allows to rapidly categorize cells without any fluorescence labels simply by analysing the shape and flow of red blood cells.
Introduction
Screening blood samples in flow cytometers is a powerful tool in diagnostics and yields a statistically meaningful analysis of the blood composition broken down to a single cell level count. Cytometry1,2 is routinely used to detect widespread blood diseases such as in haemolytic anaemias, malaria, or functional disorders in erythropoiesis.3 In the clinical context, it is used to monitor the course of a disease or the success of a therapeutic treatment. Classification of blood cells it typically achieved by cluster of differentiation protocols and immunophenotyping of cells in which biomarkers are applied to indicate cell surface molecules. In many cases several fluorescent labels are employed that bind to receptors and ligands on the cell surface and the multicolour readout of the cytometer is used to classify the cell suspension. Yet, haematological disease and many disorders generating secondary haematological changes give rise to an abnormal morphology of red blood cells impairing cell function that cannot be revealed by this surface marker approach.Microfluidics4 and optical cytometry have opened the field5,6 to a label free morphological characterization of red blood cell suspensions in flow providing a cell shape7 and contour analysis.8–10 Yet, the complex interplay of soft boundary condition with an external shear flow field introduces a challenging problem11 to identify the features which can reliably be used for diagnosis. In fact, ubiquitous stationary i.e. time-independent shapes and non-stationary shapes for example a tumbling red blood cell have been observed in experiments12,13 as well as in theoretical analysis14,15 and simulations.16,17 A potpourri of cell shapes has been reported including symmetric discocytes, parachutes, stomatocytes, elliptocytes and asymmetric slipper-shaped RBCs.12 Matching the experimentally found cell morphology with detailed theoretical models employing specific values for the mechanical moduli, has revealed essential aspects of the dynamics and shapes,18 however, a full understanding of the underlying complex physical details is often not required for diagnosis. Moreover, high throughput microfluidic experiments under the microscope only reveal a two-dimensional projection of the cell shape and extrapolation to a full three-dimensional is far from being trivial even in symmetric flow.19 Also, subtle changes in morphology and motion are often not detectable by the eye and simple image analysis. AI offers a pathway to classify cells without detailed three-dimensional modelling the cell shape20 and has the flexibility to detect small differences of both morphology and motion which evade the human eye.21
The deformation of RBCs have been studied in various conditions and external fields22 as in electric fields23 using impedance measurements24,25 or hydrodynamic fields.26 However, so far RBCs in hydrodynamic microflow have been analysed using AI methods, with very few exceptions,27 only based on AI image analysis.28 However, using still images does not capture the full dynamics of the shape transitions and motion of RBCs. In fact, the motion of RBCs is rather complex29 including tumbling,30 tank treading,31 oscillation,30 swinging,32 flipping33 and intermediary forms of motion such as vacillating breathing.34 Temporal information of RBCs has also been used in flickering analysis and applied to aging and pathological changes35,36 as well as for studies of RBC dynamics and mechanics.37,38 A video based AI classification has been used to classify tank treading from flipping motion27 of sickle cell disease samples, however, this form of motion is just one aspect of potential differences among RBCs and does not provide an end-to-end-classification of the state of a cell for medical diagnosis. Moreover, the state of the cell is often hard to identify with the bare eye and certain motion patterns or shapes are not always present. This calls for a holistic approach where the deep network uses any information available and not forcing it to focus on just one aspect, such as morphology for image analysis or motion for video analysis but let the network decide. To overcome these limitations, video classification has emerged recently39 where not just image features but also the temporal relationships between frames are learned. Initially these have been used to classify sports footage and action sequences based on YouTube videos.40 In the medical context, for example, ultrasound videos have been classified with convolutional neural networks.41
In this present work, we chemically treat red blood cells using three different chemicals to modify the viscous and elastic mechanical properties of their plasma membrane and the cytoskeleton to mimic various diseases. Unlike studies in previous work, we probe the dynamic shape transformations and flow trajectories of RBCs in a spatiotemporal varying microchannel and classify end to end the cells using TensorFlow video analysis taking the full video sequences as input and directly outputting the state of the cell being either chemically modified or native. Our analysis can differentiate between healthy, untreated red blood cells and chemically modified cells to a high accuracy and provides a powerful tool for diagnostics.
