Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Unlocking the potential of 2D nanomaterial-based biosensors in biomarker-based detection of Helicobacter pylori
Desmond
Lutomia
a,
Rachna
Poria
a,
Deepak
Kala
b,
Anupam Kumar
Singh
c,
Manoj K
Gupta
d,
Deepak
Kumar
 *e,
Ankur
Kaushal
*a and
Shagun
Gupta
*a
*e,
Ankur
Kaushal
*a and
Shagun
Gupta
*a
aDepartment of Bio-sciences and Technology, MM Engineering College, Maharishi Markandeshwar (Deemed to Be) University, Mullana, Ambala 134003, India. E-mail: shagun.gupta@mmumullana.org; ankur.kaushal@mmumullana.org
bNL-11 Centera Tetrahertz Laboratory, Institute of High Pressure Physics, Polish Academy of Sciences, 29/37 Sokolowska Street, 01142 Warsaw, Poland
cDepartment of Gasteroenterology, Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh, India
dDepartment of Chemistry, School of Basic Sciences, Central University of Haryana, Mahendergarh-123031, Haryana, India
eDepartment of Pharmaceutical Chemistry, School of Pharmaceutical Sciences, Shoolini University, Solan, Himachal Pradesh 173229, India. E-mail: guptadeepak002@gmail.com
First published on 18th November 2024
Abstract
Recent advancements in nanotechnology and biomedicine have promoted the utilization of nanomaterials for various medical applications, particularly in the detection of Helicobacter pylori infections. The colonization of the gastric mucosa by H. pylori significantly establishes the risk factors for the development of chronic gastritis, peptic ulcers, and gastric cancers. While conventional methods, both invasive and non-invasive, are available for the detection of H. pylori, they often face limitations in terms of sensitivity, specificity, cost-effectiveness, and point-of-care applications. Two-dimensional nanomaterials exhibit considerable potential in the development of robust analytical platforms tailored for point-of-care (POC) detection of H. pylori, thereby presenting streamlined and economically viable biosensing solutions for the purpose of detecting H. pylori infections. This review summarizes the primary biomarkers utilized for the detection of H. pylori infections, elucidating how 2D nanomaterials enhance biosensor efficacy and the diverse applications of biosensors coupled with 2D nanomaterials in detecting H. pylori infection. Additionally, it examines different types of biosensing platforms that harness the unique properties of 2D nanomaterials and design considerations for optimizing biosensor performance for the accurate and reliable identification of H. pylori infections. The last part explores the industrialization potential and commercial viability and challenges inherent in utilizing 2D nanomaterials in biosensing and outlines future research and development prospects in 2D nanomaterial-based biosensors for disease detection.
1. Introduction
H. pylori is a Gram-negative bacterium that colonizes the stomach in humans. This bacterium has adapted to survive in high acidic conditions of the stomach, where it colonizes the gastric mucosa, particularly the antrum and corpus regions and exists for most of its lifetime.1 Morphologically, H. pylori has a curved or spiral rod shape of 2.5–5.5 μm in length and 0.5–1.5 μm in width.2,3 It has 2–6 unipolar, sheathed lophotrichous flagella that enable mobility and chemotaxis within the viscous mucosal layer of the stomach.4 These flagella also aid adherence to gastric epithelial cells. H. pylori is microaerophilic, requiring 3–5% oxygen, 5–10% CO2, 0–10% H2, and high humidity to grow optimally.5 The bacterium possesses a unique urease enzyme that allows it to hydrolyze urea, producing ammonia and bicarbonate, which neutralize the acidic environment of the stomach and create a microenvironment favorable for bacterial survival. It also produces catalase and oxidase enzymes, which are used as biochemical factors for identification. The bacterial cell envelope consists of a cytoplasmic membrane, cell wall, and outer membrane containing lipopolysaccharides and membrane proteins. The helical cell shape, flagella, and outer membrane components are key virulence factors that promote colonization persistence in the hostile acidic stomach.This bacterium colonizes the stomach of approximately 50% of the population globally, making it one of the most ubiquitous infections worldwide.5 However, prevalence rates vary geographically with higher rates noted in developing countries compared to industrialized nations. In Africa, H. pylori infects 60–90% of adults whereas prevalence is around 20–50% in Western Europe and North America. In Asia, prevalence ranges from 22% in Japan to 87% in Bangladesh. India has an estimated H. pylori prevalence of 50–80% in adults.6,7 Low socioeconomic status and crowded living conditions correlate with higher infection risk. Primary transmission occurs through the fecal-oral and gastric-oral routes through contaminated food/water sources or close person-to-person contact. Infection is usually acquired in early childhood and without treatment, colonization persists lifelong. Given its high global prevalence, H. pylori infection is believed to play a causal role in 75–80% of gastric cancer cases worldwide.8,9 Epidemiologic patterns correspond with gastric cancer incidence in both developing and developed regions. Eradication of H. pylori can lead to regression of gastric precancerous lesions and reduce cancer risk. Hence many health organizations recommend routine testing and treatment to prevent long-term complications, especially in high-risk populations. However, reinfection after antibiotic therapy remains a challenge especially in developing countries due to continued exposure.
H. pylori colonization triggers chronic active gastritis in all infected persons but only 10–20% develop more severe complications like ulcers and cancer.8–10 Colonization initiates persistent inflammation through bacterial virulence factors and dysregulated host immune responses. Gastric epithelial cell injury arises due to direct cytotoxicity mediated by factors like the vacuolating cytotoxin A (VacA), the oncoprotein cytotoxin-associated gene A (CagA), and urease-driven pH alterations.8,11 Robust pro-inflammatory Th1-cell mediated responses further damage the mucosa.11 Concurrently, H. pylori evades immune clearance through myriad mechanisms thereby establishing persistent infection. The pattern and severity of gastritis and epithelial damage determines specific disease outcomes. Antral-predominant colonization increases acid secretion and causes duodenal ulcers. Body-predominant gastritis reduces acid levels leading to atrophic gastritis, a precursor of gastric cancer. Surface mucosal damage allows back-diffusion of acid that worsens inflammation.11 Ulcers arise when this exceeds the tissue's reparative capacity. Atrophic gastritis accompanied by mutagenic properties of H. pylori promotes oncogenic mutations in gastric stem cells over decades.12,13 The stepwise progression from chronic gastritis through precancerous lesions culminates eventually in gastric adenocarcinoma. Gastric mucosa-associated lymphoid tissue (MALT) lymphoma is another serious complication attributed to chronic H. pylori-triggered immunostimulation.14 Despite its prevalence, not all individuals infected with H. pylori develop symptomatic disease, with factors such as bacterial virulence factors, host genetics, and environmental factors playing a role in disease manifestation.
Early diagnosis and eradication of H. pylori infection is crucial to prevent progression to severe gastroduodenal complications, especially in high-risk populations allowing for timely intervention and treatment. Furthermore, early detection can help reduce the risk of transmission to others and minimize the burden of disease in affected individuals and healthcare systems. However, detection of H. pylori can be challenging as up to 50–80% of infected individuals are asymptomatic.7,15 For the detection of H. pylori, two approaches – invasive and non-invasive techniques – are utilized. Fig. 1 illustrates invasive, noninvasive, and biosensor techniques used for the diagnosis and detection of H. pylori infection. Conventional invasive tests to detect active infection require endoscopy to obtain gastric mucosal biopsies followed by culture, histology, rapid urease testing, or PCR.15 Endoscopy allows direct visualization of the stomach lining but is uncomfortable for patients, has procedure-related risks, and is expensive. Non-invasive indirect tests like the urea breath test (UBT), stool antigen test, and serology are easier and cheaper but have reduced accuracy. 13C-UBT and 14C-UBT are widely used for initial diagnosis and post-treatment confirmation; patients ingest 13C- or 14C-labeled urea which gets hydrolyzed by H. pylori's urease, releasing labeled CO2 detected in breath samples.16 Stool antigen ELISA detects H. pylori proteins in fecal samples.16 Both methods have high sensitivity and specificity but cannot gauge severity or locate lesions. Serology detects serum antibodies to H. pylori but remains positive even after clearance, limiting use for test-of-cure. Sensitivity also varies based on local infection prevalence affecting the positive predictive value. The constraints of these conventional tests include the requirement for reliably cultured bacteria, lack of quantitation, and inability to evaluate pathological changes or antibiotic susceptibility. Rapid molecular techniques like fluorescence in situ hybridization (FISH) overcome some limitations due to direct detection of H. pylori DNA/RNA in tissues or body fluids.17 But invasive sampling restricts routine applicability. Non-invasive and highly sensitive urine, saliva or blood PCR-based assays are being developed but remain experimental. Thus, while a range of methods are available for detecting active H. pylori infection, no single test meets all clinical needs regarding speed, cost, accuracy, and ability to monitor pathogenesis or response to therapy. More convenient and quantitative point-of-care tests are needed, especially in resource-limited settings with high disease burden.
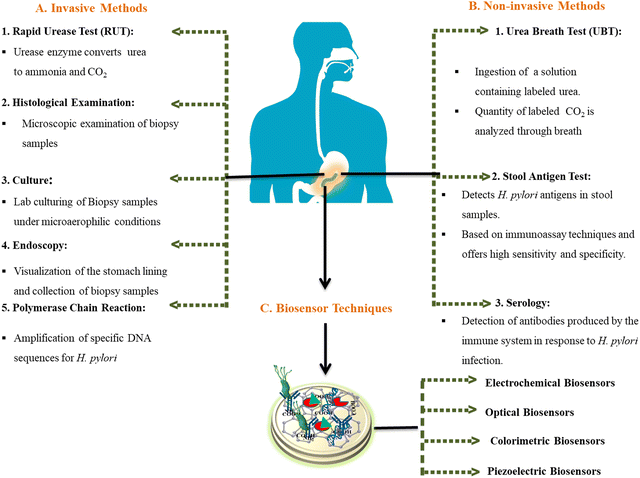 | ||
| Fig. 1 Invasive, noninvasive, and biosensor techniques used for the diagnosis and detection of H. pylori infection. | ||
The limitations of conventional techniques have driven research into nanomaterial-based biosensors for improved H. pylori detection (Table 1). Biosensors offer a non-invasive, rapid, and cost-effective alternative for H. pylori detection, enabling point-of-care testing and improving access to timely diagnosis for patients.
| Current methods | Limitation | Sensitivity (%) | Specificity (%) | Time | Ref. |
|---|---|---|---|---|---|
| Endoscopy | Uncomfortable, highly expensive, risk of infection, risk of bleeding | 95 | 99 | 7 days | 18–20 |
| Histology | Sample error (sampling can lead to false negatives) | 95 | 99 | 7–10 days | 21 |
| Urea breath test | Recent use of PPIs or antibiotics reduces the bacterial load | 90 | 95 | 1 h | 16, 22 and 23 |
| Culture | More time consuming | 58 | 99 | 7 to 10 days | 20, 24 and 25 |
| Urease test | Recent use of PPIs or antibiotics reduces the bacterial load | 95.2 | 95.1 | 1 day | 26–28 |
| Stool antigen test | If the sample is not stored properly, the antigen degrades over time | 95 | 96 | 2–4 days | 29–31 |
| Serology | Cannot differentiate between past and recent infection | 75–85 | 79–90 | More than 3 h | 20, 29 and 32 |
These biosensors incorporate a biological sensing element with a transducer to yield a detectable signal proportional to the target pathogen. 2D nanomaterials exhibit diverse properties in terms of their mechanical, chemical, and optical attributes, apart from uniqueness in their size, aspect ratio, shape, biocompatibility, and biodegradability. These varied properties afford a suitable platform for these nanomaterials for myriad applications in the healthcare sector, including drug delivery, bioimaging, tissue engineering, biosensors, and so on. However, their low dimensionality and exceptionally large aspect ratios (surface area-to-volume ratio) make these nanomaterials invaluable for uses demanding high levels of surface interactions in the nanoscale regime.33,34 Specifically, 2D nanoparticles like graphene oxide (GO) allow for efficient immobilization of biomolecules, such as antibodies or nucleic acid-based probes, to capture H. pylori cells or antigens through strong π–π stacking interactions, enhancing the specificity of detection.35 This review summarizes the overview of H. pylori infection and associated diseases, possible biomarkers of H. pylori, and how integration of 2D nanoparticles in biosensors enhances the detection of H. pylori infection. Challenges associated with biosensing technology for diagnostic purposes, recent advancements and future perspectives in the detection of H. pylori infection are covered in the last section.
