Osteoimmune-modulating and BMP-2-eluting anodised 3D printed titanium for accelerated bone regeneration†
Masood
Ali
a,
Yan
He
b,
Anna Sze Ni
Chang
c,
Alice
Wu
c,
Jingyu
Liu
d,
Yuxue
Cao
c,
Yousuf
Mohammad
ac,
Amirali
Popat
 c,
Laurie
Walsh
e,
Qingsong
Ye
*f,
Chun
Xu
c,
Laurie
Walsh
e,
Qingsong
Ye
*f,
Chun
Xu
 *e and
Tushar
Kumeria
*e and
Tushar
Kumeria
 *cg
*cg
aTherapeutics Research Group, Frazer Institute, Faculty of Medicine, University of Queensland, Brisbane, QLD 4102, Australia
bInstitute of Regenerative and Translational Medicine, Wuhan University of Science and Technology, Wuhan 430040, China
cSchool of Pharmacy, The University of Queensland, Brisbane, Queensland 4102, Australia. E-mail: t.kumeria@unsw.edu.au
dSchool of Mechanical, Medical and process Engineering, Queensland University of Technology, Brisbane, Queensland 4000, Australia
eSchool of Dentistry, The University of Queensland, Herston, Queensland 4006, Australia. E-mail: chun.xu@uq.edu.au
fCentre of Regenerative Medicine, Department of Stomatology, Renmin Hospital of Wuhan University, Wuhan 430060, China. E-mail: qingsongye@whu.edu.cn
gSchool of Materials Science and Engineering, University of New South Wales, Kensington, Sydney, New South Wales 2052, Australia
First published on 16th October 2023
Abstract
3D printing of titanium (Ti) metal has potential to transform the field of personalised orthopaedics and dental implants. However, the impacts of controlled surface topographical features of 3D printed Ti implants on their interactions with the cellular microenvironment and incorporation of biological growth factors, which are critical in guiding the integration of implants with bone, are not well studied. In the present study, we explore the role of surface topological features of 3D printed Ti implants using an anodised titania nanotube (TiNT) surface layer in guiding their immune cell interaction and ability to deliver bioactive form of growth factors. TiNT layers with precisely controlled pore diameter (between 21and 130 nm) were anodically grown on 3D printed Ti surfaces to impart a nano–micro rough topology. Immune biomarker profiles at gene and protein levels show that anodised 3D Ti surfaces with smaller pores resulted in classical activation of macrophages (M1-like), while larger pores (i.e., >100 nm) promoted alternate activation of macrophages (M2-like). The in vitro bone mineralisation studies using the conditioned media from the immunomodulatory studies elucidate a clear impact of pore diameter on bone mineralisation. The tubular structure of TiNTs was utilised as a container to incorporate recombinant human bone morphogenetic protein-2 (BMP-2) in the presence of various sugar and polymeric cryoprotectants. Sucrose offered the most sustainable release of preserved BMP-2 from TiNTs. Downstream effects of released BMP-2 on macrophages as well as bone mineralisation were assessed showing bioactivity retention of the released rhBMP-2. Overall, the TiNT surface topography in combination with controlled, sustained, and local release of bioactive growth factors can potentially enhance the osseointegration outcomes of custom 3D printed Ti implants in the clinic.
1 Introduction
Titanium (Ti) and its alloys have been the most widely utilised materials for dental and orthopaedic implants because of their proven biocompatibility and bone-matching mechanical properties.1–4 Despite wide clinical adoption, implant failures leading to corrective surgeries are mostly due to implant site infection and lack of bone integration (osseointegration).5 The role of micro- and nano-features of implants’ surface topography in promoting osseointegration is well researched and documented; however, commercial dental implants fail to generate and fabricate unique surface features that have the possibility to promote favourable conditions for osseointegration.6,7 The success of a good endosseous implant is linked to the achievement of osseointegration between the surrounding bone and the implanted material; however, such implants are incapable of drug loading and releasing due to the lack of nano-textured surfaces.1–4Generation of multi-functional nanotopography on Ti implants is made possible by an electrochemical anodisation process that generates precisely controllable nanosized tubes on the surface of the implants. Over the last decade, researchers have tuned the titania nanotubes (TiNTs) on Ti implant surfaces for release of small molecules and protein drugs for a more effective localised therapy, aimed at controlling initial bacterial invasion and promotion of bone integration. In addition, researchers have been able to tailor the immune cell interactions with TiNTs by manipulating their structural features for accelerated healing and bone regeneration.8–11 For example, Yu et al. have reported that the TiNT diameter ranging 20–70 nm promoted preosteoblast adhesion, 70 nm promoted alkaline phosphatase activity in 1–3 weeks, while 100–120 nm had any of those negligible effects.8 Brammer et al. have demonstrated the ability of a 30 nm nanotube diameter to stimulate preosteoblastic cell adhesion, whereas those measuring 70–100 nm promoted a lower number of cell adhesion with higher ALP activity and elongated morphology.9 Chamberlain et al. have reported that a 70 nm nanotube diameter produced the lowest inflammatory responses.12 Such data suggest that the TiNT diameter plays an important role in the developing bone cell behaviour. Other studies have created further modifications of these nanosized pores to impart other unique properties such as non-therapeutic antibiotic effects. The one-size-fits-all nature of the current Ti implants has been a major challenge for patients with varying diseases and implant needs. In recent years, generational advances in the metal additive manufacturing processes have enabled 3D printing of Ti and its alloys for custom built implants. The 3D printed Ti implants are mainly generated using a selective laser sintering (SLS) process. The SLS process forms a 3D structure by selective heating of metal microparticles using a laser, which means that the surface of the printed structures is covered with micron-sized spherical particulates.
Biomedical engineers have been fascinated by these surface features and have explored their interactions with bone cells. For example, Fox et al. have demonstrated improved osteoblast cells for material adhesion by simply coating the titanium scaffold surface (synthesised using selective laser sintering) with polycrystalline diamond (PCD).13,14 Losic et al. demonstrated both improved osteoblast cell adhesion and antibacterial effects on their biopolymer (chitosan and PLGA) coated TiNTs (synthesised using electrochemical anodisation) when compared to uncoated native TiNTs.15,16 Huang et al. studied the TiNT structural relationship with bone cell adhesion, demonstrating increased cellular surface protein membrane binding on TiNTs with a larger nanotube diameter, while cell proliferation and differentiation were favourable with a smaller nanotube diameter.17,18 Although such studies uncover TiNT's micron-scale topographically guided bone cell adhesion and proliferation, very little studies shed light on material-mediated immunomodulation and its effect on osteogenesis. Additionally, there is also a gap in studies exploring the loading and release of protein growth factors from TiNTs and its subsequent effect on osteogenesis.
Titanium nanotubes (TiNTs) are very well-characterised materials and various studies have investigated their interactions with immune cells and have shown their ability to modulate immune behaviour by carefully crafting surface features and chemistries.15,19–21 However, the majority of these studies have utilised a standard titanium material with smooth surface. With the rapid advent of 3D metal printing, customised Ti implants have become a reality.22–24 The 3D printed Ti implants, unlike their traditional machined counterparts, have widely distributed surface roughness, which is known to affect the behaviour of cells.24 Though attempts to modulate the cell behaviour on 3D printed Ti implants have been made, systematic characterisation of the role of micro-roughness from the inherent 3D printed Ti structure coupled with nano-roughness provided by anodized TiNTs in their surface is not well explored. Especially, a thorough investigation into the effect of the micro- and nano-roughness of anodized 3D printed Ti implants in guiding the immune cell behaviour has been missing. Furthermore, the ability of the TiNTs to incorporate and locally deliver drug payloads is well known and has been demonstrated with a number of small molecules. In recent years, researchers have also incorporated protein-based sensitive payloads like bone morphogenic protein-2 (BMP-2) into TiNTs for safer localised growth factor therapy.25–27 However, retention of activity over a long period and a sustained protein release have remained a challenge. Therefore, the systematic investigation into optimisation of the protein drug loading in the presence of FDA-approved excipients holds tremendous value and has never been reported. In this work, a sucrose-based excipient provided a sustained linear release behaviour of the bioactive form of BMP-2 from TiNTs for a period of 504 h (over 20 days), which is an impressive outcome. In addition, the activity assessment of BMP-2 released up to a 504 h period shows the long-term perseverance of its biological function on anodized 3D printed Ti implants. To conclude, the precisely tuned nano-roughness features developed on 3D printed micro-rough Ti substrates allowed us to not only control the cellular fate but also enabled long-term and sustained release of a growth factor. These aspects of our work differentiate it from the previously conducted studies that have predominantly focused on flat Ti substrates.15,20,21,28 This work concludes that the optimised surface topography in combination with controlled, sustained, and local release of bioactive forms of growth factors has potential to significantly improve osseointegration outcomes for patients implanted with custom 3D printed Ti implants in future.