Results
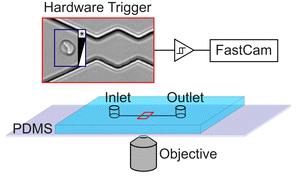
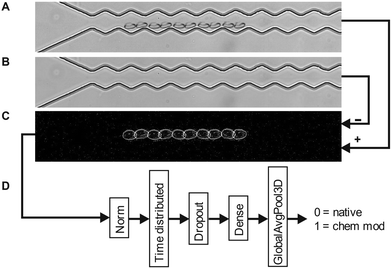
Red blood cell suspensions are diluted in a PBS buffer and injected into a PDMS based microchannel fabricated by soft lithography42,43 as shown in Fig. 1. The microfluidic setup is mounted on an inverted research microscope (Olympus, IX73) and observed in bright field. Cells that enter the periodically oscillating section of the channel are recorded with a fast camera (Phantom, VEO) at frame rates as high as 5000 FPS. To minimize storage memory, the recording of cell movies is triggered using a hardware trigger so that movies are only taken upon arrival of a cell in the region of interest. We test suspensions of native and chemically treated red blood cells in our device to modify various aspects of their viscoelastic properties and shape response in microflow to mimic various RBC diseases. Diamide (DA) has been reported to create disulphide bonds in the spectrin proteins44 and modifies the cytoskeleton network of RBCs. Glutaraldehyde (GA) and formalin (FA) include methylene bridges between amino acids and are commonly used to “fix” cells and have been shown to be a precise tool to modify the cell mechanics.45 Often mixtures of both are used because they are known to have different penetration depth. In prior experiments46 we investigated the role of confounding factors and identified the optical focus of the objective to be an important factor.47,48 To prevent the classifier to use the optical focus to distinguish between native and chemically modified, we take videos at different optical foci for all RBC suspensions. We used three different foci, one in focus and one slightly above and below the focus, respectively. The recorded dataset consists of approximately 5000 labeled video clips, each containing 250 frames. 200 videos are randomly selected from the dataset and pre-processed. The pre-processing steps include subtraction of static background as shown in Fig. 2A–C, cropping and subsampling to be able to fit all video data into the GPU memory. The preprocessed dataset is then introduced into the TensorFlow video classifier model for training. Clips being in focus and out of focus are randomly chosen for training which prevents the network to use the focus for classification.To perform training and validation, the model consists of standard layers for video classification as suggested by the TensorFlow documentation (Fig. 2D) with one numerical output: either “native” (0) or “chemically modified” (1). Training is performed by presenting the model with the pre-processed videos in random order. After each classification, the model's predicted label is compared with the true label of the video. It produces a non-zero error value if they are not the same, which is called “loss”. In simpler terms, loss is the penalty for a bad prediction. This is then used to optimise the model in a direction so that it converges towards correct classifications. Each of this training cycle where the model is adjusted is called epoch. During training the accuracy is tested against the actual training videos and against videos which were not used for training which is called “validation”. The validation provides TensorFlow with additional information how data of the training regime performs and is a measure of how good the model generalises to never seen data. After full training and simultaneous validation, the model is then tested against the test dataset, which consists exclusively of 100 videos that the model was not trained on and not been used for validation.
We conduct separate training sessions for each of the three types of chemically modified categories (native vs. FA, native vs. DA, and native vs. GA). In addition, we also train the model by asking it to just distinguish between native and anything chemically modified. We use 200 videos for each of these four cases and train the model for 100 epochs in each of these. The validation and test datasets consist of additional 100 pre-processed videos each.