2. Biomarkers associated with H. pylori infection
H. pylori utilizes a variety of virulence factors and mechanisms to induce pathogenesis while evading host immunity.4 These include both bacterial components and products that directly damage gastric tissues as well as factors that help in its persistence and proliferation.5,6 Some key general virulence attributes are the spiral shape, flagella, and urease production.4,10,11,36 The helical cell morphology along with flagella enables motility and chemotaxis allowing it to penetrate and survive within the viscous mucosal layer.8 The outer membrane lipopolysaccharides (LPS) and other surface molecules help evade phagocytosis and complement-mediated killing.8,11 The urease enzyme is a vital virulence factor for colonization as it catalyzes urea breakdown into ammonia and CO2, neutralizing gastric acid and forming a microenvironment conducive for bacterial growth. Urease is encoded by the ureA, ureB, and ureC genes and expression of the accessory genes ureI, ureE, ureF, ureG, and ureH results in the catalytically active urease enzyme.37,38 Due to the high specificity and sensitivity of the urease enzyme, it has been shown to be a significant biomarker for H. pylori infection detection.36 Studies have shown that urease activity correlates with the severity of gastritis and peptic ulcer disease (PUD), making it a valuable biomarker for disease monitoring and treatment response assessment.39H. pylori secretes two major cytotoxins (VacA and CagA) which are the major virulence factors as well as biomarkers associated specifically with H. pylori infection. The toxin vacuolating cytotoxin A (VacA) induces cellular vacuolization and injury.39 It also increases permeability of epithelial layers enabling H. pylori access to underlying tissues. Studies have shown that the VacA S1 genotype is highly associated with increased levels of interleukin-8, chronic inflammation in gastric mucosa, epithelial damage and metaplasia as compared to the VacA S2 genotype.40,41 The cytotoxin-associated gene A (CagA) protein is among the most extensively studied and occurs in 60–70% of isolates globally. It is located in the cag pathogenicity island (cagPAI), a 40 kb DNA segment encoding a type IV secretion system that forms needle-like pili.42,43 The CagA effector protein is injected via this secretion system directly into host cells where it dysregulates signaling through intramolecular interaction and causes cellular defects.42CagA by interacting with multiple host signaling molecules such as the pro-oncogenic phosphatase (SHP2) and the polarity-regulating kinase (PAR1) induces pro-inflammatory responses by activating NF-κB in a phosphorylation-dependent manner.42Fig. 2 describes the major pathogenic markers of H. pylori and the delivery of the CagA gene into the epithelial cell through the T4SS and the intramolecular interaction disrupting cellular signaling.41,42 Recent research has focused on elucidating the mechanisms underlying CagA-mediated pathogenesis and its role as a biomarker for disease risk stratification and prognosis assessment.42,44 These studies show that CagA- and cagPAI-positive strains are associated with heightened inflammation and increased risk for severe gastritis, peptic ulcers and cancer compared to cagPAI-negative isolates. A meta-analysis has associated CagA-positive strains with a 2-fold increased risk for peptic ulcer disease and 5-fold higher risk for gastric cancer compared to cagPAI-negative isolates.44CagA also causes cellular changes that may promote carcinogenesis including disruption of cell-to-cell adhesion, elongation of epithelial cells, and abnormal proliferation. Another cagPAI effector called CagL binds to and activates β1 integrin receptors on gastric cells triggering pro-inflammatory signaling.45 The cagPAI genes also induce the production of cytokines like interleukin-8 by gastric epithelial cells which drive neutrophil infiltration and chronic inflammation.42,44CagA is the first bacterial oncoprotein to be identified and remains a major virulence factor driving H. pylori-mediated injury and carcinogenesis through its multifaceted effects on gastric epithelial cells.11 Furthermore, studies have explored the use of CagA-specific antibodies and DNA probes in biosensor-based diagnostic assays for the detection of H. pylori infection, highlighting the potential clinical utility of CagA as a diagnostic biomarker.46
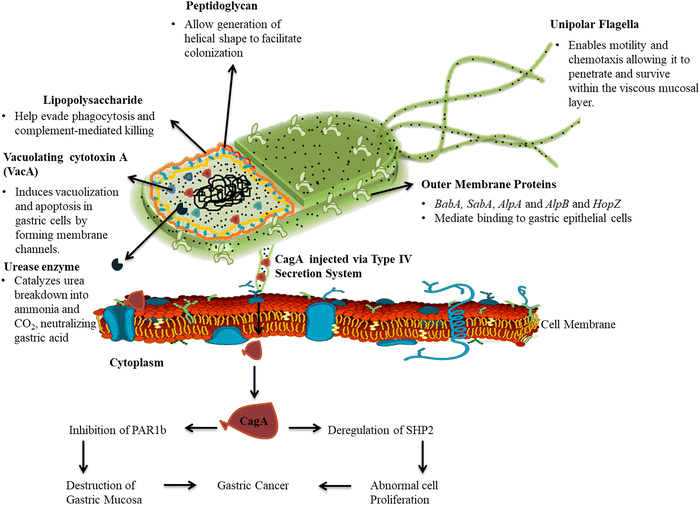 | ||
| Fig. 2 The major pathogenic markers of H. pylori and the delivery of the CagA gene into the epithelial cell through the T4SS and the intramolecular interaction disrupting cellular signaling. | ||
Other ubiquitous virulence determinants that may serve as biomarkers for pathogenic potential are adhesins like BabA, SabA, pylori AlpA, AlpB and HopZ that mediate binding to gastric epithelial cells, an essential initial step in colonization. The blood group antigen binding adhesin (BabA) is encoded by the BabA2 gene, and shows increased association with duodenal ulcer (71–83%) and gastric cancer (83–85%) compared to gastritis alone (22–59%).11,15,39,47 The outer inflammatory protein A (OipA) induces interleukin-8 secretion and neutrophil infiltration.47OipA is linked to higher inflammation severity. The dupA gene stimulates IL-12 and is variably associated with duodenal ulcers and reduced risk of gastric cancer.47 Another biomarker gene is the H. pylori cholesterol-α-glucosyltransferase (CGT) gene which is involved in the synthesis of lipopolysaccharides of the cell wall. The CGT catalyzes the conversion of membrane cholesterol to cholesteryl glucosidases which are incorporated in the cell wall enabling the bacteria to evade phagocytosis by the immune cells.47 The catalase enzyme is also secreted and plays a role in protecting the bacteria against oxidative damage by breaking down harmful reactive oxygen species. Superoxide dismutase and alkyl hydroperoxide reductase further enhance resistance to oxygen radicals.47 The arginase enzyme is also secreted and allows H. pylori to compete with host cells for L-arginine, thereby impairing T-cell function.47 Other emerging biomarkers include iceA alleles, homologues of the helicobacter outer membrane PorD protein, and fur mutations which are postulated to enhance H. pylori virulence but require more confirmatory studies.48,49 Host genetic factors like IL-1β polymorphisms combined with specific bacterial alleles are also increasingly investigated as joint biomarkers.48 Many studies have provided insights into the molecular mechanisms underlying the actions of these virulence factors and their potential as diagnostic and therapeutic targets for H. pylori-associated diseases. Recent investigations have revealed that a strategic combination of biomarkers significantly enhances the diagnostic precision for H. pylori-associated gastric and duodenal cancers. By employing isobaric tag for relative and absolute quantitation, researchers have analyzed protein expression levels in patients infected with H. pylori. The optimized integration of virulence factors OipA, BabA, and SabA achieved a predictive accuracy of 77.3% in differentiating between gastric cancer and duodenal ulcer patients.50 This combination not only improves diagnostic precision but also suggests that a protein microarray incorporating these antigens could facilitate rapid and efficient diagnosis of H. pylori-related gastric cancer. Moreover, the incorporation of these biomarkers with advanced technologies such as 2D nanomaterials underscores their potential in developing rapid diagnostic sensors that enable early detection and timely intervention.
3. Integration of 2D nanoparticles in biosensors for detection of H. pylori
3.1. Properties and synthesis of 2D nanoparticles
Nanomaterials such as graphene, graphene oxide, hexagonal boron nitride, transition metal dichalcogenides (TMDs), MXenes, layered double hydroxides, transition metal oxides, and black phosphorus have emerged as promising 2D nanomaterials for biosensing platforms. This is attributed to their exotic structural, electrical, optical, and catalytic properties originating from the quantum confinement in 2D layers with atomic-scale thickness.33,34,51,52 For instance, graphene exhibits exceptionally high electron mobility, mechanical strength, specific surface area and thermal conductivity. Its large surface area allows for efficient immobilization of biomolecules, such as antibodies or DNA probes, enhancing the sensitivity and specificity of biosensors. Moreover, the high conductivity of graphene enables rapid electron transfer, resulting in fast and accurate detection of analytes. Recent literature has demonstrated the integration of graphene-based biosensors for the detection of various biomarkers, including proteins, nucleic acids, and pathogens. For example, graphene-based biosensors have been utilized for the detection of cancer biomarkers, infectious diseases, and environmental pollutants, offering sensitive and selective detection platforms for diagnostic purposes.20 The smart health watch “GF1” is one such graphene-based commercially available product launched by Wuxi Graphene Film (a subordinate of The Sixth Elements Materials). Chemical vapor deposited graphene film acts as a touch screen conductive element in lieu of commonly used indium tin oxide glass. GF1 is a smart health monitoring watch that provides a detailed health index and scrutinizes the dynamic electrocardiogram, heart rate, temperature, and blood pressure, apart from tracking the calories burned during physical workouts.36 The integration of molybdenum disulfide (MoS2) in biosensors allows for enhanced sensitivity and selectivity, as well as improved stability and biocompatibility. Studies have highlighted the potential of MoS2-based biosensors for point-of-care diagnostics, environmental monitoring, and biomedical research. For instance, MoS2-based biosensors have been utilized to detect glucose, cholesterol, and other metabolites in clinical samples, offering rapid and accurate measurements for disease diagnosis and management.53 MXenes containing hydroxyl or fluoride terminations readily adsorb biomolecules electrostatically. Layered black phosphorus demonstrates superior anisotropic electrical properties, chemical inertness, thermal stability, and a wide bandgap optimal for field-effect biosensing. Such unique characteristics make 2D nanomaterials extremely attractive for fabricating electrochemical, optical, piezoelectric or field-effect biosensors with high sensitivity and selectivity.2D nanoparticles are generally synthesized through three broad approaches – top-down approaches, bottom-up synthesis, and hybrid methods.54,55 The top-down approach involves splitting bulk crystals into nanosheets using mechanical or liquid phase exfoliation. Mechanical exfoliation repeatedly peels off layers using adhesive tape but has very low yield. Liquid exfoliation uses ultrasonication or chemical intercalation in solvents like N-methyl-2-pyrrolidone (NMP) to produce dispersions of 2D flakes. This is simple and scalable but can damage sheets and provide limited control over thickness. The bottom-up synthesis builds 2D layers atom-by-atom via chemical vapor deposition, solvothermal synthesis, or self-assembly. While high-quality sheets are produced, these methods require high temperatures and complex setups.56–59 Hybrid techniques combine top-down and bottom-up methods by exfoliating bulk crystals and then using the nanosheets as 2D templates for further synthesis through chemical vapor deposition (CVD) or atomic layer deposition. Each approach yields nanomaterials suited for particular biosensing modalities. For instance, liquid exfoliated graphene oxide (GO) with its abundant surface oxygen groups has been widely used to develop electrochemical immunosensors and fluorescent DNA biosensors for diagnostic purposes.60 CVD-synthesized MoS2 nanosheets are integrated in field-effect transistors due to their semiconductor properties. Mechanically exfoliated boron nitride nanosheets are ideal for surface-enhanced Raman spectroscopy based detection. Thus, based on the desired transduction mechanism, suitable 2D nanomaterials can be synthesized using an optimal technique and integrated to develop highly efficient biosensors for clinical applications and diagnosis of H. pylori infection.
3.2. Surface functionalization of 2D nanoparticles for biosensing applications
Efficient functionalization and modification facilitate immobilization of biorecognition elements and improve interaction with target analytes. The utilization of 2D nanomaterials in biosensing applications transcends conventional detection methods, encompassing capabilities for real-time monitoring and point-of-care diagnostics. The high surface area and ease of functionalization of these nanomaterials significantly enhance their performance in dynamic biological environments, making them particularly suitable for continuous health monitoring applications.61 This adaptability allows for the development of biosensors that can operate effectively in complex biological matrices, thereby improving diagnostic accuracy and reliability. The clinical implications of employing functionalized 2D nanoparticles in biosensing are substantial, as they have the potential to revolutionize diagnostic practices. By facilitating early detection of diseases through enhanced sensitivity and specificity, these advanced biosensors can lead to improved patient outcomes.62,63 Moreover, the integration of 2D nanomaterials into biosensing platforms supports the implementation of personalized healthcare solutions, enabling clinicians to tailor treatments based on individual patient profiles.Two main common approaches for functionalizing 2D nanoparticles involve covalent and non-covalent strategies.64–66 Covalent approaches involve covalent attachment of biomolecules, such as antibodies, aptamers, or DNA probes, onto the nanoparticle surface forming stable bonds between surface groups like –COOH, –NH2, and –OH and biomolecules via crosslinkers, providing precise control over orientation and conformation. Non-covalent methods utilize electrostatic, π–π stacking or hydrophobic interactions for bioconjugation. These are simpler, preserve nanomaterial properties better but provide less control over binding. For example, graphene oxide contains abundant surface carboxyl and hydroxyl groups amenable to covalent tethering of enzymes, antibodies or aptamers using EDC/NHS chemistry.67 TMD nanosheets can be functionalized by silanization of surface –OH groups, allowing covalent protein attachment. MXenes with –OH, –O and –F terminations readily adsorb biomolecules non-covalently through electrostatic and hydrogen bonding. Proper bioconjugation and blockage of unused sites is vital for optimal biorecognition while minimizing non-specific binding. The literature has shown that suitable chemical or physical functionalization facilitates the immobilization of bioreceptors on 2D surfaces and enables specific, oriented binding of analytes in biosensing applications.68–72
In addition to biomolecule functionalization, surface engineering strategies have been employed to modify the properties of 2D nanoparticles for biosensing applications. One such strategy involves modifying the surfaces of 2D nanoparticles with metal nanoparticles, such as gold nanoparticles (AuNPs) or silver nanoparticles (AgNPs), to enhance their sensing capabilities. Metal-decorated 2D nanoparticles exhibit localized surface plasmon resonance (LSPR) effects, which can amplify the signal response upon target binding, leading to improved detection sensitivity.73 For instance, AuNP-modified graphene-based biosensors have been developed for the ultrasensitive detection of SARS-CoV-2 antigens in clinical samples.74 Another surface modification approach involves the introduction of defect sites or functional groups onto the nanoparticle surface to create specific binding sites for target molecules. Functionalized defect-engineered graphene has been used to selectively capture DNA sequences in biological samples, enabling the rapid and sensitive detection of the infectious pathogens in clinical samples.74 The graphene biosensor based on a field-effect transistor (FET) architecture, functionalized by phage tail spike proteins (TSPs), has been shown to accurately detect pathogens such as E. coli at the single bacterium level.75,76 Additionally, a DNA-functionalized graphene FET has been proposed for the quantitation of proteins, utilizing the ability of DNA hybridization to amplify the detection signal.77 Graphene nanosheets modified on a glassy carbon electrode have demonstrated enhanced electron transfer ability and interface adsorption capacity, leading to improved sensitivity in the detection of rutin, nitrofurazone and linagliptin.78,79 Furthermore, an armchair graphene nanoribbon (AGNR) interconnected between gold electrodes has been investigated as a detector of DNA hybridization, showing high sensitivity and accuracy in detecting changes in electrical properties upon functionalization with probe and target DNA.80,81 These surface engineering strategies offer versatile means of tailoring the properties of 2D nanoparticles for biosensing applications, allowing for the development of highly sensitive and selective biosensors for analyte detection.