2 Methods and materials
2.1 Materials
Lipopolysaccharides (LPS, Escherichia coli), SYBR® Green, cell culture medium and supplements, ammonium fluoride, ethylene glycol, and BMP-2 were purchased from Sigma Aldrich. Cell culture well plates were purchased from CORNING® (SIGMA-ALDRICH®, USA). 3D printed chips (12 × 12 mm) were purchased from Objective 3D (Australia) and anodised without any post processing. ELISA kits for IL1b and TGFb1 were purchased from Abcam (Australia). For all qRT-PCR studies, the gene primer sequences (listed in Table S1, ESI†) were purchased from Merck (Australia).2.2 Fabrication and characterisation of TiNTs on 3D titanium chips
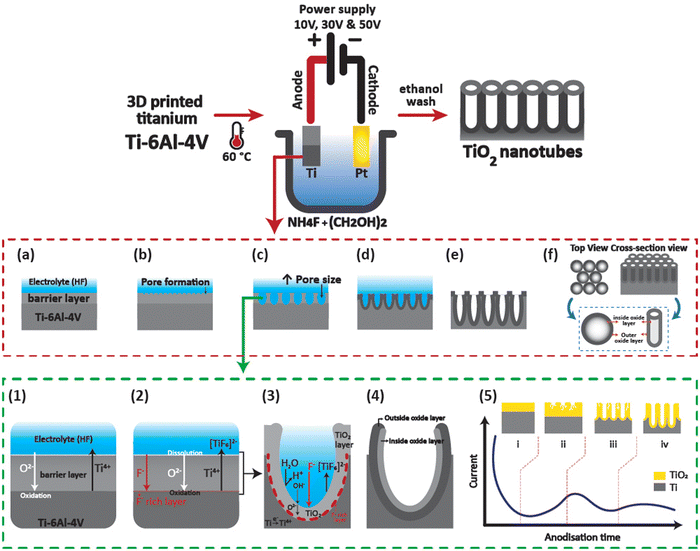
Functional hierarchical roughness on the 3D printed titanium was created by generating nanotubes of controlled dimensions on 3D Ti chips using electrochemical anodisation as described in our previous works.15 Briefly, the 3D Ti chips were sonicated in 100% ethanol, then washed with distilled water and dried with nitrogen gas. The dried chips were stored in a dry container and placed in a desiccator until further use. For anodisation, the 3D Ti chips were packed in a custom-designed holder. The anodisation electrolyte consisted of 0.1 M ammonium fluoride (NH4F) in ethylene glycol maintained at 60 °C using a hot plate (Heidolph). For creation of titanium dioxide nanotubes of diameters 21, 63, and 130 nm, applied voltages were adjusted to 10, 30, and 50 V, respectively, using a computer-controlled power supply (Keithly 2461). The samples were washed thoroughly with water and ethanol followed by storage in a vacuum desiccator until further use. It should be noted that the time of anodisation for all the samples was fixed at 45 min. The tubular structural features of the anodised 3D Ti chips were visualised using a scanning electron microscope (SEM, MIRA3, TESCAN) at 15.0 kV with a 5 nm gold coating. A schematic of the electrochemical anodisation process of 3D Ti chips is shown in Fig. 1.2.3 Cytocompatibility of macrophages on 3D TiNTs
It is important for a bone implant material to be cyto-compatible and allow the growth, attachment, and proliferation of cells on the surface. For this, RAW 264.7 macrophage cells were grown on TiNT surfaces of varying diameter. Macrophage cells (RAW264.7 cells) were seeded on TiNTs at a density of 104 cells per cm2 and their viability was assessed using the [3-(4,5-diemethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] (MTT) assay as mentioned elsewhere, with some modifications.29 After overnight incubation on the chips, LPS (1 μg mL−1) was added to the culture medium (DMEM, Life Technologies, Australia, supplemented with 10% (v/v) heat inactivated fetal bovine serum (In Vitro Technologies, Australia) and 1% (v/v) penicillin/streptomycin (P/S, Life Technologies, Australia)) to activate inflammatory macrophages and incubated for 2 h in an incubator (5% CO2 at 37 °C). Following that, the medium was replaced with a serum-free medium and incubated for a further 24 h and 48 h (5% CO2 at 37 °C). The culture medium was subsequently changed to 0.5 mg mL−1 MTT and incubated for 4 h. The medium was then replaced with dimethyl sulfoxide (DMSO) to dissolve any formazan products. The quantitative assay was performed using a microplate spectrophotometer (Infinite M200PRO TECAN) at 570 nm.In addition to cytocompatibility, morphology of macrophage cells is an important determinant of their attachment and proliferation on any implant surface. In this work, the macrophage morphology studies on TiNTs were performed using a protocol mentioned elsewhere,29 with some modifications. Briefly, macrophages were seeded on the TiNTs at a cell density of 104 cells per cm2, followed by LPS for 2 h. After 2 h, the medium was changed to a normal medium and the cells were allowed to grow for 4 h and 24 h. The cells were then fixed with 4% paraformaldehyde (PFA), followed by dehydration in graded series alcohol (30, 50, 75, 90 and 100%) and drying with the hexamethyldisilazane (HMDS) solvent and CO2. The anodised 3D Ti chips were sputter coated with gold, prior to the field emission scanning electron microscopy (MIRA3 TESCAN, SEM) imaging at 7 kV.
2.4 Macrophage polarisation on 3D TiNTs
All immunomodulation studies were performed using mouse RAW264.7 cells (macrophage cells), following a protocol recommended by the American Type culture Collection (ATCC), which was mentioned elsewhere.30 Briefly, the macrophage cells were maintained in DMEM medium, supplemented with 10% (v/v) heat inactivated fetal bovine serum and 1% (v/v) penicillin/streptomycin, in a humidified environment containing 5% CO2 at 37 °C. The cells were passaged by dislodging them from the flask surface using a scrapper after they reached 80–90% confluence, for immunomodulation studies.The expression of pro and anti-inflammatory gene in macrophage cells grown on TiNTs was quantified by a quantitative polymerase chain reaction (qRT-PCR). The RNA extraction process was reported elsewhere.30 Briefly, the macrophage cells were seeded on the 3D TiNT chips at a density of 105 cells per chip. After 24 h, the cell culture medium was replaced with a serum-free medium containing LPS (1 μg mL−1) and incubated for 2 h. After 2 h of stimulation, the medium was discarded and the cell monolayer was rinsed with PBS before adding more DMEM medium and further incubated for 12 h. Following this, the macrophage cell-conditioned medium (CM) was removed and stored at −80 °C for osteogenesis studies. The total RNA of the treated macrophage cells was extracted, and RNA (1 μg) was then reverse transcribed into cDNA using Thermo Scientific DyNAmo cDNA kit F-470. Power Up SYBR Green Master Mix (Thermo Scientific, Australia) was utilised for the qRT-PCR assay. The inflammatory genes (IL-1β, IL-6, CD86, TNF-α, and IL-18), anti-inflammatory genes (IL-1ra, CD206, arginase, and TGF-b1) and pro-angiogenic gene (VEGF) were quantified on an Applied Biosystems StepOnePlus™ Real-Time PCR machine. The relative expression was acquired by normalising the mean cycle threshold (Ct) value of each target gene with the Ct value of the housekeeper gene GAPDH. The relative gene levels were then normalized against the control group. To support the gene expression data, expression of TGFb1 (Anti) and IL1b (Pro) protein in the cell culture medium was measured by using a commercial ELISA kit (TGFb1-ab119557, IL1b-ab197742) following the protocol provided by the manufacturer.