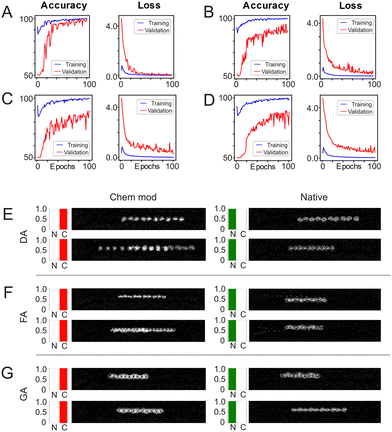
Fig. 3A–D shows the development of the accuracies and loss for classifications of DA, FA, GA chemically modified RBCs and mixed classification against native RBCs. We observe for all four cases the training accuracy converges towards 100% and the loss towards zero which means that learning overall converges. However, there are distinct differences how the four training sessions in Fig. 3A–D develop from the first to the last epoch. For RBCs chemically modified with DA (Fig. 3A) the validation accuracy rapidly reaches values close to 100% which indicates that the difference between chemically modified and native is very consistent so that learning has no need to incorporate many different shapes and motion patterns. This seems to be less the case for RBCs chemically modified with FA and GA (Fig. 3B and C) where the validation accuracy develops slower and stays at lower levels around the 90% mark. The worst performance is achieved when training against any chemically modified RBC (Fig. 3D) with a mix of FA, GA and DA modified RBC videos. This shows best for the validation loss in this case which stays at about 0.1 while any other validation loss converges towards zero. This comes as no surprise but is an excellent sanity check that shows that by creating very different behaviours and shapes by combining DA, FA and GA modified cells the algorithm finds it harder to find overall distinguishing features between native and chemically modified. After training the 4 cases (DA, FA, GA, mixed) are then tested with additional 100 videos which the classifier has not seen before. The resulting testing accuracies alongside the training and validation accuracies are shown in Table 1. The results of the testing accuracies reflect what we found during training: classification of RBCs chemically modified with DA yield the highest testing accuracy of 98%, while FA and GA reach a still respectable 93%. The detection of a mix of DA, FA and GA results in an accuracy of just 87%.
| Training (%) | Validation (%) | Test (%) | |
|---|---|---|---|
| DA | 99.5 | 99 | 98 |
| FA | 100 | 89 | 93 |
| GA | 100 | 89 | 93 |
| Mix | 98.5 | 88 | 87 |
To gain a better insight why the test accuracies between DA, FA, GA and their mix are different, we show example videos of the test dataset in Fig. 3E–G. Remember that we ran the four classification tasks separately so that we have four times a classification between chemically modified, in the 1st column, and native, in the 2nd column. The bar charts to the left of each overlay show the probability output of that video being native (N) or chem. mod. (C) as predicted by the trained model. Fig. 3E shows two examples of chemically modified RBCs with DA and two videos of native RBCs used for testing. The different lengths of the traces in the video overlays indicate different speeds of the RBCs but these do not impact on the classification result which stays robustly very close to 100%. The videos with RBCs chemically modified by FA are shown in Fig. 3F and have a slightly larger variability than the one modified by DA which explains the lower testing accuracy compared to the ones modified by DA. The last row shows the classification of RBCs chemically treated with GA (Fig. 3G). This is an interesting case where visually there is very little difference between native and chemically modified which requires the classifier to learn more subtle features and it certainly does: the classifier reaches a testing accuracy of 93%.
We tested the results shown in Fig. 3 in terms of another potential confounding factor in addition to the optical focus mentioned. We took the 6 native and 6 modified videos from the Fig. 3 output and plotted the distance of flight against the average cell distance from the centre line of zig-zag channel (Fig. 5). Both native and modified RBC values overlap in the scatter plot, indicating that the model is not biased towards the velocity or the position of the cell to classify the RBC.