Another approach involves the incorporation of 2D nanoparticles in microfluidic systems, where target molecules are selectively captured and concentrated within microfluidic channels, enabling sensitive and rapid detection. Microfluidic biosensors based on MoS2 have been developed for the immune detection of Toxoplasma gondii in serum, offering a portable and point-of-care diagnostic platform.82 The biosensor showed a detection range of 1 pg mL−1 to 10 ng mL−1 for T. gondii monoclonal antibody solutions with a sensitivity of 3.358 nm/log (mg mL−1). The biosensor demonstrated excellent specificity and clinical characteristics when tested with rabies virus, pseudorabies virus, and T. gondii serum, indicating its potential for biomedical applications.82 Integration of biosensing platforms leverages the unique properties of 2D nanoparticles to achieve sensitive, selective, and rapid detection of infectious pathogens such as H. pylori biomarkers, paving the way for the development of next-generation POC diagnostic devices for the early diagnosis and monitoring of H. pylori-related diseases.
3.3. Biosensing platforms for H. pylori detection
Biosensors are qualitative and semiquantitative analytical devices that translate a biological recognition reaction into a measurable signal. They integrate biological components like enzymes, antibodies, nucleic acid, and cells with a physicochemical transducer.83 Upon interaction with the analyte, the biorecognition element undergoes a specific biochemical reaction, leading to a change in the physicochemical properties of the transducer, which is then converted into an electrical, optical, or mechanical signal. This signal is subsequently amplified, processed, and displayed by a readout system, allowing for the quantitative analysis of the target analyte. These features have revolutionized biosensors as the point-of-care detection technique for various infections by offering rapid, sensitive, and specific diagnostic capabilities. The development of advanced biosensing platforms for the detection of H. pylori infection represents a crucial bridge between scientific innovation and clinical practice. Multiplexing capabilities of these platforms significantly enhance diagnostic precision by allowing simultaneous detection of multiple biomarkers, offering a more detailed analysis of a patient's condition.50 This approach facilitates the development of personalized therapeutic strategies tailored to individual biomarker profiles. Clinically, these biosensing technologies hold promise for delivering rapid and highly reliable diagnostics, leading to more effective patient management and improved health outcomes.84–86 Furthermore, their adaptability for point-of-care use positions these biosensors as invaluable tools in resource-constrained environments where conventional diagnostic infrastructure may be unavailable.20,87Recent studies revealed that several electrochemical, optical, field-effect transistor and piezoelectric biosensors have been developed for point-of-care detection of H. pylori infection. Immunosensors utilizing H. pylori antibodies as biorecognition elements are integrated into formats like enzyme-linked immunosorbent assay (ELISA), chemiluminescence, and fluorescence platforms. DNA biosensors incorporating ssDNA probes complementary to H. pylori 16S rRNA or 23S rRNA provide rapid hybridization-based analysis. Aptasensors using synthetic aptamers selected against H. pylori whole cells or specific antigens offer high selectivity. Emerging 2D nanomaterials have significantly improved the performance of biosensors for H. pylori detection. Recent literature has evaluated the performance of different biosensor systems in terms of sensitivity, specificity, selectivity, and limit of detection. These 2D nanoparticle-based biosensors demonstrated high selectivity, good stability, reproducibility, and cost-effectiveness for early detection of H. pylori.87–95 These biosensors offer superior performance compared to conventional methods such as enzyme-linked immunosorbent assay (ELISA) and polymerase chain reaction (PCR), which typically have higher LODs and longer assay times. Table 2 shows a comparison of performance parameters among 2D nanoparticle-based biosensors, biosensors with other nanomaterials, and conventional diagnosis methods. 2D nanoparticles have been integrated into optical, colorimetric and electrochemical biosensing platforms as well as field-effect transistors and piezoelectric sensors due to their unique properties and ease of functionalization.27,85,86,96 The integration of 2D nanoparticles into these sensing platforms has further advanced their detection capabilities by enhancing sensitivity, specificity, and selectivity.
| Features | 2D Nanoparticle-based biosensors | Biosensors with other nanomaterials | Conventional diagnosis methods (RUT, UBT, culture, endoscopy, PCR) | Ref. |
|---|---|---|---|---|
| Fabrication and integration processes | Complex fabrication, specialized equipment and expertise required. Integration involves functionalization of 2D nanoparticles with recognition elements | Fabrication may vary based on nanomaterials used, involving specialized equipment. Integration requires functionalization of nanomaterials with recognition elements | Standardized procedures, requiring specialized equipment and trained personnel | 29, 64 and 66 |
| Stability | Moderate stability, susceptible to degradation over time due to environmental factors and biofouling | Moderate stability, may experience degradation over time | Stable under appropriate conditions | 69 and 97–99 |
| Durability | Moderate durability, may degrade over prolonged use | Moderate durability, may degrade over prolonged use | Durable, can be used repeatedly | 28 and 100 |
| Scalability | Limited scalability due to complex fabrication processes and material availability | Moderate scalability, depending on nanomaterial synthesis and functionalization techniques | Scalable for mass production | 101 and 102 |
| Reproducibility | Variable reproducibility due to batch-to-batch variations in nanoparticle synthesis and functionalization | Variable reproducibility, influenced by nanomaterial synthesis and functionalization methods | High reproducibility, standardized procedures | 103–105 |
| Toxicity | Concerns regarding potential toxicity of 2D nanoparticles, necessitating thorough biocompatibility testing | Potential toxicity of other nanomaterials, necessitating biocompatibility assessment | Generally safe, but potential risks associated with invasive procedures | 98 and 106–108 |
| Biocompatibility | Requires thorough assessment of biocompatibility due to potential interactions with biological systems | Requires assessment of biocompatibility based on specific nanomaterials used | Biocompatible, suitable for use in biological systems | 100, 101 and 109 |
| Detection time | Rapid detection, often within minutes to hours | Rapid detection, similar to 2D nanoparticle-based biosensors | Variable detection time, depending on the method used: RUT (1–24 hours), UBT (30 minutes), culture (3–14 days), endoscopy (same day), PCR (3–12 hours) | 87, 93, 101, 110 and 111 |
| Limit of detection | Low limit of detection, down to picomolar or femtomolar levels | Low limit of detection, similar to 2D nanoparticle-based biosensors | Variable limit of detection, influenced by the sensitivity of the method used: RUT (qualitative), UBT (qualitative), culture (102 CFU mL−1), endoscopy (qualitative), PCR (102 CFU mL−1) | 24, 28, 29, 87, 88, 101, 103, 112 and 113 |
| Selectivity | High selectivity, can distinguish the target analyte from interfering substances | Variable selectivity, influenced by nanomaterial properties and surface functionalization | High selectivity based on specific biomolecular interactions | 90, 101, 103, 114 and 115 |
| Sensitivity | High sensitivity, capable of detecting low concentrations of the target analyte (92–98%) | Variable sensitivity, influenced by nanomaterial properties and detection method (88–96%) | Variable sensitivity, depending on the method used: RUT (70–90%), UBT (90–98%), PCR (94–98%), ELISA (80–90%) | 9, 15, 17, 24, 86, 101, 103 and 116–118 |
| Specificity | High specificity, able to accurately identify the target analyte amid complex biological matrices (94–99%) | Variable specificity influenced by nanomaterial properties and recognition elements (90–97%) | High specificity, based on specific biomolecular interactions: RUT (80–95%), UBT (92–98%), PCR (95–99%), ELISA (85–95%) | 9, 14–17, 24, 36, 86, 99, 101, 103 and 116–118 |
| Material and production cost | Moderate to high upfront costs, but cost-efficient in large-scale production due to scalable manufacturing techniques. These materials are typically cheaper than conventional ones once production is scaled up | Similar initial cost as the 2D nanomaterial-based biosensors | Typically high due to expensive reagents, specialized lab equipment, and complex infrastructure requirements | 119–121 |
| Design and maintenance cost | Compact and portable designs possible, with low to moderate cost of maintenance. These biosensors are often portable, disposable, or require minimal maintenance, reducing long-term upkeep | Similar design and cost considerations as 2D nanoparticle-based biosensors, with varying costs depending on the nanomaterial and fabrication processes | Standardized design and costs based on established protocols, with endoscopy and PCR being more expensive than RUT and UBT. High maintenance cost requiring regular calibration, expensive reagents, and highly trained personnel | 17, 27, 98 and 121–124 |
| Result consistency | Variable consistency due to batch-to-batch variations in nanoparticle synthesis and functionalization | Variable consistency, influenced by nanomaterial properties | Varying consistency: RUT (88–95%), UBT (90–95%), culture (90–98%), endoscopy (90–95%), PCR (95–99%) | 28, 101, 114 and 115 |
| Acceptability to the public | Moderate acceptability due to the novelty of nanomaterial-based biosensors and potential concerns about toxicity | Similar acceptability challenges as 2D nanoparticle-based biosensors, with varying public perception based on the nanomaterial and potential toxicity concerns | Conventional diagnostic methods are generally well-accepted by the public, with the exception of invasive procedures like endoscopy | 17, 118, 125 and 126 |
| Commercialization | Moderate commercialization potential, with several companies and startups exploring nanomaterial-based biosensors | Similar commercialization potential as 2D nanoparticle-based biosensors, with varying market readiness based on the nanomaterial and sensing mechanism | Widely commercialized and available in clinical settings, with varying degrees of accessibility and affordability | 127–130 |
| Real-world applications | Potential for rapid and decentralized diagnostics during disease outbreaks, with the ability to scale up production and deployment | Similar potential applications as 2D nanoparticle-based biosensors, with varying suitability based on the nanomaterial and sensing mechanism | Conventional diagnostic methods can be deployed during epidemics, but may face challenges in terms of accessibility, turnaround times, and logistics | 29, 53, 89, 90, 100, 101, 103, 106, 115 and 127–145 |
In a recent study, Mirzaei et al. introduced a novel Hsp60 biosensor with enhanced sensitivity and selectivity, developed using a nanocomposite with an aptamer-recognition surface for enhancing H. pylori detection.146 The biosensor was constructed by modifying a glassy carbon electrode with a nanocomposite composed of carbon quantum dots, gold nanoparticles, and polythiophene. The integration of these materials provided the biosensor with high stability, a large surface area, excellent electrical conductivity, and compatibility with biological systems. Electrochemical techniques, including square wave voltammetry (SWV) and electrochemical impedance spectroscopy (EIS), were used to detect the interaction of Hsp60 with immobilized aptamers, resulting in a decrease in peak current and increased resistance. The aptasensor demonstrated a detection limit of 7.38 nM and a linear detection range of 0.01 to 0.25 μM.146 It also exhibited high reproducibility, selectivity, rapid response, and stability for detecting H. pylori, highlighting its potential for biomedical, bioengineering, and clinical applications.