2.5 Osteogenic differentiation of rBMSCs
Rat bone marrow mesenchymal stromal cells (pre-osteoblast cell line, rBMSCs) were extracted from rat femurs with ethics approval from the Ethics Committee of the University of Queensland (Ethics number 2018/AE000433). The extraction process was mentioned elsewhere.31 Only rBMSCs of passage 2–4 were used in the osteogenesis experiments. To investigate the effect of cytokines secreted by macrophages stimulated through the surface nanotopography of the TiNTs on the osteogenic differentiation of rBMSCs, qRT-PCR and Alizarin Red S staining were performed. An established set of five osteogenic differentiation genes (OCN, OPN, RUNX2, IBSP, and Col1A) was quantified through the following protocol. The undifferentiated rBMSCs were seeded at a density of 106 cells per well in a 24-well plate. After incubation for 24 h, the culture medium was replaced with implant surface topography-stimulated macrophage-derived CM (134 nm NT and 3D Ti) followed by 3 days of incubation, a protocol mentioned elsewhere.31,32 The total extracted RNA of the rBMSCs was reverse transcribed into cDNA using a Thermo Scientific Maxima First Strand cDNA Synthesis Kit with a 1000 ng RNA sample per well, and the qPCR was operated by using a SimpliAmp Thermal Cycler (Life Technologies, Australia). The osteogenic-related genes were tested on an Applied Biosystems StepOnePlus Real-Time PCR system. The relative expression was calculated by normalizing the mean cycle threshold (Ct) value of each target gene with the Ct value of the housekeeper gene (GAPDH).Mineralised nodule formation is a hallmark of osteogenic differentiation rBMSC. In this work, Alizarine Red S staining was performed to visualise the mineralised nodules in rBMSCs seeded in a 24-well plate for 14 days in the presence of CM. The undifferentiated rBMSCs were seeded at a density of 106 cells per well in a 24-well plate. After incubation for 24 h, the culture medium was replaced with implant surface topography-stimulated macrophage-derived CM (134 nm NT and 3D Ti) followed by 14 days of incubation, replenishing the media every 3 days, following an established protocol mentioned elsewhere31,32 After 2 weeks, the cells were washed with water and stained in a 1 mL of 2% Alizarin Red S solution (pH 4.1, 20 μg mL−1) for 20 min. The samples were air dried, and images were acquired using a light microscope (Olympus CKX41).
2.6 Loading and release of BMP-2 from TiNTs
The large open pore volume of TiNTs prepared at 50 V was utilized to load protein growth factors. To obtain the highest possible protein bioactivity retention of the released payload from TiNTs, we assessed co-loading of lysozyme (Lyso) as a model protein with a variety of commonly used sugar- (dextrose, sucrose, and maltose) and polymer-based (polyvinyl alcohol, polyvinyl pyrrolidone, and chitosan) cryo-protectants. To start, a 5 mg mL−1 lysozyme solution was prepared in either a 1 M aqueous solution of sugar-based cryoprotectants or a 0.25% w/v aqueous solution of polymeric cryoprotectants. The loading process involved dropping 10 μL of Lyso solutions onto the anodised part of the 3D Ti chips followed by vacuum infiltration for 15 min. This step was repeated 10 times in total for all the Lyso-cryoprotectant mixtures to obtain 500 μg of total loading of the Lyso per 3D Ti chip. After the 10th time, the chips were flash frozen in liquid nitrogen and lyophilised. The protein-loaded lyophilised TiNT chips were stored at −20 °C until further use.To understand the Lyso release and activity retention, we incubated the Lyso-loaded chips into 2 mL phosphate buffer solution at pH 7.4. A 1 mL aliquot was aspirated at defined time points and the amount of total Lyso released was measured using a Micro BCA™ Protein Assay Kit (Catalog number: 23235, Thermo-Fisher, USA) according to the supplied protocol. The activity of the released Lyso was determined using a well-established bacteria digestion assay. For this, a Micrococcus lysodeikticus suspension with optical density (o.d.) value close to 0.7 (a.u.) at 450 nm was prepared in a 0.1 M phosphate citrate per citrate buffer at pH 5.8. The activity of the released Lyso was measured by recording the kinetics of digestion of Micrococcus lysodeikticus over 5 min. Briefly, 92 μL of bacterial suspension was pipetted out into a low volume 96-well microplate and 8 μL of calibration standard or release aliquot was added to this suspension and the OD values at 450 nm read immediately for 5 min. The slope of the digestion curve was plotted against the concentration to prepare a calibration curve. The amount of active Lyso in the release aliquot was calculated by fitting the unknown slope values into the calibration curve.
After determining which excipient was best for preserving protein bioactivity, BMP-2 was loaded into TiNTs using a similar process as Lyso using a 100 μg mL−1 solution of BMP-2 in 1 M sucrose or 0.25% wt/v PVA. Briefly, 10 μL BMP-2 solution with or without cryoprotectants was added to the TiNT surface and vacuum infiltrated for 15 min. For BMP-2 loading, this step was repeated 5 times in total to achieve 2.5 μg protein loading per chip. The final chips were then frozen immediately using liquid nitrogen and lyophilised. Like the Lyso-loaded TiNTs, the chips after BMP-2 loading and lyophilisation were stored at −20 °C until use.
For the release study, the TiNT chips were placed in a 24-well plate. 1 mL phosphate buffer saline (PBS, pH 7.4) was added to each well and incubated at 37 °C. The well plates were placed securely on a shaker (330 rpm, Multi Reax, Heidolph Instruments, Germany). The supernatant PBS solution was collected from each well, at specified time points (2 h, 5 h, and days 1, 3, 5, 7, 9, 18, and 21). The release profile was quantified using a BMP-2 ELISA kit from Invitrogen as per the manufacturer's instruction. The release aliquots were frozen for future use.