Discussion
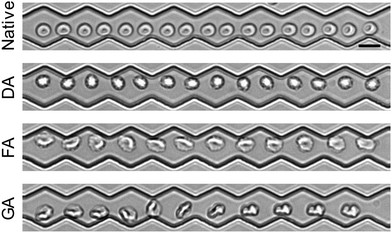
AI based video classification was initially developed on everyday actions such as cycling or sports because of the vast amount of data on social media such as YouTube.49 In medicine video classification was first applied to pose estimation50 and then to predict or diagnose illnesses based on behaviour such as autism51 or cerebral palsy.52 More recently ultrasound videos have been classified with machine learning for example to detect cancer53 or illnesses of the heart.54 While single images of RBCs have been classified with machine learning in the past,20,28 video classification is extremely rare and to our knowledge we are the first who have presented a machine learning end-to-end approach which classifies healthy and diseased RBCs directly from video. In this context Darrin et al. present classification of different RBC motions such as tank-treading and flipping which could then be used to detect the sickle cell disease based on motion. However, in our experiments these features occur rarely. Unlike in Darrin et al. where sickle cell shaped cells are used, that are simple to be identified by their shape, in our study it is generally difficult to judge the state of a cell (native/chemically modified) by the naked eye based on a micrograph or motion. However, some representative examples are shown in Fig. 4 to indicate different shapes and motions that we observe. Native cells are often flowing relatively “smoothly” through the channels, adapting their shape and undergoing a “gentle” tank-treading-like motion. Generally, for chemically modified cells a tumbling motion can be observed more often. In contrast to Darrin et al. we do not try to classify motion patterns first, but let the AI directly detect the modified cells. This allows the AI to choose any other feature which is not directly related to cell-motion. For example, for DA and FA modified cells, echinocytes are more frequently observed, displaying an irregular, spiked cell surface which can be also learned by our AI as it is not limited to motion analysis. AI image analysis has been extensively used in the context of RBCs but also here an intermediate step is usually applied: while video analysis naturally used motion as an intermediate step image analysis uses morphology as an intermediate step and then infer from the morphology to the illness of the RBC.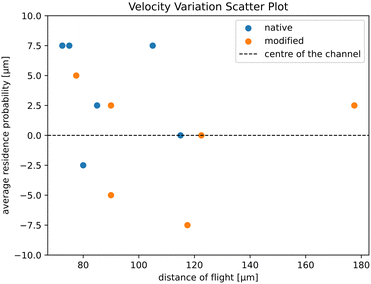 | ||
| Fig. 5 Scatter plot of showing the RBC residence probability20 of the RBC (distance from the channel centre line) against the distance of flight (which is proportional to the cell velocity). In the plot we used all RBCs shown in Fig. 3. The channel width at its widest section is 20 μm. | ||
Finally, the work by O’Connor et al.55 is not specifically about red blood cells but shows a different approach to ours where they first classify the cells and then for temporal sequence learning they use long-term-short-term memory (LSTM).55
We achieve an accuracy of 98%, 93% and 93% for DA, FA GA respectively. For the mix of all chemically modified we reached an accuracy of 87%. For this mixture we shuffled all the frames in each test video in order to understand the role of the temporal factors in the video classification and reached an accuracy of 81%. The study of Darrin et al. achieved a high accuracy of 97% between two RBC motion patterns. However, in a pre-processing step they discarded already 97% of unreliable cell sequences and used the remaining 3% for classification. In contrast, we only discarded clips with empty frames but otherwise drew a random selection from our pool.
It should be also stressed that a direct comparison between their performance and our performance is strictly not possible because we ultimately train our classifiers to detect different categories: motion detection vs. detection of a certain chemical modification.
Conclusions
Our label free microfluidic cytometer can distinguish to high accuracy between native and various chemically modified RBC suspensions. The different protocols that have been applied in this study, control the cell mechanics in multiple ways. While FA interconnects amino acids by methylene bridges in proteins in the cell membrane and potentially in the cytosol, DA induces the formation of disulphide bonds in spectrin proteins in the plasma-membrane. In this way various changes of viscoelastic moduli have been tested in this model system to mimic known changes of cell mechanics as they occur in haematological diseases and secondary disorders that affect the mechanical properties such as diabetes mellitus or malaria. In the future we plan to use our validated video AI based cytometer to widespread diseases such as diabetes mellitus and malaria.In our study we have used a stock video classifier as suggested by TensorFlow to show how classification can be achieved out of the box by approaches recommended by the industry. Our goal was to demonstrate the feasibility of this approach also in an industrial environment. For this reason, we also used open-source pre-trained models to achieve very fast training.56
Material and methods
Microfluidic device preparation
Microfluidic channels are created through the process of soft lithography, which involves several steps.29,30 First, the channel structure is designed using computer-aided design (CAD) software and transferred onto a chromium mask (ML&C GmbH). The channel itself is zigzag shaped with 20 periods of 20 μm length, an amplitude of 20 μm and a narrowness of 10 μm. The field of view of the camera is 230 μm × 40 μm and covers nine periods.Next, the pattern from the mask is transferred onto a silicon wafer coated with a 10 μm layer of SU8-3010 photoresist (Microchem, SU8 3000 series) using a mask aligner (MA6, Süss MicroTec). After development with Microposit™EC Solvent, the structured SU8 layer serves as a template for creating PDMS (polydimethylsiloxane) moulds. The PDMS (Sylgard™ 184 Silicone Elastomer Kit) is poured onto the template and cured for four hours at 75 °C. The ratio of elastomer base to curing agent used is 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.