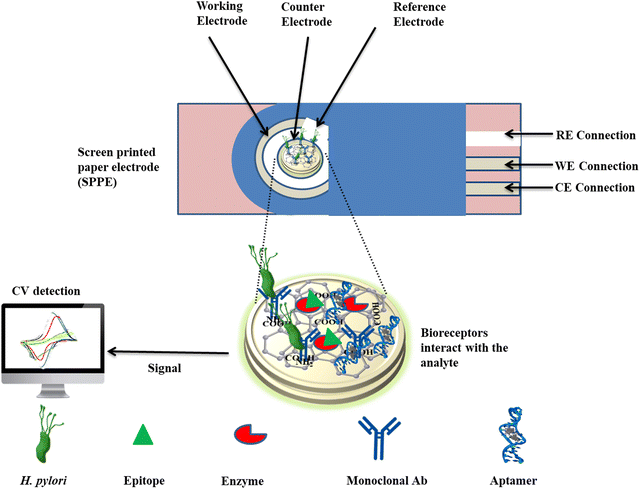
Saxena et al. conducted research on a molecularly imprinted polymer (MIP)-based electrochemical biosensor to detect the VacA antigen.86 This was achieved by polymerizing the template VacA antigen on a paper electrode. Notably, the VacA antigen utilized MIP coated on a silicon dioxide (SiO2) substrate integrated into a screen-printed paper electrode (SPPE) platform (VacA-MIP/SiO2/SPPE sensor) exhibited exceptional sensitivity (0.304 mA ng−1 mL−1) and an exceptionally low limit of detection (0.01 ng mL−1) within a linear range of 0.01–100 ng mL−1.86 Furthermore, the sensor demonstrated successful determination of the VacA antigen in the presence of various potential interferents, thereby underscoring its robustness and specificity. In another study, a nanohybrid-based immunosensor was developed to detect the H. pylori BabA antigen. This biosensor utilized an electrode loaded with palladium nanoparticles (Pd) through electrodeposition to create rGO and poly(3,4-ethylenedioxythiophene) (PEDOT). This nanohybrid platform enhanced the surface area and electroconductivity for the immobilization of antibodies and the BabA antigen.85 The immunosensor demonstrated the capability to detect H. pylori antigens in a linear range of 0.2–20 ng mL−1 with an LOD of 0.2 ng mL−1. Computational simulations confirmed stable antigen–antibody interactions, emphasizing the sensor's functionality in the electrochemical detection of H. pylori. This immunosensor exhibited high specificity, sensitivity, and reproducibility, making it a promising tool for accurate detection of the H. pylori BabA antigen.
Furthermore, a label-free electrochemical immunosensor was fabricated for rapid detection of H. pylori. This sensor was based on the combination of titanium oxide, carboxylated multi-walled carbon nanotubes, and polyindole carboxylic acid composites.147 The sensor demonstrated an LOD of 0.1 ng mL−1 in a dynamic linear range of 0.1–8.0 ng mL−1, having high sensitivity and selectivity. The study involved testing for H. pylori detection in five human stool specimens, showing good results with suitable accuracy.147,148 The analysis of these studies highlights the potential of electrochemical biosensors for rapid, accurate, and timely detection of H. pylori infection. The rapid and accurate detection of H. pylori infection facilitated by these electrochemical biosensors has led to timely therapeutic interventions, ultimately improving patient outcomes and reducing the risk of associated complications. In another study, a titanium carbide (Ti3C2Tx) MXene-based electrochemical biosensor was developed for the detection of antibodies in serum samples. The biosensor exhibited higher sensitivity with an LOD of 0.1 ng mL−1 and better selectivity compared to conventional serological tests like ELISA.79 The superior performance was attributed to the exceptional conductivity, high bandgap, large surface area, and tunable surface chemistry of Ti3C2Tx MXene, enabling efficient antibody immobilization and signal transduction. Furthermore, an immunosensor was developed for CagA antigen detection. The gold electrode was subsequently modified by platinum nanoparticles, poly(3,4-ethylenedioxythiophene) (PEDOT), and reduced graphene oxide (rGO) for immobilization of the CagA antigen on this electrode. The fabricated device was used for immunosensing through interaction between antigen and antibodies present in serum samples. The linear range of the biosensor was calculated as 0.1 ng mL−1 to 30 ng mL−1 and LOD as 0.1 ng mL−1.80
Similar to immuno-based biosensors, researchers have developed nucleic acid based sensors, such as DNA or aptamer based biosensors, to detect H. pylori. For example, the DNA-based sensors rely on the hybridization of the single-stranded DNA probe with its complementary DNA target sequence. These DNA-based biosensing platforms are advantageous due to their high chemical stability and ease of modification, and have attracted the focus of many researchers developing biosensors aimed at detecting H. pylori.149 The ability to tailor the DNA probes and transduce hybridization events into measurable outputs has led to widespread exploration of DNA biosensors for detection of H. pylori. Hajihosseini et al. developed a sensitive electrochemical DNA biosensor for detecting H. pylori utilizing a single-stranded DNA probe immobilized on a graphene oxide/gold (GO/Au) nanocomposite modified glassy carbon electrode. The biosensor demonstrated a linear range of 60 pM to 600 pM with an LOD of 27 pM to detect H. pylori.150
del Pozo et al. employed a surface-based approach to study DNA-based electrochemical interactions with redox-active osmium complexes containing 1,10-phenanthroline-5,6-dione ligands. In this approach, gold electrodes were functionalized with DNA probes and osmium reporter molecules like [Os(bpy)2(phen-dione)]3+/2+ enabling accumulation within the DNA layer. Voltammetric charge quantified the bound osmium complexes, exploiting the phen-dione quinone's low-potential redox activity for electrochemical DNA sensing. A single-stranded H. pylori DNA probe was linked with thiol and immobilized on gold, and hybridization with its complement allowed osmium accumulation within the dsDNA. The platform enabled quantification of H. pylori sequences ranging from 5 to 20 pM, with a detection limit of 6 pM.151 In another study, Asadzadeh-Firouzabadi et al. demonstrated a novel electrochemical DNA hybridization biosensor with a signal range from 20.0 to 410.0 nM for H. pylori complementary DNA, achieving a detection limit of 7.2 nM. This sensing platform utilized chlorogenic acid (CGA) as the electroactive indicator. The detection mechanism of the sensor relied on interaction of CGA with an immobilized 18-mer H. pylori alkanethiol DNA probe and its hybridized form on a self-assembled gold electrode.84
In addition to DNA, researchers employ small oligonucleotides known as aptamers for creating nucleic acid-based electrochemical biosensors. These aptamers exhibit exceptional binding affinity for their targets; they are selected from randomized libraries and are capable of recognizing and binding to particular protein regions with high specificity. For example, Yadav and colleagues developed an aptasensor by utilizing graphitic carbon nitride (g-C3N4) with 3-(aminopropyl)triethoxysilane (APTES). H. pylori CagA-specific aptamers were covalently conjugated to the modified electrode enabling the detection of CagA with 1.98 μA ng mL−1 cm−1 sensitivity and 0.1–160 ng mL−1 linear range. The aptasensor demonstrated an LOD of 0.017 ng mL−1. In this study, the aptasensor developed exhibited exceptional specificity toward the H. pylori CagA toxin, enabling highly stable gastric cancer detection for point-of-care applications.152
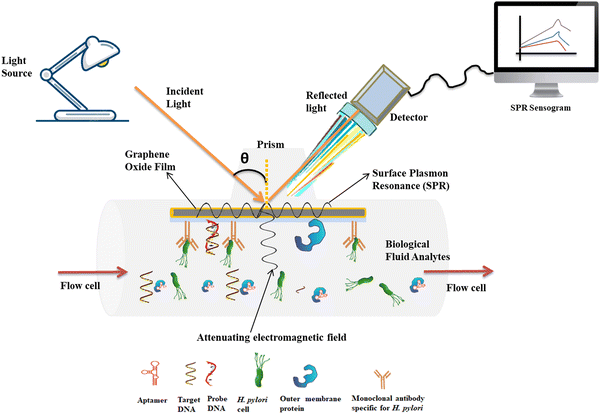 | ||
| Fig. 4 Advanced design of optical label-free biosensors for detection of H. pylori biomarkers in biological fluids. | ||
In addition to plasmonic biosensors, advancements in photonic and photonic crystal biosensors have enabled highly sensitive and selective detection of infections. Photonic biosensors exploit the principles of light propagation and modulation within photonic structures, such as waveguides, resonators, and gratings, to detect changes in the refractive index or optical properties induced by biomolecular interactions. For instance, photonic crystal biosensors utilize periodic nanostructures with tailored optical properties to achieve enhanced sensitivity and specificity in detecting infectious agents.147 Studies have demonstrated the application of photonic crystal biosensors for the label-free detection of bacteria, viruses, and toxins in clinical samples, offering advantages such as rapid analysis, low sample consumption, and compatibility with microfluidic systems. These photonic biosensors hold promise for point-of-care diagnostics, environmental monitoring, and biosecurity applications, facilitating early detection and containment of infectious diseases.
Furthermore, the integration of advanced materials, such as graphene, quantum dots, and metal nanoparticles, into optical biosensors has led to significant improvements in sensitivity, signal amplification, and multiplexed detection capabilities. Graphene-based biosensors, for example, offer unique optical and electrical properties that enable highly sensitive detection of biomolecular interactions.153 Similarly, quantum dot-based biosensors utilize semiconductor nanoparticles with tunable optical properties to achieve ultrasensitive detection of biomolecules. In a study, a FRET (fluorescence resonance energy transfer) based biosensor was developed for sensitive determination of H. pylori. Two types of oligonucleotide probes were labeled with cadmium telluride (CdTe) QDs (the donor) and 5-carboxytetramethylrhodamine (the acceptor) respectively. Following the donor's photoexcitation, the donor and acceptor's proximity is used to measure FRET. The probes were not able to bind in the absence of target analytes and, subsequently, there was no emission. The fabricated biosensor was simple, rapid, and highly efficient for the detection of homogeneous DNA.154 Israa et al. developed an optical biosensor based on a multi-mode no core fiber to determine the minimum inhibitory concentrations (MICs) of antibiotics against H. pylori. In this study, the biosensor used an inline Mach-Zehnder interferometer to measure the bacteria in the BHI broth turbidity stabilizer and compared the results with traditional methods.125 The developed optical biosensor was presented as a facile, rapid, and effective method for bacterial detection in comparison to the conventional methods.
Shahrashoob et al. reported a novel optics-based biosensor to detect a H. pylori urease-labeled probe. The probe was immobilized on APTES-activated glass-bound gold nanoparticles. The hybridization between the probe and the target analyte resulted in a decrease in the optical density corresponding to the analyte concentration. This optics-based approach achieved a remarkable limit of detection of 0.5 nM for H. pylori.155 In a recent study, Rong et al. proposed a novel optical biosensor utilizing porous silicon (PSi) nanomaterial and photonic Tamm plasmon polariton (TPP) surface states to detect H. pylori CagA. The findings of this study demonstrated the biosensor's high sensitivity of 100 pm (ng mL−1)−1, low detection limit of 0.01 ng mL−1, and excellent specificity with a positive-to-negative ratio exceeding six. These promising performance metrics suggest the potential of the TPP biosensor for reliable diagnosis of H. pylori infection in both laboratory and point-of-care testing settings.156
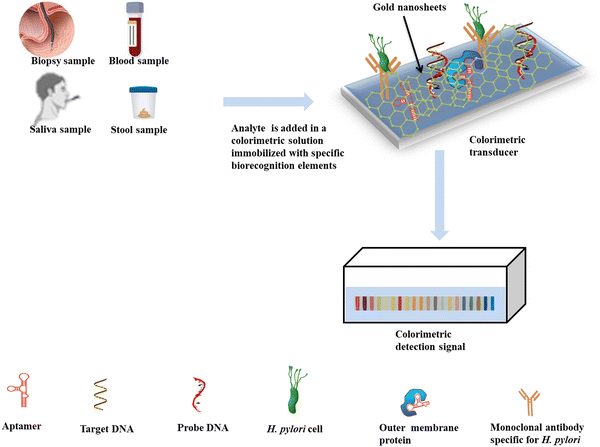
Recently, Suaifan et al. conducted a study focusing on the creation of a colorimetric nanomaterial-based paper biosensor specifically designed for the detection of H. pylori using extracellular proteases as biomarkers.159 This innovative biosensor employs a unique substrate that is functionalized with magnetic nanobeads and integrated into a gold sensing platform. The biosensor demonstrated a limit of detection (LOD) of 100 CFU mL−1,159 while also exhibiting notable selectivity and stability. Furthermore, its efficacy in detecting H. pylori in clinical specimens was evaluated, yielding promising results regarding both sensitivity and specificity.
Fei et al. developed a colorimetric biosensing assay to detect H. pylori in stool samples. The biosensing assay utilized oligonucleotide probes impregnated with gold nanoparticles together with aptamers specific for H. pylori.160 The aggregation and deaggregation helped to achieve the detection process with a detection limit of 25 CFU mL−1. This assay offers a promising method for clinical field detection of H. pylori, as it is noninvasive and can be used for both qualitative and quantitative detection. As research in this field continues to progress, we can expect even more sensitive, selective, and user-friendly colorimetric biosensors for the rapid and accurate detection of H. pylori and other infectious diseases, ultimately contributing to improved patient outcomes and public health management.