2.7 Statistical analysis
All experiments were performed in triplicate independently and the data were presented as mean ± standard deviation (SD). A Student's – t test was used to compare between two different groups. A one-way analysis of variance (ANOVA) was used for group analysis, from a previously followed protocol.33 The statistical differences were considered significant if the p value was <0.01. GraphPad Prism was used for all the statistical calculations.3 Results
3.1 Structure of TiNTs on 3D printed Ti chips
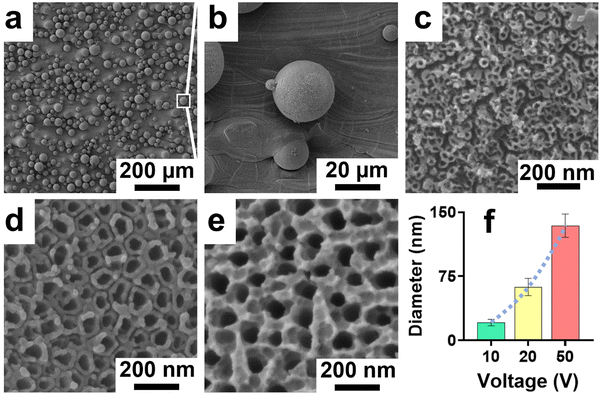
The structure of the nanotubes fabricated on the 3D Ti chips was assessed using SEM. The surface of the 3D printed Ti implant chips at low magnification showed microspheres on the surface imparting a microrough morphology to their surface (Fig. 2a and b). This microroughness is from the selective laser sintering (SLS)-based manufacturing of the 3D Ti implant chips. The SLS process uses a high energy laser to sinter microspheres of Ti to form the desired 3D geometry (square shaped chips in this case). The microspheres on the surface were firmly fused to the underlying 3D printed layers, suggesting a stable and highly interconnected micro-rough surface.34 Tubular titanium dioxide (TiNTs) can be seen on the microrough surface of the 3D Ti implant chips after electrochemical anodisation. By altering the anodisation voltage between 10, 20, and 50 V, TiNTs of 20.8 ± 3.8 nm, 62.5 ± 10.1 nm, and 134.2 ± 13.7 nm, respectively, were generated (Fig. 2c–e). As expected, a graph presenting tube diameter against anodisation voltage shows an exponential increase (Fig. 2f).343.2 Cytocompatibility of macrophages on TiNTs
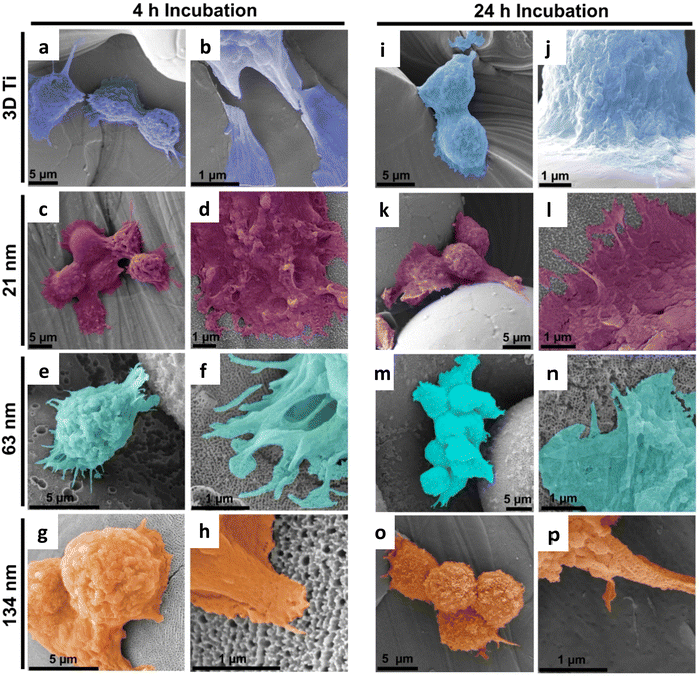
Cytocompatibility of TiNTs on 3D Ti chips was investigated using macrophage cells via MTT assay. The cell viability on TiNTs with varying tube diameters was greater than 90%, alluding to their high cytocompatibility (ESI,† Fig. S1). It should be noted that cell viability measurements using MTT assay were carried out for the immunomodulation experiments to account for any cell death after LPS treatment.In addition to macrophage viability assessment, their attachment and proliferation on the TiNTs on 3D Ti chips was assessed using scanning electron microscopy (SEM). The SEM images in Fig. 3 show that macrophage cells attached, spread, and grew well on all of the tested samples. The cells on 3D Ti control (non-anodised) and all the three TiNT samples can be seen as spherical structures with filopodia spreading and reaching out to nearby rough structures. At 4 h time point, macrophages cultured on TiNTs were spherical in morphology with numerous microvilli or small protrusions (left panel in Fig. 3). The interaction between the cells and the titanium surface is evident from cell spreading, ruffled cell membranes and extended filopodia. After 24 h of cell culture (right panel in Fig. 3), macrophages on the 3D Ti surface remain spherical in morphology. The cells on TiNTs with 21 and 63 nm average pore diameter were observed to form thin lamellar, flat, and widely adherent to disc like pattern with spherical cells still remaining. Macrophages on the 134 nm TiNTs can be seen transitioning into an elongated morphology, and cell clustering is evident from multiple cells in close proximity to each other. Behaviours, such as cell spreading, filopodial extensions and cell clustering, are signs of macrophage attachment and activation.12 Macrophages growing on native Ti extended long filopodia and appeared to colonize larger areas within 4 h and 24 h in comparison to macrophages seeded on derivatized Ti (Fig. 3), which were also circular shaped and formed shorter filopodia. This result confirmed that macrophages growing on derivatized Ti are mostly inactive.
3.3 Immunomodulation of macrophages via TiNT morphology
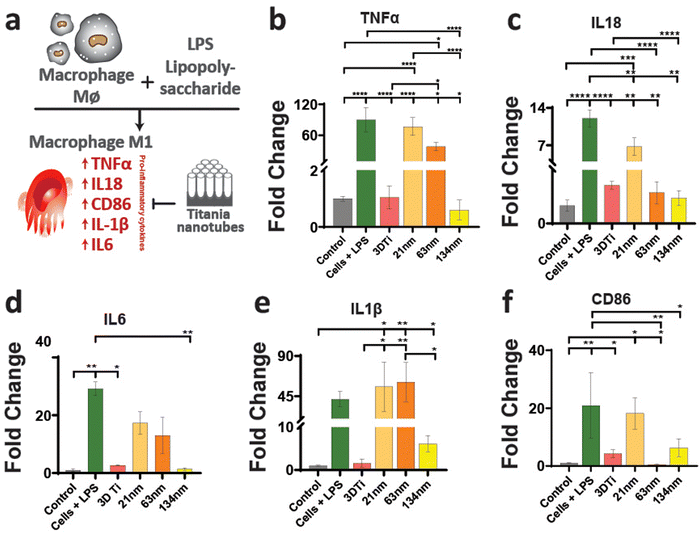
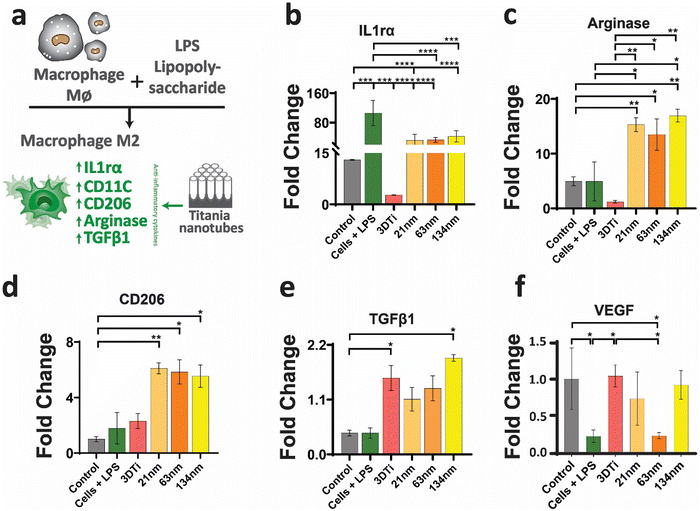
After establishing macrophage viability and attachment on TiNTs, immunomodulation and phenotypic polarisation of macrophages were investigated by measuring the gene expression levels after stimulation with LPS. Expression levels of five pro-inflammatory genes (TNF-α, IL-6, IL-18, IL-1β, and CD86) are shown in Fig. 4. Compared to control (grey bar) the cells treated with LPS (green bar) showed a sharp increase in the expression of the selected pro-inflammatory genes. The cells on non-anodised 3D Ti surfaces showed a reduced expression of these genes. The expression of these genes on anodised TiNT surfaces shows an inverse relationship with tube diameter (i.e., the highest on 21 nm and the lowest on 134 nm tubes). Interestingly, 3D Ti surfaces consistently had lower pro-inflammatory gene expression levels in comparison to TiNTs with diameters 21 nm and 63 nm, whereas the TiNTs had significantly reduced levels of four (TNF-α, IL-6, IL-18, and CD86) out of total five selected pro-inflammatory genes.The gene expression levels of four key anti-inflammatory genes (IL1ra, arginase, TGFb1, and CD206) and a pro-angiogenic marker (VEGF) are presented in Fig. 5. The data show that the cells grown on TiNT surfaces consistently show higher levels of the selected genes, compared to non-anodised 3D Ti and other controls.
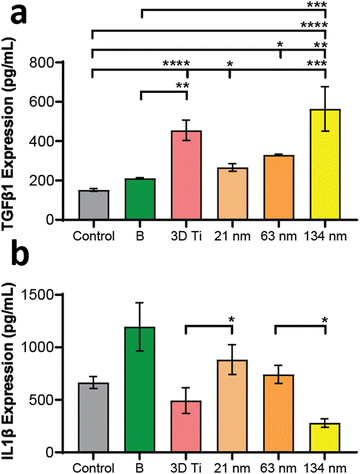
In addition to gene expression, extracellular concentration of one pro-inflammatory and one anti-inflammatory protein transcribed by a gene in each panel was also measured. The protein expression levels, measured by ELISA, matched the gene expression data, wherein the TGF-b1 (anti-inflammatory) levels (Fig. 6a) increased with increasing TiNT diameter, while IL-1β (pro-inflammatory) levels (Fig. 6b) showed an opposite trend. Taken together, the gene expression and ELISA results demonstrate surface guided polarisation of macrophages with a clear indication that smaller nanotubes lead to more pro-inflammatory classical activation (i.e., M1 polarisation) of macrophages, whereas the TiNTs with larger pore diameter (>100 nm) alternatively activate (M2 polarisation) the macrophages.