To establish connections for the inlet and outlet of the channels, holes are punched into the cured PDMS moulds, allowing for tubing attachment. Finally, the PDMS mould is covalently bonded to a microscope slide using oxygen plasma.
RBC preparation
For our experiments, we prepared native and three different chemically modified red blood cell samples using the following procedure. Whole blood was purchased from Cambridge Bioscience from screened, healthy donors (Research Donors, Cambridge Bioscience) in accordance with the general principles set out in the Declaration of Helsinki. Samples were washed 3× in a phosphate-buffered saline solution (PBS 1×, pH 7.4, 330 mOsm L−1, Gibco Life Technologies). After each wash, the sample was centrifuged at 2500 rpm for 5 minutes using the mini spin plus centrifuge (Eppendorf), and the white buffy coat and supernatant were carefully discarded.For the chemically modified RBC experiments, we employed a combination of chemicals to achieve the desired modifications. Initially, we prepared a solution by mixing 5 μL of a 37% formaldehyde solution (final concentration of 0.37% formaldehyde, Sigma-Aldrich) with 485 μL of PBS. We then added 10 μL of the RBC pellet to this formaldehyde solution and incubated it for 10 minutes at room temperature. After incubation, the cell suspension underwent three thorough washes to eliminate any residual formaldehyde.
In addition to the formaldehyde treatment, we employed two other chemicals. Firstly, to induce oxidative stress, we created a premixed solution of 10 μL of 20 mM diamide solution and 180 μL of PBS (final concentration of 1 mM diamide). We then added 10 μL of the RBC pellet to this diamide solution and incubated it for 30 minutes at 37 °C. Subsequently, the cell suspension underwent three washes to remove any residual diamide.
Secondly, to facilitate crosslinking, we created a premixed solution of 20 μL of 25% glutaraldehyde with 470 μL of PBS (final concentration of 1% glutaraldehyde). We then added 10 μL of the RBC pellet to this glutaraldehyde solution and incubated it for 30 minutes at room temperature. Following the glutaraldehyde treatment, the cell suspension underwent three additional washes to ensure the proper removal of any unbound or excess glutaraldehyde.
To prevent cell sedimentation during the experiments, we suspended the cells in a density-matched solution using OptiPrep Density Gradient Medium (Sigma Life Science). OptiPrep is a sterile non-ionic solution containing 60% (w/v) iodixanol in water. Furthermore, to prevent cell adhesion to each other and the microchannel walls, we incorporated bovine serum albumin (BSA, Ameresco) in the suspension. To achieve this, we weighed 40 mg of BSA and added 3035 μL of PBS. The BSA mass concentration of this solution was 10 mg mL−1. We then mixed it with 945 μL of OptiPrep solution. After thoroughly mixing the solution, it was degassed for at least 15 minutes before use.
For the experiments, 5 μL of either the native or the different chemically modified cell pellet, after the washing steps, were resuspended in 995 μL of the density solution. This created samples with a haematocrit of Ht = 0.5% and a density of the solution ρ = 1.080 g mL−1. All experiments were conducted on the same day as the blood collection to maintain the freshness and viability of the cells.
The rbcs are injected into the channel using a pressure driven system with a pressure drop of 2 kPa.
TensorFlow and Python code
TensorFlow 2.11.0 compiled from source and Keras 2.11 is installed on an Intel Xeon E5630 2.53 GHz computer with 24 GB RAM and an NVIDIA GTX1070 graphics card. This system is running Ubuntu 22.04 LTS and is configured with CUDA 11.8 and cuDNN 8.6.The python code is available under https://zenodo.org/record/8126539. To train our model to be able to distinguish between native and chemically modified RBCs, we implemented a random selection process: where in total 200 videos were chosen, with an equal split of 100 videos from the native category and 100 videos from the chemically modified category. The videos were labelled as 1 for native and 0 for the chemically modified and then pre-processed.