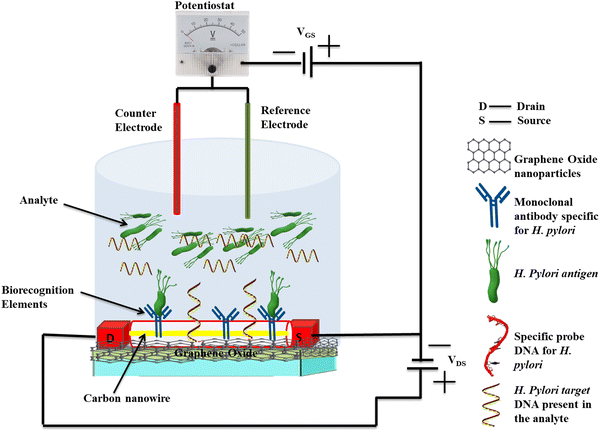
Researchers have also explored the integration of 2D nanoparticle-based FET biosensors with microfluidic and lab-on-chip devices to enhance their performance and facilitate point-of-care (POC) diagnostics. A microfluidic chip-based FET biosensor has been developed to quantify the presence of pathogenic biomarkers.163 These biosensors have been shown to utilize complementary metal oxide or carbon semiconductor as the channel material in the FET device, and specific biomarkers such as H. pylori CagA or BabA can be immobilized on its surface. The binding of biomarkers will induce a change in the electrical properties of the metal oxide or carbon channel, resulting in a measurable change in the source–drain current (Fig. 6). These integrated biosensors exhibit high sensitivity, with a lower detection limit and they demonstrate excellent performance in the analysis of clinical samples.163,164 The advancements in 2D nanoparticle-based FET biosensors offer several advantages, including label-free detection, high sensitivity, and the potential for real-time monitoring. As research in this field continues to progress, we can expect even more sensitive, selective, and user-friendly FET biosensors for the rapid and accurate detection of H. pylori and other infectious diseases, ultimately contributing to improved patient outcomes and public health management.
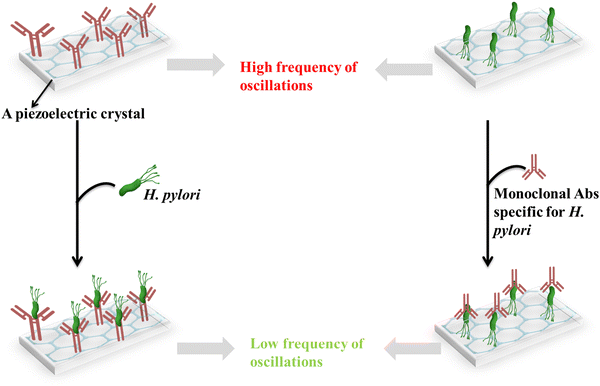
In a reported study, an enzymatically amplified piezoelectric immunosensor was developed for the sensitive detection of antibodies against the H. pylori pathogen. In this sensor, a sandwich assay format on AT-cut quartz crystal resonators operating at 10 MHz frequency was employed. Recombinant H. pylori antigen proteins were then immobilized onto the quartz crystal surface serving as capture probes for the corresponding antibodies present in patient serum samples. It was reported that the study employed anti-human IgG secondary antibody probes specifically binding to the human antibodies previously captured on the antigen-modified surface, forming sandwiched immunocomplexes. This secondary antibody binding event significantly improved the positive-to-negative signal ratio, which was increased to 4.1–5.0. Moreover, the horseradish peroxidase enzyme was conjugated with anti-IgG probes to achieve an additional signal amplification. This enzymatic amplification strategy resulted in a striking enhancement of sensitivity, enabling reliable positive detection even in serum samples diluted up to 1/100. Through this multi-pronged approach, the piezoelectric immunosensor overcomes the obstacles posed by low antibody levels in clinical specimens, paving the way for efficient serological diagnosis of H. pylori infections.123Fig. 7 shows a piezoelectric immunosensor for the determination of H. pylori and monoclonal Abs specific for H. pylori.
3.4. Design considerations for optimizing biosensor performance
The primary design goal for biosensors intended for the clinical detection of H. pylori infections is to achieve high sensitivity, specificity, selectivity, and reliability. Several interdependent factors influence biosensor's analytical performance and require careful optimization during the design phase. One critical aspect is the selection and immobilization of biorecognition elements onto the biosensor surface.64,66 Optimizing the design of biosensors for H. pylori detection plays a pivotal role in translating scientific innovations into effective clinical outcomes. Emerging research underscores the crucial role of choosing suitable transducers and bio-recognition elements, as these components directly impact the sensitivity and specificity of the biosensors. For example, surface plasmon resonance (SPR) biosensors have exhibited exceptional sensitivity, capable of detecting H. pylori at concentrations as low as 200 CFU mL−1, being significantly more sensitive compared to conventional diagnostic approaches like the rapid urease test (RUT) that require much higher bacterial concentrations to achieve reliable detection.166,167 For H. pylori detection, antibodies, aptamers, and DNA probes specific to H. pylori antigens or DNA sequences are commonly used as biorecognition elements. Antibodies, though highly specific, can suffer from batch-to-batch variability, instability and high costs. Aptamers are increasingly preferred as robust, reusable synthetic receptors but require stringent selection strategies.70,168 DNA/PNA probes enable excellent specificity but are unsuitable for detecting proteins/whole cells.169 For instance, a recent study reported the development of an ultrasensitive electrochemical aptasensor for H. pylori detection. This sensor is based on surface modified graphene oxide doped gold nanoparticles conjugated polythiophene. The linear behavior of the modified electrode against a wide range of concentrations of H. Pylori, shows the reliability of the developed biosensor, with a limit of detection (LOD) of 0.0080 μM for impedance and 0.0067 μM for square wave voltammetry curves.110 The optimized immobilization strategy of the aptamer on the polythiophene (PTP)-modified electrode resulted in enhanced sensitivity and stability of the biosensor, enabling the rapid and reliable detection of H. pylori antigens in clinical samples.110 Surface chemistries utilized for bioconjugation also impact orientation, steric hindrance and bioactivity.Furthermore, the choice of the transducer substrate influences biosensor sensitivity. Electrochemical transducers, such as screen-printed electrodes (SPEs) and field-effect transistors (FETs), are commonly employed for H. pylori detection due to their high sensitivity, low cost, and compatibility with miniaturized devices for point-of-care testing. Recent advancements in signal amplification techniques, such as enzymatic amplification, nanoparticle-based amplification, and signal enhancement strategies, have further improved the detection limits and dynamic range of biosensors for H. pylori detection. For example, Chen et al. developed a MoS2-based biosensor for exosome detection, where signal amplification was achieved through the use of horseradish peroxidase (HRP)-exosome probes and the tetramethylbenzidine (TMB) substrate, resulting in significantly enhanced sensitivity and specificity compared to conventional methods.93
In addition, minimizing non-specific binding and matrix interference from complex clinical samples is another critical aspect in biosensor design. Immobilizing biorecognition elements in oriented configurations helps reduce steric hindrances. Incorporating blocking agents like BSA, ethanolamine, and pyrene layers prevents non-specific adsorption. Wang et al. developed a 3D reduced graphene oxide-polypyrrole immunosensor using aryldiazonium salt grafts to create antifouling surfaces, enhancing specificity against interfering proteins.92 2D nanomaterials can initially be more expensive than conventional materials. However, these costs can decrease as mass production and miniaturization become more feasible, leading to potential economies of scale.119 In addition, maintenance costs for nanomaterial-based biosensors tend to be lower than for traditional equipment, as they are typically more robust, portable, and easier to use, particularly in resource-limited settings. Surface functionalization with zwitterionic polymers like poly(carboxybetaine) creates highly antifouling yet biocompatible biosensor interfaces.90 Other antifouling materials include polyethylene glycol, dextran, and polyoxazoline.90 The use of these materials increases the upfront costs associated with developing the biosensors. Advanced designs employing microfluidic, paper-based, and lab-on-chip devices will minimize sample volumes, improve reaction kinetics and enable instrument-free operation. A recent 3D-printed microfluidic electrochemical cytosensor detected cancer cells directly from gastric biopsy specimens without complex labeling.91 Similarly, a low-cost paper-based vertical flow assay incorporated antibody-conjugated carbon nanotubes for visual immunoassay.132 Smart design strategies like these that synergistically optimize biorecognition elements, transducers, surface chemistry and device architecture are key to realizing high-performance biosensors. These designs can be integrated for robust H. pylori diagnosis at the point-of-care.
3.5. Industrial and commercialization viability of 2D nanomaterial-based biosensors
The industrialization potential and commercial viability of 2D nanomaterial-based biosensors for H. pylori detection are significant, especially given the growing demand for rapid, sensitive, and cost-efficient diagnostic tools in healthcare. The use of 2D nanomaterials in biosensors can dramatically lower the detection limits for H. pylori, which is essential for early diagnosis and treatment of gastrointestinal disorders.125,151 A nanohybrid-based immunosensor was developed for the detection of the H. pylori BabA antigen.85 The reduced graphene oxide/poly(3,4-ethylenedioxythiophene) immunosensor developed demonstrated a strong ability to detect H. pylori antigens over a linear detection range of 0.2 to 20 ng mL−1, with a low limit of detection of 0.2 ng mL−1.85 This sensitivity highlights its potential for industrial and commercialization purposes enhancing accurate and prompt diagnosis of H. pylori-related infections. Additionally, the simplicity and affordability of electrochemical biosensors made with 2D materials make them attractive alternatives to conventional diagnostic methods, which are often complicated and require specialized skills.20,120,170 Zhang et al. introduced a novel glucose sensor utilizing enzyme-modified whole-graphene solution-gated transistors (SGGTs).171 The sensor operates through the detection of hydrogen peroxide (H2O2) produced during glucose oxidation, which occurs near the gate. This innovative biosensor achieves detection limits below 0.5 μM, demonstrating excellent selectivity for non-invasive glucose monitoring in bodily fluids.171 The transconductance of these devices is significantly higher – by two orders of magnitude – compared to that of traditional silicon field-effect transistors, which is a critical factor for their heightened sensitivity.95,162,171 Additionally, the fabrication process is straightforward and cost-effective, enhancing commercialization potential for practical application in biosensing technologies.Point-of-care testing is gaining traction in healthcare settings, particularly in under-resourced areas, due to its portable and user-friendly nature, which allows for immediate results. This trend is bolstered by investments from governments and health organizations aimed at advancing disease detection and management technologies, thereby expanding the biosensor market. The biosensor sector is projected to grow significantly, with estimates suggesting a market size of USD 24.12 billion in 2024, potentially reaching USD 36.55 billion by 2029, and reflecting a compound annual growth rate (CAGR) of 8.67% during this period.172 The rise in chronic diseases, including peptic ulcers, diabetes, and cancer, underscores the necessity for continuous monitoring of H. pylori infection, blood glucose levels, and cancer cell proliferation.93,157,173 This demand drives the development of implantable and wearable biosensors capable of providing accurate real-time monitoring. Specifically, the need for reliable biosensors for H. pylori detection is critical, as early diagnosis and ongoing management of gastritis are vital for preventing severe complications associated with the infection.
The widespread commercialization and adoption of 2D nanomaterial-based biosensors face challenges, including the lack of standardized safety and efficacy guidelines for nanomaterials, which complicates market entry due to the demands of the regulatory bodies for comprehensive risk assessments.114,121 Although 2D nanomaterials enhance sensor performance, their synthesis and integration can be costly, limiting accessibility, particularly in resource-limited settings. To ensure practical applications, issues such as the long-term stability, biocompatibility, and consistent performance of nanomaterials must be addressed. Furthermore, the competitive biosensor market requires new entrants to demonstrate clear advantages over existing technologies to gain traction and secure a foothold in the industry.
4. Challenges and future perspectives
4.1. Challenges in fabrication, integration and biocompatibility
The integration of 2D nanoparticles in biosensors for the detection of H. pylori and other infectious diseases has shown promising results, but it is not without challenges. Different studies have shown that researchers encounter notable challenges during the fabrication and integration processes. The controlled synthesis and functionalization of 2D nanoparticles is one of the significant challenges that nanosensing technology is facing. These nanomaterials often exhibit diverse physical and chemical properties which make it difficult to achieve consistent and reproducible characteristics. For instance, the fabrication of GO and MoS2 nanosheets in biosensors displayed varying degrees of oxidation, defect density, and surface functional groups, which impacted their interaction with biorecognition elements and target analytes.174,175 This means that multi-step oxidation processes are required to improve the reproducibility and performance of the biosensor with such nanomaterials. Studies have shown that the integration of 2D nanoparticles into some biosensing platforms often requires complex fabrication processes and specialized equipment. Savannah et al. investigated the development of a graphene-field effect biosensor to enable real-time, quantitative detection of the Zika virus.176 The use of graphene nanosheets offered advantages such as high surface area and excellent biocompatibility, but challenges arose concerning the reproducibility of the fabrication process and the optimization of graphene nanosheet dispersion in the biosensor.Another challenge lies in the effective immobilization and orientation of biorecognition elements on the surface of 2D nanoparticles. Improper immobilization can lead to reduced binding affinity, steric hindrance, and decreased sensitivity of the biosensor. The integration of MoS2 nanosheets with a specific DNA aptamer encountered challenges in achieving optimal orientation and spacing of the aptamer on the MoS2 surface, which initially resulted in reduced target binding and signal transduction.174,175 In addition to fabrication challenges, the integration of 2D nanoparticles in biosensors for infectious disease detection can be a challenge due to biocompatibility, stability, and potential toxicity properties of the 2D nanomaterial in question.100,102,104,114,130 Nanomaterials may interact with biological systems in unexpected ways, potentially causing adverse effects or interfering with the performance of the biosensor. Extensive testing and optimization are required to ensure the biosafety and long-term stability of these biosensor systems, especially for in vivo applications.