3.4 Immunomodulation induced bone mineralisation in rBMSCs
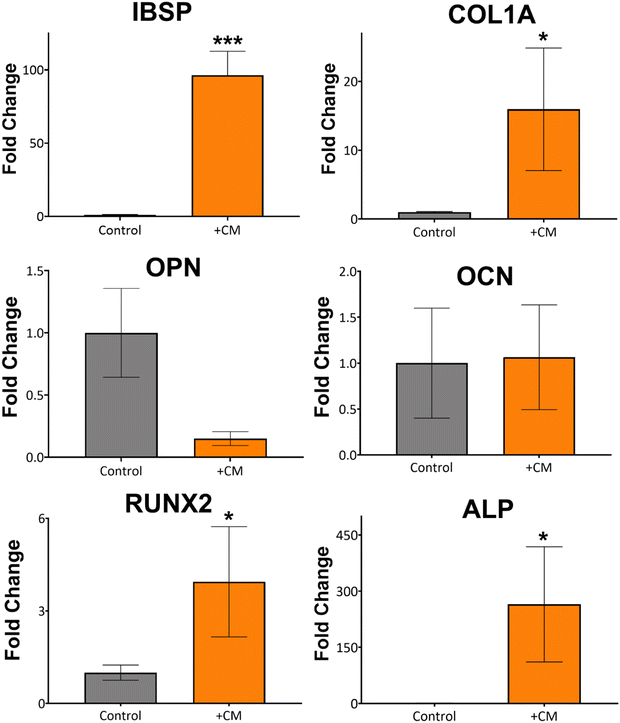
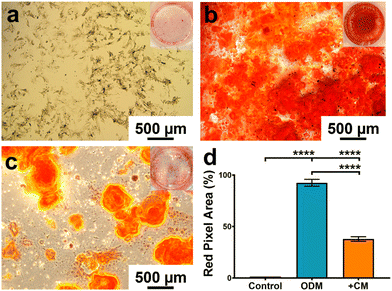
Based on the macrophage polarisation results elucidated by the gene and protein expression data, culture medium from 134 nm TiNTs, with proven surface mediated anti-inflammatory properties, was used to induce bone mineralisation in vitro. For this, rBMSCs were treated with a conditioned medium for 3 days and gene expression levels of six osteogenic factors were measured (shown in Fig. 7). Four of the key genes, IBSP, COL1A, RUNX2, and ALP were significantly overexpressed in rBMSCs treated with the macrophage-conditioned medium, while OPN and OCN were not affected. Overall, gene expression data confirm that treatment with 134 nm TiNT-conditioned medium stimulated the rBMSCs to express osteogenic factors compared to unstimulated rBMSCs.To support the gene expression data, we carried out in vitro biomineralisation using the Alizarin Red assay. The digital photographs and microscopy images of the Alizarin Red assay data shown in Fig. 8 demonstrate that untreated rBMSCs show minimal biomineralisation (Fig. 8a), while the rBMSCs cultured in the osteogenic differentiation medium (ODM; Fig. 8b) show the highest biomineralisation with large amounts of calcinated nodule formation. The rBMSCs treated with CM (Fig. 8c) lead to significantly higher biomineralisation compared to untreated rBMSCs (Fig. 8a). However, the amount of biomineralisation is considerably lower compared to ODM cultured cells. Percentage of the red pixel area normalised to the full image area of the untreated, ODM-, and CM-treated rBMSCs is presented in Fig. 8d with 92% and 38% area stained by Alizarin Red for ODM-and CM-treated samples, compared to only 1.4% area stained for untreated control. Therefore, it can be inferred that the cytokine cocktail in the CM derived from the nanotube-stimulated macrophages has prompted the unstimulated rBMSCs towards osteogenesis.
3.5 BMP-2 release from TiNTs
The large open pore diameter of TiNTs (134 nm) was utilized for the loading and release of protein growth factor studies. Although most nanocarriers allow for loading and release of protein-based drugs, their bioactivity retention is often poor for a variety of reasons. Past studies have shown proteolytic inactivation and degradation during the loading, lyophilisation and release of therapeutics from TiNTs.35,36 Therefore, to obtain the highest possible bioactivity retention of the protein payload released from TiNTs, first, the protein loading was optimized using lysozyme (Lyso) as a model protein37 in the presence of a variety of commonly used sugar (dextrose, sucrose, and maltose) and polymeric (polyvinyl alcohol, polyvinyl pyrrolidone, and chitosan) excipients.The lysozyme bioactivity retention was firstly examined by loading it naked into the TiNTs and studying its loading and release profiles. The lysozyme showed excellent burst release within the first hour and then gradually released completely within 5 h (ESI,† Fig. S2). However, only half (56.13 ± 10.45%) of the total released lysozyme from the TiNTs retained its bioactivity (ESI,† Fig. S2). To improve the bioactivity of lysozyme, we then spiked it with three polymers and sugars, separately. The bioactivity retention values of the lysozyme spiked with the respective polymers were shown to be quite low, 23.22 ± 13.1% for PVA, 10.99 ± 2.16% for chitosan and 20.41 ± 5.80% for PVP, alluding to its poor protein stabilising properties (ESI,† Fig. S3a–c). On the other hand, although an initial burst release of lysozyme was observed when spiked with the three sugars, the bioactivity values of the released lysozyme were 77.56 ± 0.1% for trehalose, 20.95 ± 0.24% for dextrose and 96.93 ± 0.59% for sucrose (ESI,† Fig. S4a–c). The bioactivity retention of trehalose spike lysozyme was comparatively higher than that of a study conducted by Zhang et al.18 (∼65%). Data for the near-complete bioactivity retention of lysozyme spiked with sucrose also aligned with data obtained by Nema et al., alluding to its superior protein stabilising properties.38 The protein bioactivity retention data agreed with the deduction made by Wang et al., where disaccharides (trehalose, sucrose) were more effective than monosaccharides (dextrose) in stabilising proteins.39 Therefore, sucrose was chosen to be loaded with BMP-2 into TiNTs to study the stabilising effects it had on BMP-2.