During this pre-processing stage, we applied background subtraction by utilizing the OpenCV create Background Subtractor MOG2 algorithm. This algorithm leverages Gaussian mixture-based background/foreground segmentation, incorporating a history of 100 and a varThreshold of 10 to effectively separate the foreground objects from the background.
To ensure consistency, address the presence of empty frames in some recorded videos, and optimize the efficiency of the model training process, we subsampled each video down to 10 frames. This reduction was accomplished by starting from frame #50 and incrementing by 10 up to frame #140. Consequently, the processed videos assumed a shape of (10, 132, 800, 3), containing 10 frames at a height of 132 pixels, a width of 800 pixels, and 3 colours per channel. We subsequently converted these videos into tensors, along with their corresponding labels. The time of flight for all experiments was 20 ms.
For our neural network model, we employed the Keras sequential model as suggested in the TensorFlow tutorial documentation57 for video classification, which consisted of five layers. The pre-trained EfficientNetB0 model was employed for transfer learning approach, with its convolutional base used for feature extraction. Its base layers are frozen to prevent it from retraining. The following classification layers are then added to map the extracted features to the output classes: The rescaling layer was used to normalize the pixel values, ensuring consistent input across frames. The TimeDistributed layer applied the EfficientNetB0 model independently to each frame, capturing temporal dependencies. The dropout layer was introduced to mitigate overfitting, preventing the model from relying too heavily on specific features. The dense layer facilitated the learning of higher-level representations, capturing complex relationships between input frames and labels. Finally, the GlobalAveragePooling3D layer summarized the learned features into a concise representation, allowing for efficient analysis and classification. We trained the classifier end-to-end using Adam optimizer with Sparse Categorical Crossentropy loss function.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The works was supported by the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 813786 (EVOdrops). Additionally, the authors acknowledge support from the UK Engineering and Physical Sciences Research Council (EPSRC) via grant EP/P018882/1.References
- P. A. Arndt and G. Garratty, Transfus. Med. Rev., 2010, 24, 172–194 CrossRef PubMed.
- H. M. Shapiro, Practical Flow Cytometry, Wiley, 2003 Search PubMed.
- G. Moore, G. Knight and A. D. Blann, Haematology, Oxford University Press, 3rd edn, 2021 Search PubMed.
- A. Abay, S. M. Recktenwald, T. John, L. Kaestner and C. Wagner, Soft Matter, 2020, 16, 534–543 RSC.
- A. M. Forsyth, J. Wan, W. D. Ristenpart and H. A. Stone, Microvasc. Res., 2010, 80, 37–43 CrossRef CAS PubMed.
- M. Faivre, C. Renoux, A. Bessaa, L. Da Costa, P. Joly, A. Gauthier and P. Connes, Front. Physiol., 2020, 11, 1–10 Search PubMed.
- G. Tomaiuolo and S. Guido, Microvasc. Res., 2011, 82, 35–41 CrossRef PubMed.
- A. Merlo, S. Losserand, F. Yaya, P. Connes, M. Faivre, S. Lorthois, C. Minetti, E. Nader, T. Podgorski, C. Renoux, G. Coupier and E. Franceschini, Biophys. J., 2023, 122, 360–373 CrossRef CAS PubMed.
- G. Tomaiuolo, M. Barra, V. Preziosi, A. Cassinese, B. Rotoli and S. Guido, Lab Chip, 2011, 11, 449–454 RSC.
- M. Levant and V. Steinberg, Phys. Rev. E, 2016, 94, 062412 CrossRef PubMed.
- G. Tomaiuolo, M. Simeone, V. Martinelli, B. Rotoli and S. Guido, Soft Matter, 2009, 5, 3736 RSC.
- M. Abkarian, M. Faivre, R. Horton, K. Smistrup, C. A. Best-Popescu and H. A. Stone, Biomed. Mater., 2008, 3, 034011 CrossRef PubMed.
- S. Braunmüller, L. Schmid and T. Franke, J. Phys.: Condens. Matter, 2011, 23, 184116 CrossRef PubMed.
- C. Misbah, J. Phys.: Conf. Ser., 2012, 392, 012005 CrossRef.
- D. A. Fedosov, M. Peltomäki and G. Gompper, Soft Matter, 2014, 10, 4258–4267 RSC.
- H. Noguchi and G. Gompper, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 14159–14164 CrossRef CAS PubMed.