In addition, biocompatibility presents a significant challenge in the development of 2D nanomaterial-based biosensors, as these materials can elicit immune responses or exhibit cytotoxic effects when in contact with biological systems.109,177 To address these concerns, one effective approach is to functionalize nanomaterials with biocompatible polymers or biomolecules, enhancing their compatibility with biological components while reducing toxicity. Over the past years, polymer-based biosensors have demonstrated success in detecting various analytes, including glucose, proteases, and hydrogen peroxide.178 Recent research has highlighted their application in the detection of H. pylori. For instance, Saber Mirzaei et al. developed an ultrasensitive electrochemical aptasensor utilizing surface-modified graphene oxide combined with gold nanoparticles and conjugated polythiophene polymer for H. pylori detection.110 This innovative biosensor exhibited impressive limits of detection, reaching 0.0080 μM for impedance measurements and 0.0067 μM for square wave voltammetry (SWV). Surface modifications with biocompatible polymers can protect nanomaterials from direct exposure, further enhancing their safety in medical applications.
4.2. Challenges related to stability, reproducibility and sensitivity
The stability and reproducibility of 2D nanoparticles in biosensing platforms still remain a challenge. 2D nanomaterials often exhibit high surface energy and reactivity, which makes them susceptible to degradation or alteration over time. Investigations on the stability of graphene oxide nanosheets used in an electrochemical biosensor found that the nanosheets underwent structural changes and oxidation over time, leading to a gradual decrease in the sensitivity and accuracy of the biosensor.114,130,174 In this study, the use of protective coatings and encapsulation techniques enhanced the long-term stability of the GO nanosheets, but these approaches added complexity and cost to the fabrication process. Challenges with the reproducibility of the physicochemical properties of 2D nanoparticles can significantly impact the performance and reliability of biosensors. The reproducibility of biosensor fabrication using 2D nanoparticles is also hindered by variations in material properties, synthesis methods, and environmental factors.126,129,130,153,160 This significantly affects the acceptability and commercialization of biosensing technologies during epidemics. Additionally, the dispersion of 2D nanoparticles within sensing matrices poses a significant challenge, as agglomeration can compromise the performance of the biosensor and result in unreliable detection outcomes.179 The variations in the size, thickness, and defect density of the MoS2 nanosheets led to inconsistencies in the sensitivity and selectivity of the biosensors developed using such nanosheets. Exposure of 2D nanoparticle-based biosensors to harsh environmental conditions such as temperature fluctuations, humidity, and potential contaminants can affect their performance and reliability and result in loss of sensitivity over prolonged use. Some 2D nanoparticles exhibit high magnetic properties which poses high chances of contamination with other magnetic particles during exposure to the environment. Several studies have highlighted that the instability of graphene-based nanomaterials in physiological environments can lead to degradation and loss of sensing functionality over time.100,180–183Wu et al. investigated the stability of a surface-enhanced Raman scattering (SERS) biosensor based on silver nanoparticles and found that the SERS biosensor exhibited a gradual decrease in sensitivity and signal intensity when exposed to high temperatures or humid conditions, potentially limiting its practical applications in resource-limited settings or field-based diagnostics.184 Furthermore, different studies have noted that unreliable or inconsistent performance due to stability, scalability, and reproducibility related issues can lead to inaccurate diagnoses, potentially compromising public health efforts and undermining the confidence of healthcare professionals and regulatory authorities in these biosensing platforms. The challenges together are multifaceted and require concerted efforts from multidisciplinary teams to unlock the full potential of these advanced biosensing platforms, enabling rapid and accurate disease detection during epidemics and enhancing public health preparedness on a global scale.
4.3. Strategies for improving biosensor performance
Recent advancements in biosensors for the detection of infectious diseases, particularly targeting H. pylori, have seen a surge in interest due to the pressing need for rapid and accurate diagnostic tools. The optimization of biosensor performance for the detection of H. pylori is essential for enhancing diagnostic accuracy and clinical applicability. Recent studies have highlighted several strategies that can significantly improve biosensor functionality, bridging scientific advancements with real-world clinical implications.87,166 Researchers have been actively exploring strategies to enhance the performance of biosensors, including surface functionalization, bioconjugation, hybrid nanostructures, and nanocomposites. These approaches aim to improve sensitivity, selectivity, and integration with microfluidic and lab-on-chip systems, ultimately enabling more efficient and reliable H. pylori detection. Surface functionalization and bioconjugation have emerged as powerful strategies to improve biosensor performance. A number of studies have demonstrated the promising potential of graphene oxide nanosheet-based electrochemical biosensors in detecting specific bacterial strains with high sensitivity and selectivity. Hajihosseini et al. developed biosensors utilizing GO-based platforms that could sensitively and selectively detect H. pylori. These biosensors demonstrated exceptional performance, with detection limits as low as 27.0 pM for H. pylori.150 Additionally, Duan et al. demonstrated the potential of a GO-based aptasensor for detecting Salmonella typhimurium, with a detection limit of 100 CFU mL−1.185 This aptamer-functionalized GO biosensor exhibited remarkable stability and reusability, making it a promising platform for point-of-care diagnostics.Another strategy involves the development of hybrid nanostructures and nanocomposites, which combine the unique properties of different nanomaterials to create synergistic effects. Saxena et al. reported the fabrication of an electrochemical immunosensor, which detected the VacA gene, on a gold electrode decorated with graphitic carbon nitride/zinc oxide.185 The g-C3N4/ZnO nanocomposite was functionalized with specific antibodies, enabling selective capture and detection of anti-H. pylori VacA antigens in patient serum samples. The combination of graphitic carbon nitride with zinc oxide enhanced the performance of the biosensor which showed a sensitivity of 0.3 μA ng−1 mL−1 in a linear detection range of 0.1 to 12.8 ng mL−1.86,117 Integrating biosensors with microfluidic and lab-on-chip systems has also emerged as a promising strategy for enhancing biosensor's ability to detect H. pylori and other infectious agents. These integrated systems offer several advantages, including reduced sample and reagent volumes, automated sample handling, and the potential for multiplexed analysis. Yao Xie et al. demonstrated the development of a microfluidic electrochemical biosensor for the detection of gastric cancer biomarkers. In this study, an electrochemical microfluidic chip that combines multiple biomarkers, including carbohydrate antigen 19-9 (CA19-9), H. pylori CagA protein (H.P.), P53 oncoprotein (P53), pepsinogen I (PG I), and PG-II, for the early detection of gastric cancer was developed.186 The developed sensor offered better sensitivity compared to enzyme-linked immunosorbent assay results.
In addition, the development of robust encapsulation or surface modification techniques to improve the stability and durability of 2D nanoparticles in biosensors is a promising strategy to improve biosensor performance. Studies have demonstrated the use of a polymer-based encapsulation method to enhance the long-term stability and reproducibility of GO nanosheets in an electrochemical biosensor for the detection of H. pylori antibodies.117,150,187 The encapsulated GO nanosheets exhibited minimal degradation and consistent performance over an extended period, offering a potential solution for commercialization and widespread adoption. Furthermore, the development of standardized protocols and quality control measures for the synthesis, characterization, and integration of 2D nanoparticles in biosensors is crucial for ensuring reproducibility and consistent performance. Collaborative efforts among research institutions, industry partners, and regulatory bodies are essential to establish guidelines and best practices for the fabrication and validation of these biosensing platforms, facilitating their acceptability and commercialization during epidemics.
4.4. Future directions and conclusions
The future of 2D nanoparticle-based biosensors for the detection of H. pylori holds immense potential, driven by advancements in multiplexed detection, point-of-care (POC) applications, integration with wearable and implantable devices, CRISPR-Cas technology, and the incorporation of machine learning (ML) and artificial intelligence (AI) algorithms. One significant direction for future research involves the development of multiplexed biosensors capable of simultaneously detecting multiple biomarkers associated with H. pylori infection. By incorporating a variety of recognition elements, such as antibodies, aptamers, and DNA probes, into the biosensor platform, researchers can enhance the specificity and accuracy of detection while enabling comprehensive profiling of the pathogen's biomolecular signatures. For example, recent studies have demonstrated the feasibility of multiplexed detection using arrays of functionalized graphene-based nanomaterials, enabling the simultaneous detection of H. pylori antigens, DNA sequences, and metabolic biomarkers.188–190 This multiplexed approach not only streamlines diagnostic workflows but also facilitates personalized treatment strategies tailored to individual patient profiles, thereby improving clinical outcomes and reducing healthcare costs associated with H. pylori-related diseases.Furthermore, the integration of 2D nanoparticle-based biosensors with POC devices represents a paradigm shift in biosensing technology, enabling rapid and decentralized diagnosis of H. pylori infection at the point of need. POC biosensors leverage miniaturized and portable platforms, such as microfluidic chips and smartphone-based devices, to enable real-time analysis of clinical samples in resource-limited settings. For instance, recent advancements in microfluidic-based biosensors have enabled the development of compact and user-friendly POC devices for the detection of pathogenic bacterial cells, antigens and DNA in saliva, blood, and stool samples.186,191,192 These POC biosensors offer significant advantages in terms of accessibility, affordability, and scalability, making them invaluable tools for community-based screening, outbreak surveillance, and remote monitoring of H. pylori infection. In addition, wearable and implantable devices integrated with biosensors hold promise for continuous and non-invasive monitoring of H. pylori infection dynamics in real-time. Wearable biosensors, such as smart patches and wearable bands, can detect biomarkers associated with H. pylori infection from bodily fluids, sweat, or breath, providing valuable insights into disease progression and treatment response.193 Implantable biosensors, on the other hand, offer long-term monitoring capabilities by interfacing directly with the host's physiological environment, enabling early detection of H. pylori-related complications, such as gastric ulcers and gastric cancer.194,195 These integrated diagnostic strategies pave the way for personalized and preemptive healthcare interventions, ultimately improving patient outcomes and reducing the burden of H. pylori-related diseases on healthcare systems worldwide.
CRISPR-Cas technology also offers promising results for biosensor-based disease diagnosis. This technology involves precision in targeting specific DNA or RNA sequences which enables highly sensitive and specific detection of pathogens or genetic mutations associated with diseases.196 A CRISPR/Cas12a-assisted array has been developed for the analysis of H. pylori DNA in saliva.197 This method utilizes the CRISPR/Cas12a system to detect specific genes of H. pylori, such as the 16S rDNA, CagA, and VacA genes.22,197,198 The assay process can be performed at a single temperature in about 30 minutes, making it rapid and convenient. The sensitivity of the method is 2 copies per μL and the LOD is about 50 copies.199 The CRISPR/Cas12a system has been shown to have high specificity for target recognition. The detection results can be displayed using lateral flow strips, allowing for easy interpretation.22,196–199 This method is simple and cost-effective, and can be widely applied in various environments, including primary hospitals, community clinics, and POC settings.
Moreover, the integration of ML and AI algorithms into 2D nanoparticle-based biosensors promises to revolutionize disease diagnosis by enhancing the accuracy, speed, and reliability of detection. ML algorithms leverage vast datasets of biosensor measurements to learn complex patterns and relationships between input signals and target analytes, enabling robust and adaptive detection algorithms. For example, recent studies have demonstrated the application of ML algorithms for data analysis and pattern recognition in graphene-based biosensors for disease detection, achieving high sensitivity and specificity compared to traditional signal processing techniques.200,201 Furthermore, AI-based diagnosis holds potential in overcoming major challenges faced by present biosensing technology, such as sensor drift, background noise, and interferences from complex biological matrices. Similarly, development of advanced models such as the convolutional neural network (CNN) model that could accurately classify the presence of disease biomarkers will revolutionize the detection techniques for H. pylori infections. By integrating ML and AI algorithms into biosensor platforms, researchers can develop smart diagnostic systems capable of real-time error correction, self-calibration, and adaptive signal processing, thus improving the reliability and reproducibility of H. pylori detection in diverse clinical settings.202 Additionally, the use of AI-based diagnosis opens new avenues for predictive analytics, disease modeling, and precision medicine, enabling proactive management of H. pylori infection and its associated complications.
Beyond H. pylori detection, these future technologies hold promise for addressing a wide range of healthcare challenges such as other infectious pathogens, cancer biomarkers, environmental pollutants, chronic conditions, personalized medicine, and biological warfare agents. For example, multiplexed biosensors and POC devices can be adapted to detect other infectious agents, such as bacteria, viruses (SARS-CoV-2 antigens), and parasites, enabling rapid diagnosis and surveillance of emerging infectious diseases.173,203–206 Wearable and implantable biosensors offer opportunities for continuous monitoring of biomarkers associated with chronic conditions, such as diabetes, cardiovascular disease, and cancer, enabling early detection of disease progression and timely intervention. Moreover, the integration of 2D nanomaterials with AI amplifies the potential of telemedicine. AI-powered systems optimize the release of therapeutic agents in drug delivery systems and improve the precision of disease detection through data analysis and modeling. Real-time data analysis allows for personalized healthcare by analyzing individual patient data, predicting disease risk, and optimizing treatment strategies based on patient-specific profiles, enabling immediate adjustments and tailored interventions. This convergence of 2D nanomaterials and AI fosters dynamic synergy, offering the prospect of elevated patient outcomes, reduced side effects, and the expedited development of cutting-edge healthcare solutions. Collectively, these future technologies have the potential to transform healthcare delivery, improve patient outcomes, and mitigate the global burden of disease, making them indispensable tools in the fight against H. pylori and beyond. Addressing the challenges associated with 2D nanoparticle-based biosensors is crucial for their widespread acceptance and commercialization as frontline diagnostic tools during public health crises. Commercialization of 2D-nanostructure-based biomedical products, as of today, has been lagging in its technologic development owing to multifaceted problems, not exclusively monetary or managerial but more of fundamental, including manufacturing at large scale (with), quality control (with precision in size and shape), and biosafety issues. However, overcoming these challenges requires interdisciplinary collaborations between materials scientists, engineers, bio-scientists and healthcare professionals to ensure the development of practical and deployable biosensing solutions tailored to the needs of epidemic management. Additionally, regulatory frameworks and quality assurance protocols must be established to guarantee the safety, efficacy, and reliability of 2D nanoparticle-based biosensors for use in clinical settings.