The release profiles of BMP-2 with or without stabilisers from TiNTs were quantified using an ELISA kit, and the absorbance was measured using a spectrophotometer. TiNTs with a payload of sucrose/BMP-2 solution showed a release profile extending to 21 days with the highest protein bioactivity retention of 96.3%, whereas for BMP-2 without any stabilisers the release stopped after 9 days (ESI,† Fig. S5). The data suggest that sucrose, besides acting as a stabiliser and a cryoprotectant, facilitated longer release of BMP-2 from TiNTs. These data agree with various studies conducted investigating the stabilising effect of saccharides on sensitive proteins.38,40 The data also showed controlled and sustained diffusion of BMP-2 during the release study, indicative of effective loading of the proteins into the deeper parts of the nanotubes.41
3.6 Osteogenic, osteoclastogenic and fibrotic effects of BMP2 released from 3D TiNTs on macrophages
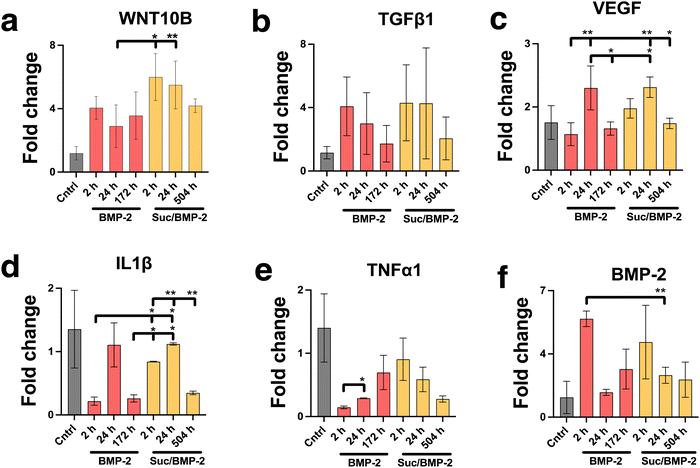
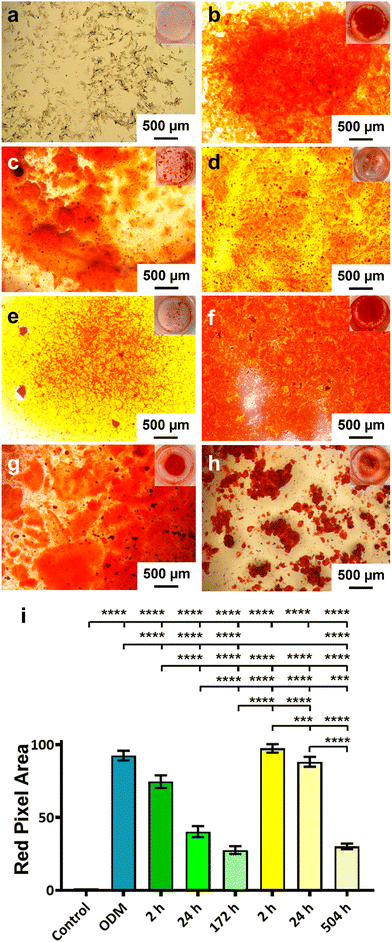
The activity of the BMP-2 released from TiNTs was measured by assessing the osteogenic, osteoclastogenic and fibrotic activities, respectively, using macrophages treated with BMP-2 release aliquot at different time points in the release profile. Macrophage differentiation upon treatment with BMP-2 released from 134 nm TiNTs was studied through measurement of gene expression levels of three osteogenic factors (WNT10B, TGFβ1 and VEGF), an osteoclastogenic factor (IL-1β) and a fibrogenic factor (TNFα). The data (Fig. 9) suggest that treatment with released BMP-2 upregulated the expression of osteogenic factors, while downregulated the expression of osteoclastogenic and fibrogenic factors. The balance between fibrogenesis and osteogenesis plays a vital role in determining the outcome of bone biomaterials, by either forming new bone or creating a fibrotic capsule.Fig. 10 shows images of biomineralisation in rBMSCs under control conditions and upon treatment with BMP-2 released from TiNTs (134 nm). Fig. 10a shows a representative digital photograph of the well and a microscope image of Alizarin red assay of untreated rBMSCs and Fig. 10b shows the corresponding Alizarin red assay image of osteogenic differentiation medium-treated rBMSCs, while Fig. 10c–e show representative Alizarin red assay images of rBMSCs treated with three time-point samples (2, 24, and 172 h) of BMP-2 released from 134 nm TiNTs loaded in the absence of a cryoprotectant. Similarly, Alizarin red images of rBMSCs treated with Suc-BMP-2 (loaded in the presence of sucrose as a cryoprotectant) released from TiNTs at 2, 24, and 504 h release time points are shown in Fig. 10f–h. Areas of red pixels extracted from the Alizarin red data corresponding to Fig. 10a–h are presented in Fig. 10i. A clear trend of a decrease in the amount of biomineralised nodule formation can be seen when treated with both BMP-2 (Fig. 10c–e) and Suc-BMP-2 (Fig. 10f–h) at increasing release-time points. The area of red pixels in the case of 2 h time point Suc-BMP-2-treated rBMSCs was higher than those of the ODM and 2 h BMP-2 treatments. A significantly higher red pixel area was measured for 24 h suc-BMP-2 treated rBMSCs compared to the same 24 h time-point aliquot of BMP-2. It is worth mentioning that the red pixel area for rBMSCs treated with a 172 h aliquot of BMP-2 was comparable to that treated with a 504 h release aliquot of Suc-BMP-2. These results clearly indicate a greater bioactivity retention by Suc-BMP-2 compared to BMP-2 released from TiNTs.
4 Discussion
The layers of highly ordered and vertically aligned titania nanotube arrays were formed using electrochemical anodisation, a process that produces nanostructures on the surfaces of metal-based implants.34 The diameter of TiNTs increased with the applied current density as it changed the electrochemical etching rate as reported previously.42In order to assess macrophage viability on the nanotubular and 3D Ti surfaces, the cells were stimulated to elicit a proinflammatory response and then allowed to grow on the previously mentioned surfaces. As such, a contaminated environment was created to mimic the nature of a compromised implant, by exposing the cells to LPS, recognised by the innate immune system as a sign of infection.43 The macrophage morphology produced cytoplasmic elongations and cells spread on the titania surface, especially upon stimulation by LPS, which agrees with multiple studies.44,45 It must be noted that the SEM images in Fig. 3 are representative images of a larger scanned area. SEM was performed to examine the adhesion of RAW 264.7 cells (macrophages), specifically examining the filopodial extension as well as spreading of macrophage cells on the TiNT surface (with/without TiNTs). Even though we have not examined macrophage cell adhesion speed on the titania surface upon stimulation by LPS, where past results have demonstrated morphological changes associated with macrophage phenotype polarisation,45,46 our pro-inflammatory genetic profile studies have exhibited a protective effect against LPS-induced morphological changes (demonstrated by a macrophage gene expression profile on TiNTs with a 134 nm diameter). This is represented by the low expression of TNFα, IL-6, IL18 and IL-1β genes and higher expression of IL-1ra, TGF-b1 and VEGF genes, in LPS-induced macrophages compared to unstimulated macrophages when grown on 134 nm TiNTs and 3D Ti surfaces. Not only did the nanotubes of 134 nm significantly down-regulate the gene expression of all the previously mentioned pro-inflammatory cytokines and decreased their extracellular production (IL-1β), but also increased the production of anti-inflammatory cytokine production, which is in agreement with multiple other studies.45,46 In previous studies, pore diameter has been correlated to the surface roughness. Therefore, it is worth noting that the immune modulation via tuning of the diameter of TiNTs can be attributed to the surface roughness. Surface roughness of TiNTs of different dimensions has been correlated with their diameters on flat surfaces in previous studies.47–50 The micro-roughness on the 3D printed substrates makes it challenging to accurately quantify and correlate the surface roughness of the TiNT substrates to their diameter. Therefore, only the TiNT diameter was correlated with immune function of the cells. Macrophages are classically activated by bacterial endotoxin LPS and some chemokines, displaying either of the phenotypes, known to be involved in type I inflammation responses or pro bone healing processes.3 For example, exposure to IL-6 cytokines has led to activating the M1 profile of macrophages,51 whereas BMP-2 exposure has led to an alternatively activated M2 profile,52 which suggests that macrophages can convert to different phenotypes due to their high diversity and plasticity. This transition has created an osteoimmune environment that is conducive to osteogenesis. Furthermore, our studies have also demonstrated the role of BMP-2 in a macrophage morphological shift towards osteo-specific phenotype through the expression of osteogenic factors (BMP-2, WNT10B, TGFβ1 and VEGF), lower expressions of both osteoclastogenic (IL-1β) and fibrosis-enhancing factor (TNFα), respectively.
The nanotubular structures can act as drug reservoirs and protect drugs from harsher conditions such as proteolytic degradation and facilitate their sustained release at the same time.37 However, bioactivity retention for most protein growth factors as well as control over their release tend to be poor. Sugar- and polymer-based stabilisers and excipients have been successfully used in the past to protect the encapsulated proteins and maintain sustained release.53 In this work, six different excipients were explored including trehalose, dextrose, sucrose, PVA, chitosan and PVP. Our data demonstrated that sucrose had the highest bioactivity retention (96.3%) and were more effective in protecting the susceptible protein loaded onto the TiNTs, compared to other sugar- and polymer-based excipients. We have also shown a prolonged release of BMP-2 when co-administered with sucrose from the TiNT structures for 504 h compared to BMP-2 release only for 216 h. This suggests that such long nanotubular structures provide more protein storage space, and secondly the cylindrical shape of nanotubes strengthens their resistance to drug release to maintain a sustained release profile, which is also in agreement with other studies.54
As mentioned earlier, the bone remodeling process is a highly complicated, multidisciplinary environment. The effect of a combination of a positive osteoimmune environment and bone morphogenetic protein in switching the osteoblast cells towards mineralisaton was shown in our studies. Collagen 1A (COL1A) is an early osteoblast differentiation marker that is associated with early osteoid formation,55 OPN is a middle-stage osteogenic differentiation marker that participates in late ECM mineralisation,56 and OCN is a marker of osteoblast differentiation during the late-stage and signifies the beginning of mineralisation.57,58 Consistent with past reports,54,59 our in vitro results exhibit higher expression of three osteoblast differentiation marker genes (COL1A, OPN and OCN), which also suggests that the combination of an osteoimmune environment (macrophage medium from 134 nm TiNTs) and BMP-2 has the ability to shift the osteoblast phenotype towards mineralisation. These results are suggestive of the synergistic effects of nanotopography and BMP-2 on osteogenesis-related gene expression and the bone healing phase. Interestingly, we have also observed an inhibitory effect of the unfavourable osteoimmune environment (LPS-induced macrophages) on osteoblast differentiation, as evidenced by lower levels of osteogenic gene markers and mineralisation, indicating the synergistic role of pro-inflammatory cytokines in osteogenesis.51,60–62
In conclusion, we have successfully fabricated TiNTs on a 3D printed titanium surface, which displayed a drug protective effect and a sustained drug release profile, respectively. The modified TiNT surface facilitated macrophage cell adhesion, proliferation and differentiation, in vitro, with continuously enhanced gene expressions related to a pro healing phenotype. Furthermore, in vitro osteoblast experiment has shown the osteogenic ability guided by a favourable osteoimmune environment created by macrophage TiNT surfaces. Our study suggests that TiNT surfaces on 3D printed titanium nanotubes possess the ability to inhibit macrophage polarisation and enhance implant osseointegration, which provides a promising strategy for designing future implants.