- J. Mauer, S. Mendez, L. Lanotte, F. Nicoud, M. Abkarian, G. Gompper and D. A. Fedosov, Phys. Rev. Lett., 2018, 121, 118103 CrossRef CAS PubMed.
- S. Atwell, C. Badens, A. Charrier, E. Helfer and A. Viallat, Front. Physiol., 2022, 12 DOI:10.3389/fphys.2021.775584.
- B. Kaoui, G. Biros and C. Misbah, Phys. Rev. Lett., 2009, 103, 188101 CrossRef PubMed.
- S. M. Recktenwald, M. G. M. Lopes, S. Peter, S. Hof, G. Simionato, K. Peikert, A. Hermann, A. Danek, K. van Bentum, H. Eichler, C. Wagner, S. Quint and L. Kaestner, Front. Physiol., 2022, 13 DOI:10.3389/fphys.2022.884690.
- P. S. Clegg, Soft Matter, 2021, 17, 3991–4005 RSC.
- K. Matthews, E. S. Lamoureux, M.-E. Myrand-Lapierre, S. P. Duffy and H. Ma, Lab Chip, 2022, 22, 1254–1274 RSC.
- Y. Zheng, J. Nguyen, C. Wang and Y. Sun, Lab Chip, 2013, 13, 3275 RSC.
- Y. Man, D. Maji, R. An, S. P. Ahuja, J. A. Little, M. A. Suster, P. Mohseni and U. A. Gurkan, Lab Chip, 2021, 21, 1036–1048 RSC.
- R. Reale, A. De Ninno, T. Nepi, P. Bisegna and F. Caselli, IEEE Trans. Biomed. Eng., 2023, 70, 565–572 Search PubMed.
- S. Braunmüller, L. Schmid, E. Sackmann and T. Franke, Soft Matter, 2012, 8, 11240 RSC.
- M. Darrin, A. Samudre, M. Sahun, S. Atwell, C. Badens, A. Charrier, E. Helfer, A. Viallat, V. Cohen-Addad and S. Giffard-Roisin, Sci. Rep., 2023, 13, 745 CrossRef CAS PubMed.
- A. Kihm, L. Kaestner, C. Wagner and S. Quint, PLoS Comput. Biol., 2018, 14, e1006278 CrossRef PubMed.
- J. Dupire, M. Socol and A. Viallat, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 20808–20813 CrossRef CAS PubMed.
- J. M. Skotheim and T. W. Secomb, Phys. Rev. Lett., 2007, 98, 078301 CrossRef CAS PubMed.
- T. Fischer and H. Schmid-Schönbein, in Red Cell Rheology, Springer Berlin Heidelberg, Berlin, Heidelberg, 1978, pp. 347–361 Search PubMed.
- M. Abkarian, M. Faivre and A. Viallat, Phys. Rev. Lett., 2007, 98, 188302 CrossRef PubMed.
- A. Viallat and M. Abkarian, Int J. Lab. Hematol., 2014, 36, 237–243 CrossRef CAS PubMed.
- C. Misbah, Phys. Rev. Lett., 2006, 96, 028104 CrossRef PubMed.
- K. Fricke and E. Sackmann, Variation of frequency spectrum of the erythrocyte flickering caused by aging, osmolarity, temperature and pathological changes, 1984, vol. 803 Search PubMed.
- H. Strey, M. Peterson and E. Sackmann, Biophys. J., 1995, 69, 478–488 CrossRef CAS PubMed.
- Y. Z. Yoon, H. Hong, A. Brown, C. K. Dong, J. K. Dae, V. L. Lew and P. Cicuta, Biophys. J., 2009, 97, 1606–1615 CrossRef CAS PubMed.
- G. Popescu, Y. Park, W. Choi, R. R. Dasari, M. S. Feld and K. Badizadegan, Blood Cells, Mol., Dis., 2008, 41, 10–16 CrossRef CAS PubMed.
- A. Karpathy, G. Toderici, S. Shetty, T. Leung, R. Sukthankar and L. Fei-Fei, in 2014 IEEE Conference on Computer Vision and Pattern Recognition, IEEE, 2014, pp. 1725–1732 Search PubMed.
- J. Carreira and A. Zisserman, in 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), IEEE, 2017, vol. 2017-Janua, pp. 4724–4733 Search PubMed.