Data availability
No data was used for the research described in the article.Conflicts of interest
There are no conflicts to declare.References
- R. Bücker, M. Azevedo-Vethacke, C. Groll, D. Garten, C. Josenhans, S. Suerbaum and S. Schreiber, Helicobacter pylori colonization critically depends on postprandial gastric conditions, Sci. Rep., 2012, 2, 994 CrossRef.
- C. S. Goodwin, R. K. McCulloch, J. A. Armstrong and S. H. Wee, Unusual Cellular Fatty Acids and Distinctive Ultrastructure in a New Spiral Bacterium (Campylobacter Pyloridis) from the Human Gastric Mucosa, J. Med. Microbiol., 1985, 19, 257–267 CrossRef CAS.
- D. M. Jones, A. Curry and A. J. Fox, An Ultrastructural Study of the Gastric Campylobacter-like Organism ‘Campylobacter pyloridis’, Microbiology, 1985, 131, 2335–2341 CAS.
- S. Moens and J. Vanderleyden, Functions of Bacterial Flagella, Crit. Rev. Microbiol., 1996, 22, 67–100 CrossRef CAS.
- S. Hirukawa, H. Sagara, S. Kaneto, T. Kondo, K. Kiga, T. Sanada, H. Kiyono and H. Mimuro, Characterization of morphological conversion of Helicobacter pylori under anaerobic conditions, Microbiol. Immunol., 2018, 62, 221–228 CrossRef CAS PubMed.
- J. K. Y. Hooi, W. Y. Lai, W. K. Ng, M. M. Y. Suen, F. E. Underwood, D. Tanyingoh, P. Malfertheiner, D. Y. Graham, V. W. S. Wong, J. C. Y. Wu, F. K. L. Chan, J. J. Y. Sung, G. G. Kaplan and S. C. Ng, Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis, Gastroenterology, 2017, 153, 420–429 CrossRef.
- Y. Li, H. Choi, K. Leung, F. Jiang, D. Y. Graham and W. K. Leung, Global prevalence of Helicobacter pylori infection between 1980 and 2022: a systematic review and meta-analysis, Lancet Gastroenterol. Hepatol., 2023, 8, 553–564 CrossRef CAS.
- L. E. Wroblewski, R. M. Peek and K. T. Wilson, Helicobacter pylori and Gastric Cancer: Factors That Modulate Disease Risk, Clin. Microbiol. Rev., 2010, 23, 713–739 CrossRef CAS.
- N. Uemura, S. Okamoto, S. Yamamoto, N. Matsumura, S. Yamaguchi, M. Yamakido, K. Taniyama, N. Sasaki and R. J. Schlemper, Helicobacter pylori infection and the development of gastric cancer, N. Engl. J. Med., 2001, 345, 784 CrossRef CAS PubMed.
- C. Dunne, Factors that mediate colonization of the human stomach by Helicobacter pylori, World J. Gastroenterol., 2014, 20, 5610 CrossRef CAS PubMed.
- B. M. Roesler, E. M. A. Rabelo-Gonçalves and J. M. R. Zeitune, Virulence Factors of Helicobacter pylori: A Review, Clin. Med. Insights: Gastroenterol., 2014, 7, S13760 Search PubMed.
- N. Uemura, S. Okamoto, S. Yamamoto, N. Matsumura, S. Yamaguchi, M. Yamakido, K. Taniyama, N. Sasaki and R. J. Schlemper, Helicobacter pylori infection and the development of gastric cancer, N. Engl. J. Med., 2001, 345, 784 CrossRef CAS PubMed.
- S. C. Shah, M. B. Piazuelo, E. J. Kuipers and D. Li, AGA Clinical Practice Update on the Diagnosis and Management of Atrophic Gastritis: Expert Review, Gastroenterology, 2021, 161, 1325–1332 CrossRef CAS PubMed.
- B. Guevara and A. G. Cogdill, Helicobacter pylori: A Review of Current Diagnostic and Management Strategies, Dig. Dis. Sci., 2020, 65, 1917–1931 CrossRef.
- P. Malfertheiner, M. C. Camargo, E. El-Omar, J.-M. Liou, R. Peek, C. Schulz, S. I. Smith and S. Suerbaum, Helicobacter pylori infection, Nat. Rev. Dis. Prim., 2023, 9, 19 CrossRef PubMed.
- M. Ferwana, Accuracy of urea breath test in Helicobacter pylori infection: Meta-analysis, World J. Gastroenterol., 2015, 21, 1305 CrossRef.
- O. C. Aktepe, Five methods for detection of Helicobacter pylori in the Turkish population, World J. Gastroenterol., 2011, 17, 5172 CrossRef CAS PubMed.
- D. T. Quach, R. Aoki, A. Iga, Q. D. Le, T. Kawamura, K. Yamashita, S. Tanaka, M. Yoshihara and T. Hiyama, Diagnostic Accuracy of H. pylori Status by Conventional Endoscopy: Time-Trend Change After Eradication and Impact of Endoscopic Image Quality, Front. Med., 2022, 8, 830730 CrossRef.
- M. P. Dore and G. M. Pes, What Is New in Helicobacter pylori Diagnosis. An Overview, J. Clin. Med., 2021, 10, 2091 CrossRef CAS PubMed.
- D. S. Bordin, I. N. Voynovan, D. N. Andreev and I. V. Maev, Current Helicobacter pylori Diagnostics, Diagnostics, 2021, 11, 1458 CrossRef CAS PubMed.
- J. Y. Lee and N. Kim, Diagnosis of Helicobacter pylori by invasive test: histology, Ann. Transl. Med., 2015, 3, 10 Search PubMed.
- B. Dai, A. Xiang, D. Qu, G. Chen, L. Wang, W. Wang, D. Zhai, L. Wang and Z. Lu, Rapid and Sensitive Assay of Helicobacter pylori With One-Tube RPA-CRISPR/Cas12 by Portable Array Detector for Visible Analysis of Thermostatic Nucleic Acid Amplification, Front. Microbiol., 2022, 13, 858247 CrossRef PubMed.
- M. M. Ahmad, D. S. Ahmed, A. H. M. Rowshon, S. C. Dhar, M. Rahman, M. Hasan, C. Beglinger, N. Gyr and A. K. A. Khan, Long-Term Re-Infection Rate after Helicobacter pylori Eradication in Bangladeshi Adults, Digestion, 2007, 75, 173–176 Search PubMed.
- L. P. Andersen and T. Wadström, Helicobacter pylori: Physiology and Genetics, 2001 Search PubMed.
- N. Albertson, I. Wenngren and J. E. Sjöström, Growth and survival of Helicobacter pylori in defined medium and susceptibility to Brij 78, J. Clin. Microbiol., 1998, 36, 1232 CrossRef CAS PubMed.
- A. Shiotani, M. P. Dore and D. Y. Graham, Helicobacter pylori, Springer, Japan, Tokyo, 2016, pp. 143–155 Search PubMed.
- Y. Kohli, T. Kato, S. Ito, H. Miyazi, T. Azuma, K. Nagata, H. Tsuruta, S. Matsui, K. Oka and M. Nakamura, Diagnostic System for Helicobacter pylori Urease Based on a pHsensitive Field Effect Transistor, Dig. Endosc., 1995, 7, 27–34 CrossRef.
- A. B. Adamopoulos, G. S. Stergiou, G. N. Sakizlis, D. G. Tiniakos, E. G. Nasothimiou, D. K. Sioutis and A. D. Achimastos, Diagnostic value of rapid urease test and urea breath test for Helicobacter pylori detection in patients with Billroth II gastrectomy: a prospective controlled trial, Dig. Liver Dis., 2009, 41, 4–8 CrossRef CAS.
- N. Vakil and D. Vaira, Non-invasive tests for the diagnosis of H. pylori infection, Rev. Gastroenterol. Disord., 2004, 4, 1–6 Search PubMed.
- G. Oderda, Detection of Helicobacter pylori in stool specimens by non-invasive antigen enzyme immunoassay in children: multicentre Italian study, BMJ, 2000, 320, 347–348 CrossRef CAS.
- X. Zhou, J. Su, G. Xu and G. Zhang, Accuracy of stool antigen test for the diagnosis of Helicobacter pylori infection in children: A meta-analysis, Clin. Res. Hepatol. Gastroenterol., 2014, 38, 629–638 CrossRef CAS PubMed.
- R. A. Hussein, M. T. S. Al-Ouqaili and Y. H. Majeed, Detection of Helicobacter Pylori infection by invasive and non-invasive techniques in patients with gastrointestinal diseases from Iraq: A validation study, PLoS One, 2021, 16, e0256393 CrossRef CAS.
- Z. Rafiei-Sarmazdeh, S. Morteza Zahedi-Dizaji and A. Kafi Kang, Nanostructures, IntechOpen, 2020 Search PubMed.
- T. Hu, X. Mei, Y. Wang, X. Weng, R. Liang and M. Wei, Two-dimensional nanomaterials: fascinating materials in biomedical field, Sci. Bull., 2019, 64, 1707–1727 CrossRef CAS PubMed.
- V. K. Vaishnav, K. Prasad, R. Yadav, A. Aharwar and B. Nath Tiwary, Recent Advances in Biosensor Technology, Bentham Science Publishers, 2023, pp. 45–77 Search PubMed.
- D. Y. Graham and M. Miftahussurur, Helicobacter pylori urease for diagnosis of Helicobacter pylori infection: A mini review, J. Adv. Res., 2018, 13, 51–57 CrossRef CAS.
- V. Cussac, R. L. Ferrero and A. Labigne, Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions, J. Bacteriol., 1992, 174, 2466–2473 CrossRef CAS.
- A. Labigne, V. Cussac and P. Courcoux, Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity, J. Bacteriol., 1991, 173, 1920–1931 CrossRef CAS.
- H. C. Sharndama and I. E. Mba, Helicobacter pylori: an up-to-date overview on the virulence and pathogenesis mechanisms, Brazilian J. Microbiol., 2022, 53, 33–50 CrossRef CAS.
- M. Sugimoto and Y. Yamaoka, The association of vacA genotype and Helicobacter pylori-related disease in Latin American and African populations, Clin. Microbiol. Infect., 2009, 15, 835–842 CrossRef CAS PubMed.
- M. Karbalaei, A. Talebi Bezmin Abadi and M. Keikha, Clinical relevance of the cagA and vacA s1m1 status and antibiotic resistance in Helicobacter pylori: a systematic review and meta-analysis, BMC Infect. Dis., 2022, 22, 573 CrossRef.
- M. Hatakeyama, Structure and function of Helicobacter pylori CagA, the first-identified bacterial protein involved in human cancer, Proc. Jpn. Acad., Ser. B, 2017, 93, 196–219 CrossRef CAS PubMed.
- L. Jeyamani, J. Jayarajan, V. Leelakrishnan and M. Swaminathan, CagA and VacA genes of Helicobacter pylori and their clinical relevance, Indian J. Pathol. Microbiol., 2018, 61, 66 CrossRef.
- T. L. Cover, D. B. Lacy and M. D. Ohi, The Helicobacter pylori Cag Type IV Secretion System, Trends Microbiol., 2020, 28, 682–695 CrossRef CAS PubMed.
- J. M. Noto and R. M. Peek, The Helicobacter pylori cag Pathogenicity Island, Methods Mol. Biol., 2012, 921, 41–50 CrossRef CAS.
- K. Saxena, B. T. Murti, P.-K. Yang, B. D. Malhotra, N. Chauhan and U. Jain, Fabrication of a Molecularly Imprinted Nano-Interface-Based Electrochemical Biosensor for the Detection of CagA Virulence Factors of H. pylori, Biosensors, 2022, 12, 1066 CrossRef CAS.
- J. S. Vital, L. Tanoeiro, R. Lopes-Oliveira and F. F. Vale, Biomarker Characterization and Prediction of Virulence and Antibiotic Resistance from Helicobacter pylori Next Generation Sequencing Data, Biomolecules, 2022, 12, 691 CrossRef CAS PubMed.
- M. Oleastro, R. Cordeiro, A. Menard and J. P. Gomes, Allelic Diversity among Helicobacter pylori Outer Membrane Protein Genes homB and homA Generated by Recombination, J. Bacteriol., 2010, 192, 3961–3968 CrossRef CAS.
- X. Huang, D. Zhaomin, Q. Zhang, W. Li, B. Wang and M. Li, Relationship between the iceA gene of Helicobacter pylori and clinical outcomes, Ther. Clin. Risk Manag., 2016, 12, 1085–1092 CrossRef PubMed.
- Y.-L. Su, H.-L. Huang, B.-S. Huang, P.-C. Chen, C.-S. Chen, H.-L. Wang, P.-H. Lin, M.-S. Chieh, J.-J. Wu, J.-C. Yang and L.-P. Chow, Combination of OipA, BabA, and SabA as candidate biomarkers for predicting Helicobacter pylori-related gastric cancer, Sci. Rep., 2016, 6, 36442 CrossRef CAS PubMed.