The knowledge gained from this study will contribute towards better understanding the interaction between implants and immune cells, thus facilitating rationally engineering the nanotopographical surface of 3D printed titanium accordingly. The current study has demonstrated the effect of cytokine cocktails, secreted by macrophage cells upon stimulation by TiNTs with various diameters, on the osteogenic differentiation of osteoblast cells. In future studies, thorough investigation into attachment, proliferation, and differentiation of osteoblast cells directly seeded on TiNT substrates would be useful as shown in other studies.19,63,64 Finally, new methods of measuring surface roughness of 3D printed Ti will allow an accurate correlation of surface roughness with the structural features of TiNTs, which is believed to be key driver of immune modulation on TiNTs as demonstrated in past works on TiNTs grown on flat titanium implant substrates.47
Author contributions
Masood Ali wrote the original manuscript draft. Masood Ali, Anna Sze Ni Chang, Jingyu Liu, and Alice Wu, conducted the experiments, analysed data, developed methodology, and carried out manuscript drafting and editing. Amirali Popat, Yousuf Mohammad, and Laurie Walsh provided support with manuscript editing and resources. Yan (Helen) He, Chun Xu, Tushar Kumeria and Qingsong (Adam) Ye conceptualised the idea, supervised and oversaw the experiments, developed methodology, analysed data, and carried out drafting, editing of the manuscript, submission, and management of the article.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This study was performed under a grant provided by the Australian Dental Research Foundation (ADRF) and the University of Queensland Health and Behavioural Science Faculty Seed Grant. The 3D TiNTs were prepared at the School of Pharmacy UQ. All in vitro experiments were performed at the research labs at the School of Dentistry UQ.References
- T.-K. Ahn, D. H. Lee, T.-S. Kim, G. C. Jang, S. Choi, J. B. Oh, G. Ye and S. Lee, in Novel Biomaterials for Regenerative Medicine, ed. H. J. Chun, K. Park, C.-H. Kim and G. Khang, Springer Singapore, Singapore, 2018, pp. 355–368 DOI:10.1007/978-981-13-0947-2_19.
- K. Gulati, M. Prideaux, M. Kogawa, L. Lima-Marques, G. J. Atkins, D. M. Findlay and D. Losic, J. Tissue Eng. Regener. Med., 2017, 11, 3313–3325 CrossRef CAS PubMed.
- Z. Chen, T. Klein, R. Z. Murray, R. Crawford, J. Chang, C. Wu and Y. Xiao, Materials Today, 2016, 19, 304–321 CrossRef CAS.
- M. Nair and E. Elizabeth, J. Nanosci. Nanotechnol., 2015, 15, 939–955 CrossRef CAS PubMed.
- D. Schwartz-Arad, N. Kidron and E. Dolev, J. Periodontol., 2005, 76, 1431–1435 CrossRef PubMed.
- A. Bachhuka, B. Delalat, S. R. Ghaemi, S. Gronthos, N. H. Voelcker and K. Vasilev, Nanoscale, 2017, 9, 14248–14258 RSC.
- A. Bachhuka, J. D. Hayball, L. E. Smith and K. Vasilev, ACS Appl. Mater. Interfaces, 2017, 9, 5874–5884 CrossRef CAS.
- W. Q. Yu, X. Q. Jiang, F. Q. Zhang and L. Xu, J. Biomed. Mater. Res., Part A, 2010, 94, 1012–1022 CrossRef.
- K. S. Brammer, S. Oh, C. J. Cobb, L. M. Bjursten, H. V. D. Heyde and S. Jin, Acta Biomater., 2009, 5, 3215–3223 CrossRef CAS PubMed.
- J. Park, S. Bauer, K. von der Mark and P. Schmuki, Nano Lett., 2007, 7, 1686–1691 CrossRef CAS PubMed.
- S. Oh, K. S. Brammer, Y. S. Li, D. Teng, A. J. Engler, S. Chien and S. Jin, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 2130–2135 CrossRef CAS PubMed.
- L. M. Chamberlain, K. S. Brammer, G. W. Johnston, S. Chien and S. Jin, J. Biomater. Nanobiotechnol., 2011, 2(3), 8 Search PubMed.
- A. Rifai, N. Tran, V. Leitch, M. A. Booth, R. Williams and K. Fox, ACS Appl. Bio Mater., 2021, 4, 7509–7516 CrossRef CAS PubMed.
- M. Harrison, L. Pope, P. Sherrell, A. Stacey, P. Tran and K. Fox, Mater. Sci. Eng., C, 2021, 130, 1–9 Search PubMed.
- T. Kumeria, H. Mon, M. S. Aw, K. Gulati, A. Santos, H. J. Griesser and D. Losic, Colloids Surf., B, 2015, 130, 255–263 CrossRef CAS PubMed.
- K. Gulati, S. Ramakrishnan, M. S. Aw, G. J. Atkins, D. M. Findlay and D. Losic, Acta Biomater., 2012, 8, 449–456 CrossRef CAS.
- Y. Li, S. Wang, Y. Dong, P. Mu, Y. Yang, X. Liu, C. Lin and Q. Huang, Bioact. Mater., 2020, 5, 1062–1070 Search PubMed.
- Y. Zhang, X. Wang, Y. Li, J. Liang, P. Jiang, Q. Huang, Y. Yang, H. Duan, X. Dong, G. Rui and C. Lin, Regener. Biomater., 2022, 9, rbac046 CrossRef CAS.
- Z. Liu, Y. Liu, S. Liu, D. Wang, J. Jin, L. Sun, Q. Wang and Z. Yi, Mater. Des., 2021, 207, 109831 CrossRef CAS.
- T. Guo, S. Ivanovski and K. Gulati, Mater. Des., 2022, 223, 111110 CrossRef CAS.
- A. Jayasree, N. T. Raveendran, T. Guo, S. Ivanovski and K. Gulati, Mater. Today Adv., 2022, 15, 100256 CrossRef CAS.
- J. F. Destino, N. A. Dudukovic, M. A. Johnson, D. T. Nguyen, T. D. Yee, G. C. Egan, A. M. Sawvel, W. A. Steele, T. F. Baumann, E. B. Duoss, T. Suratwala and R. Dylla-Spears, Adv. Mater. Technol., 2018, 3, 1700323 CrossRef.
- H.-T. Chunate, J. Khamwannah, A. A. A. Aliyu, S. Tantavisut, C. Puncreobutr, A. Khamkongkaeo, C. Tongyam, K. Tumkhanon, T. Phetrattanarangsi, T. Chanamuangkon, T. Sitthiwanit, D. Decha-umphai, P. Pongjirawish and B. Lohwongwatana, Materials, 2021, 14, 6576 CrossRef CAS.
- J.-H. Kim, M.-Y. Kim, J. C. Knowles, S. Choi, H. Kang, S.-H. Park, S.-M. Park, H.-W. Kim, J.-T. Park, J.-H. Lee and H.-H. Lee, Dent. Mater., 2020, 36, 945–958 CrossRef CAS PubMed.