- J. P. Howard, J. Tan, M. J. Shun-Shin, D. Mahdi, A. N. Nowbar, A. D. Arnold, Y. Ahmad, P. McCartney, M. Zolgharni, N. W. F. Linton, N. Sutaria, B. Rana, J. Mayet, D. Rueckert, G. D. Cole and D. P. Francis, J. Med. Artif. Intell., 2020, 3, 4 CrossRef PubMed.
- Y. Xia and G. M. Whitesides, Annu. Rev. Mater. Sci., 1998, 28, 153–184 CrossRef CAS.
- D. Qin, Y. Xia and G. M. Whitesides, Nat. Protoc., 2010, 5, 491–502 CrossRef CAS PubMed.
- A. M. Forsyth, J. Wan, W. D. Ristenpart and H. A. Stone, Microvasc. Res., 2010, 80, 37–43 CrossRef CAS PubMed.
- A. Abay, G. Simionato, R. Chachanidze, A. Bogdanova, L. Hertz, P. Bianchi, E. van den Akker, M. von Lindern, M. Leonetti, G. Minetti, C. Wagner and L. Kaestner, Front. Physiol., 2019, 10, 514 CrossRef PubMed.
- A. Link, I. L. Pardo, B. Porr and T. Franke, RSC Adv., 2023, 13, 28576–28582 RSC.
- V. Rizzuto, A. Mencattini, B. Álvarez-González, D. Di Giuseppe, E. Martinelli, D. Beneitez-Pastor, M. del, M. Mañú-Pereira, M. J. Lopez-Martinez and J. Samitier, Sci. Rep., 2021, 11, 13553 CrossRef CAS PubMed.
- J. R. Zech, M. A. Badgeley, M. Liu, A. B. Costa, J. J. Titano and E. K. Oermann, PLoS Med., 2018, 15, e1002683 CrossRef PubMed.
- S. Abu-El-Haija, N. Kothari, J. Lee, P. Natsev, G. Toderici, B. Varadarajan and S. Vijayanarasimhan, arXiv, 2016, preprint, arXiv:1609.08675 DOI:10.48550/arXiv:1609.08675.
- A. Mathis, P. Mamidanna, K. M. Cury, T. Abe, V. N. Murthy, M. W. Mathis and M. Bethge, Nat. Neurosci., 2018, 21, 1281–1289 CrossRef CAS PubMed.
- H. Abbas, F. Garberson, E. Glover and D. P. Wall, J. Am. Med. Assoc., 2018, 25, 1000–1007 Search PubMed.
- H. Haberfehlner, S. S. van de Ven, S. A. van der Burg, F. Huber, S. Georgievska, I. Aleo, J. Harlaar, L. A. Bonouvrié, M. M. van der Krogt and A. I. Buizer, Front Robot AI, 2023, 10 DOI:10.3389/frobt.2023.1108114.
- C. Chen, Y. Wang, J. Niu, X. Liu, Q. Li and X. Gong, IEEE Trans Med Imaging, 2021, 40, 2439–2451 Search PubMed.
- J. W. Hughes, N. Yuan, B. He, J. Ouyang, J. Ebinger, P. Botting, J. Lee, J. Theurer, J. E. Tooley, K. Nieman, M. P. Lungren, D. H. Liang, I. Schnittger, J. H. Chen, E. A. Ashley, S. Cheng, D. Ouyang and J. Y. Zou, EBioMedicine, 2021, 73, 103613 CrossRef CAS PubMed.
- T. O’Connor, A. Anand, B. Andemariam and B. Javidi, Biomed. Opt. Express, 2020, 11, 4491 CrossRef PubMed.
- P. Rajpurkar, A. Park, J. Irvin, C. Chute, M. Bereket, D. Mastrodicasa, C. P. Langlotz, M. P. Lungren, A. Y. Ng and B. N. Patel, Sci. Rep., 2020, 10, 3958 CrossRef CAS PubMed.
- TensorFlow documentation for video classification, https://www.tensorflow.org/tutorials/load_data/video, accessed 21 November 2023.
Footnote |
| † The python code with companion files. See https://doi.org/10.5281/zenodo.8126539. |
| This journal is © The Royal Society of Chemistry 2024 |