- C. Xu, L. Xiao, Y. Cao, Y. He, C. Lei, Y. Xiao, W. Sun, S. Ahadian, X. Zhou, A. Khademhosseini and Q. Ye, Nano Res., 2020, 13, 2323–2331 CrossRef CAS.
- C. Xu, J. Xu, L. Xiao, Z. Li, Y. Xiao, M. Dargusch, C. Lei, Y. He and Q. Ye, RSC Adv., 2018, 8, 16503–16512 RSC.
- Y. Gu, L. Wei, Z. Zhang, J. Van Dessel, R. B. Driesen, I. Lambrichts, R. Jacobs, L. Tian, Y. Sun, Y. Liu and C. Politis, Mater. Des., 2022, 215, 110443 CrossRef CAS.
- Y. Hu, K. Cai, Z. Luo, D. Xu, D. Xie, Y. Huang, W. Yang and P. Liu, Acta Biomater., 2012, 8, 439–448 CrossRef CAS PubMed.
- S. J. Sun, W. Q. Yu, Y. L. Zhang, X. Q. Jiang and F. Q. Zhang, Cell Proliferation, 2013, 46, 685–694 CrossRef CAS.
- Y. Cao, L. Xiao, Y. Cao, A. Nanda, C. Xu and Q. Ye, Biochem. Biophys. Res. Commun., 2019, 512, 889–895 CrossRef CAS.
- C. Xu, Y. Cao, C. Lei, Z. Li, T. Kumeria, A. K. Meka, J. Xu, J. Liu, C. Yan, L. Luo, A. Khademhosseini, A. Popat, Y. He and Q. Ye, ACS Appl. Nano Mater., 2020, 3, 1457–1467 CrossRef CAS.
- Z. Chen, A. Bachhuka, S. Han, F. Wei, S. Lu, R. M. Visalakshan, K. Vasilev and Y. Xiao, ACS Nano, 2017, 11, 4494–4506 CrossRef CAS.
- D. L. Vaux, F. Fidler and G. Cumming, EMBO Rep., 2012, 13, 291–296 CrossRef CAS PubMed.
- J. Qin, D. Yang, S. Maher, L. Lima-Marques, Y. Zhou, Y. Chen, G. J. Atkins and D. Losic, J. Mater. Chem. B, 2018, 6, 3136–3144 RSC.
- W. Feng, Z. Geng, Z. Li, Z. Cui, S. Zhu, Y. Liang, Y. Liu, R. Wang and X. Yang, Mater. Sci. Eng., C, 2016, 62, 105–112 CrossRef CAS.
- S. Maher, G. Kaur, L. Lima-Marques, A. Evdokiou and D. Losic, ACS Appl. Mater. Interfaces, 2017, 9, 29562–29570 CrossRef CAS PubMed.
- N. Thamwattana, P. Sarapat and Y. Chan, J. Phys.: Condens. Matter, 2019, 31, 265901 CrossRef CAS PubMed.
- S. Nema and K. E. Avis, J. Parenter. Sci. Technol., 1993, 47, 76–83 CAS.
- W. Wang, Int. J. Pharm., 2000, 203, 1–60 CrossRef CAS.
- J. Zhao, S. Wang, J. Bao, X. Sun, X. Zhang, X. Zhang, D. Ye, J. Wei, C. Liu, X. Jiang, G. Shen and Z. Zhang, PLoS One, 2013, 8, e54645 CrossRef CAS PubMed.
- Y. Li, Y. Song, A. Ma and C. Li, BioMed Res. Int., 2019, 2019, 5697250 Search PubMed.
- N. K. Awad, S. L. Edwards and Y. S. Morsi, Mater. Sci. Eng., C, 2017, 76, 1401–1412 CrossRef CAS PubMed.
- M. J. Fenton and D. T. Golenbock, J. Leukocyte Biol., 1998, 64, 25–32 CrossRef CAS PubMed.
- L. M. Chamberlain, K. S. Brammer, G. W. Johnston, S. Chien and S. Jin, J. Biomater. Nanobiotechnol., 2011, 2(3), 8 Search PubMed.
- P. Neacsu, A. Mazare, A. Cimpean, J. Park, M. Costache, P. Schmuki and I. Demetrescu, Int. J. Biochem. Cell Biol., 2014, 55, 187–195 CrossRef CAS.
- K. M. Ainslie, S. L. Tao, K. C. Popat, H. Daniels, V. Hardev, C. A. Grimes and T. A. Desai, J. Biomed. Mater. Res., Part A, 2009, 91, 647–655 CrossRef.
- P. R. L. Dabare, A. Bachhuka, J. Y. Quek, L. F. Marsal, J. Hayball and K. Vasilev, Small Sci., 2023, 2300080 CrossRef.
- M. Nouri-Goushki, A. Isaakidou, B. I. M. Eijkel, M. Minneboo, Q. Liu, P. E. Boukany, M. J. Mirzaali, L. E. Fratila-Apachitei and A. A. Zadpoor, Nanoscale, 2021, 13, 14304–14315 RSC.
- M. Shayan, J. Padmanabhan, A. H. Morris, B. Cheung, R. Smith, J. Schroers and T. R. Kyriakides, Acta Biomater., 2018, 75, 427–438 CrossRef CAS.
- T. Tylek, C. Blum, A. Hrynevich, K. Schlegelmilch, T. Schilling, P. D. Dalton and J. Groll, Biofabrication, 2020, 12, 025007 CrossRef CAS.
- R.-L. Huang, Y. Sun, C.-K. Ho, K. Liu, Q.-Q. Tang, Y. Xie and Q. Li, Cell Death Dis., 2018, 9, 144 CrossRef.
- F. Wei, Y. Zhou, J. Wang, C. Liu and Y. Xiao, Tissue Eng., Part A, 2018, 24, 584–594 CrossRef CAS.
- I. El Bialy, W. Jiskoot and M. Reza Nejadnik, Pharm. Res., 2017, 34, 1152–1170 CrossRef CAS.
- Y. Li, Y. Song, A. Ma and C. Li, BioMed Res. Int., 2019, 2019, 5697250 Search PubMed.
- M. Khan, N. Donos, V. Salih and P. Brett, Bone, 2012, 50, 1–8 CrossRef CAS.
- J. Zhou, B. Li, S. Lu, L. Zhang and Y. Han, ACS Appl. Mater. Interfaces, 2013, 5, 5358–5365 CrossRef CAS.
- V. Vijayan, S. Gupta and S. Gupta, BioFactors, 2017, 43, 558–566 CrossRef CAS PubMed.
- X. Zhao, T. Wang, S. Qian, X. Liu, J. Sun and B. Li, Int. J. Mol. Sci., 2016, 17, 292 CrossRef.
- C. Bayram, M. Demirbilek, N. Çalışkan, M. E. Demirbilek and E. B. Denkbcş, J. Biomed. Nanotechnol., 2012, 8, 482–490 CrossRef CAS.
- X. Li, Z.-Y. Zhou, Y.-Y. Zhang and H.-L. Yang, PLoS One, 2016, 11, e0154677 CrossRef.
- C. B. Sullivan, R. M. Porter, C. H. Evans, T. Ritter, G. Shaw, F. Barry and J. M. Murphy, Stem Cell Res. Ther., 2014, 5, 104 CrossRef.
- D. C. Lacey, P. J. Simmons, S. E. Graves and J. A. Hamilton, Osteoarthritis Cartilage, 2009, 17, 735–742 CrossRef CAS.
- D. W. Zhao, B. Ren, H. W. Wang, X. Zhang, M. Z. Yu, L. Cheng, Y. H. Sang, S. S. Cao, F. M. Thieringer, D. Zhang, Y. Wan and C. Liu, Bone Jt. Res., 2021, 10, 411–424 CrossRef.
- Y. Fu, Z. Jing, T. Chen, X. Xu, X. Wang, M. Ren, Y. Wu, T. Wu, Y. Li, H. Zhang, P. Ji and S. Yang, J. Nanobiotechnol., 2023, 21, 229 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb01029e |
| This journal is © The Royal Society of Chemistry 2024 |