- C. Anichini, W. Czepa, D. Pakulski, A. Aliprandi, A. Ciesielski and P. Samorì, Chemical sensing with 2D materials, Chem. Soc. Rev., 2018, 47, 4860–4908 RSC.
- J. Li, H. Zeng, Z. Zeng, Y. Zeng and T. Xie, Promising graphene-based nanomaterials and their biomedical applications and potential risks: A comprehensive review, ACS Biomater. Sci. Eng., 2021, 7, 5363–5396 CrossRef CAS PubMed.
- J. Wang, L. Sui, J. Huang, L. Miao, Y. Nie, K. Wang, Z. Yang, Q. Huang, X. Gong, Y. Nan and K. Ai, MoS2-based nanocomposites for cancer diagnosis and therapy, Bioact. Mater., 2021, 6, 4209–4242 CAS.
- J. Wang, G. Li and L. Li, Two-dimensional Materials – Synthesis, Characterization and Potential Applications, InTech, 2016 Search PubMed.
- Y. Chen, Z. Fan, Z. Zhang, W. Niu, C. Li, N. Yang, B. Chen and H. Zhang, Two-Dimensional Metal Nanomaterials: Synthesis, Properties, and Applications, Chem. Rev., 2018, 118, 6409–6455 CrossRef CAS PubMed.
- P. Nguyen Tri, C. Ouellet-Plamondon, S. Rtimi, A. A. Assadi and T. A. Nguyen, Noble Metal-Metal Oxide Hybrid Nanoparticles, Elsevier, 2019, pp. 51–63 Search PubMed.
- G. Habibullah, J. Viktorova and T. Ruml, Current Strategies for Noble Metal Nanoparticle Synthesis, Nanoscale Res. Lett., 2021, 16, 47 CrossRef CAS.
- R. Indiarto, L. P. A. Indriana, R. Andoyo, E. Subroto and B. Nurhadi, Bottom–up nanoparticle synthesis: a review of techniques, polyphenol-based core materials, and their properties, Eur. Food Res. Technol., 2022, 248, 1–24 CrossRef CAS.
- A. Triphati and T. B. Pirzadah, Synthesis of Bionanomaterials for Biomedical Applications, Elsevier, 2023, pp. 57–76 Search PubMed.
- T. Lage, R. O. Rodrigues, S. Catarino, J. Gallo, M. Bañobre-López and G. Minas, Graphene-Based Magnetic Nanoparticles for Theranostics: An Overview for Their Potential in Clinical Application, Nanomaterials, 2021, 11, 1073 CrossRef CAS.
- Z. Li, X. Li, M. Jian, G. S. Geleta and Z. Wang, Two-Dimensional Layered Nanomaterial-Based Electrochemical Biosensors for Detecting Microbial Toxins, Toxins, 2019, 12, 20 CrossRef PubMed.
- G. Sanità, B. Carrese and A. Lamberti, Nanoparticle Surface Functionalization: How to Improve Biocompatibility and Cellular Internalization, Front. Mol. Biosci., 2020, 7, 587012 CrossRef.
- A. Shakeri, S. Khan and T. F. Didar, Conventional and emerging strategies for the fabrication and functionalization of PDMS-based microfluidic devices, Lab Chip, 2021, 21, 3053–3075 RSC.
- W. Putzbach and N. Ronkainen, Immobilization Techniques in the Fabrication of Nanomaterial-Based Electrochemical Biosensors: A Review, Sensors, 2013, 13, 4811–4840 CrossRef CAS.
- Y. Zhou, Y. Fang and R. Ramasamy, Non-Covalent Functionalization of Carbon Nanotubes for Electrochemical Biosensor Development, Sensors, 2019, 19, 392 CrossRef PubMed.
- E. B. Aydin, M. Aydin and M. K. Sezginturk, New Developments in Nanosensors for Pharmaceutical Analysis, Elsevier, 2019, pp. 47–78 Search PubMed.
- D. Kala, T. K. Sharma, S. Gupta, V. Verma, A. Thakur, A. Kaushal, A. V. Trukhanov and S. V. Trukhanov, Graphene Oxide Nanoparticles Modified Paper Electrode as a Biosensing Platform for Detection of the htrA Gene of O. tsutsugamushi, Sensors, 2021, 21, 4366 CrossRef CAS.
- F. Löscher, T. Ruckstuhl, T. Jaworek, G. Wegner and S. Seeger, Immobilization of Biomolecules on Langmuir−Blodgett Films of Regenerative Cellulose Derivatives, Langmuir, 1998, 14, 2786–2789 CrossRef.
- A. Sassolas, L. J. Blum and B. D. Leca-Bouvier, Immobilization strategies to develop enzymatic biosensors, Biotechnol. Adv., 2012, 30, 489–511 CrossRef CAS PubMed.
- S. Khan, B. Burciu, C. D. M. Filipe, Y. Li, K. Dellinger and T. F. Didar, DNAzyme-Based Biosensors: Immobilization Strategies, Applications, and Future Prospective, ACS Nano, 2021, 15, 13943–13969 CrossRef CAS.
- V. Kathiresan, D. Thirumalai, T. Rajarathinam, M. Yeom, J. Lee, S. Kim, J.-H. Yoon and S.-C. Chang, A simple one-step electrochemical deposition of bioinspired nanocomposite for the non-enzymatic detection of dopamine, J. Anal. Sci. Technol., 2021, 12, 5 CrossRef CAS.
- R. Ahmad, O. S. Wolfbeis, Y.-B. Hahn, H. N. Alshareef, L. Torsi and K. N. Salama, Deposition of nanomaterials: A crucial step in biosensor fabrication, Mater. Today Commun., 2018, 17, 289–321 CrossRef CAS.
- S. Palani, J. P. Kenison, S. Sabuncu, T. Huang, F. Civitci, S. Esener and X. Nan, Multispectral Localized Surface Plasmon Resonance (msLSPR) Reveals and Overcomes Spectral and Sensing Heterogeneities of Single Gold Nanoparticles, ACS Nano, 2023, 17, 2266–2278 CrossRef CAS.
- V. Kumar, S. Mishra, R. Sharma, J. Agarwal, U. Ghoshal, T. Khanna, L. K. Sharma, S. K. Verma, P. Mishra and S. Tiwari, Development of RNA-Based Assay for Rapid Detection of SARS-CoV-2 in Clinical Samples, Intervirology, 2022, 65, 181–187 CrossRef CAS.
- Y. Huang, X. Dong, Y. Liu, L.-J. Li and P. Chen, Graphene-based biosensors for detection of bacteria and their metabolic activities, J. Mater. Chem., 2011, 21, 12358 RSC.
- D. Kwong Hong Tsang, T. J. Lieberthal, C. Watts, I. E. Dunlop, S. Ramadan, A. E. del Rio Hernandez and N. Klein, Chemically Functionalised Graphene FET Biosensor for the Label-free Sensing of Exosomes, Sci. Rep., 2019, 9, 13946 CrossRef PubMed.
- L. Chen, G. Li, A. Yang, J. Wu, F. Yan and H. Ju, A DNA-functionalized graphene field-effect transistor for quantitation of vascular endothelial growth factor, Sensors Actuators B Chem., 2022, 351, 130964 CrossRef CAS.
- H. Du, J. Ye, J. Zhang, X. Huang and C. Yu, Graphene Nanosheets Modified Glassy Carbon Electrode as a Highly Sensitive and Selective Voltammetric Sensor for Rutin, Electroanalysis, 2010, 22, 2399–2406 CrossRef CAS.
- A. M. Haredy, S. M. Derayea, A. A. Gahlan, M. A. Omar and G. A. Saleh, Graphene oxide modified glassy carbon electrode for determination of linagliptin in dosage form, biological fluids, and rats’ feces using square wave voltammetry, Arab. J. Chem., 2022, 15, 103663 CrossRef CAS.
- S. Bagherzadeh-Nobari and R. Kalantarinejad, Real-time label-free detection of DNA hybridization using a functionalized graphene field effect transistor: a theoretical study, J. Nanoparticle Res., 2021, 23, 185 CrossRef CAS PubMed.
- J. Yamaguchi, H. Hayashi, H. Jippo, A. Shiotari, M. Ohtomo, M. Sakakura, N. Hieda, N. Aratani, M. Ohfuchi, Y. Sugimoto, H. Yamada and S. Sato, Small bandgap in atomically precise 17-atom-wide armchair-edged graphene nanoribbons, Commun. Mater., 2020, 1, 36 Search PubMed.
- H. Chen, B. Luo, S. Wu, S. Shi, Q. Dai, Z. Peng and M. Zhao, Microfluidic Biosensor Based on Molybdenum Disulfide (MoS2) Modified Thin-Core Microfiber for Immune Detection of Toxoplasma gondii, Sensors, 2023, 23, 5218 CAS.
- A. K. Bhatia and S. Dewangan, Handbook of Biomolecules, Elsevier, 2023, pp. 259–274 Search PubMed.
- A. Asadzadeh-Firouzabadi, H. R. Zare and N. Nasirizadeh, Electrochemical Biosensor for Detection of Target DNA Sequence and Single-Base Mismatch Related to Helicobacter Pylori Using Chlorogenic Acid as Hybridization Indicator, J. Electrochem. Soc., 2016, 163, B43–B48 CAS.
- S. Gupta, U. Jain, B. T. Murti, A. D. Putri, A. Tiwari and N. Chauhan, Nanohybrid-based immunosensor prepared for Helicobacter pylori BabA antigen detection through immobilized antibody assembly with @ Pdnano/rGO/PEDOT sensing platform, Sci. Rep., 2020, 10, 21217 CrossRef CAS PubMed.
- K. Saxena, N. Chauhan, B. D. Malhotra and U. Jain, A molecularly imprinted polymer-based electrochemical biosensor for detection of VacA virulence factor of H. pylori causing gastric cancer, Process Biochem., 2023, 130, 87–95 CrossRef CAS.
- H. Jaradat, B. M. Hryniewicz, I. A. Pašti, T. L. Valério, A. Al-Hamry, L. F. Marchesi, M. Vidotti and O. Kanoun, Detection of H. pylori outer membrane protein (HopQ) biomarker using electrochemical impedimetric immunosensor with polypyrrole nanotubes and carbon nanotubes nanocomposite on screen-printed carbon electrode, Biosens. Bioelectron., 2024, 249, 115937 CrossRef CAS.
- T. B. A. Akib, S. Mostufa, M. M. Rana, M. B. Hossain and M. R. Islam, A performance comparison of heterostructure surface plasmon resonance biosensor for the diagnosis of novel coronavirus SARS-CoV-2, Opt. Quantum Electron., 2023, 55, 448 CrossRef CAS PubMed.
- T. N. Ghosh, D. R. Rotake and S. G. Singh, 2D vanadium disulfide nanosheets assisted ultrasensitive, rapid, and label-free electrochemical quantification of cancer biomarker (MMP-2), Nanotechnology, 2023, 34, 395501 CrossRef CAS PubMed.
- C. Rodriguez-Emmenegger, E. Brynda, T. Riedel, M. Houska, V. Šubr, A. B. Alles, E. Hasan, J. E. Gautrot and W. T. S. Huck, Polymer Brushes Showing Non-Fouling in Blood Plasma Challenge the Currently Accepted Design of Protein Resistant Surfaces, Macromol. Rapid Commun., 2011, 32, 952–957 CrossRef CAS PubMed.
- H. Chen, Z. Wang, Z. Liu, Q. Niu, X. Wang, Z. Miao, H. Zhang, J. Wei, M. Wan and C. Mao, Construction of 3D electrochemical cytosensor by layer-by-layer assembly for ultra-sensitive detection of cancer cells, Sensors Actuators B Chem., 2021, 329, 128995 CrossRef CAS.
- R. Wang, H. You, Y. Zhang, Z. Li, Y. Ding, Q. Qin, H. Wang, J. Sun, Y. Jia, F. Liu, J. Ma and X. Cheng, Constructing (reduced) graphene oxide enhanced polypyrrole/ceramic composite membranes for water remediation, J. Memb. Sci., 2022, 659, 120815 CrossRef CAS.
- W. Zhang, Z. Tian, S. Yang, J. Rich, S. Zhao, M. Klingeborn, P.-H. Huang, Z. Li, A. Stout, Q. Murphy, E. Patz, S. Zhang, G. Liu and T. J. Huang, Electrochemical micro-aptasensors for exosome detection based on hybridization chain reaction amplification, Microsystems Nanoeng., 2021, 7, 63 CrossRef CAS.
- M. T. Hwang, M. Heiranian, Y. Kim, S. You, J. Leem, A. Taqieddin, V. Faramarzi, Y. Jing, I. Park, A. M. van der Zande, S. Nam, N. R. Aluru and R. Bashir, Ultrasensitive detection of nucleic acids using deformed graphene channel field effect biosensors, Nat. Commun., 2020, 11, 1543 CrossRef CAS.
- J. Wang, D. Chen, W. Huang, N. Yang, Q. Yuan and Y. Yang, Aptamer-functionalized field-effect transistor biosensors for disease diagnosis and environmental monitoring, Exploration, 2023, 3, 20210027 CrossRef PubMed.
- L. Wang, K. Cui, P. Wang, M. Pei and W. Guo, A sensitive electrochemical DNA sensor for detecting Helicobacter pylori based on accordion-like Ti3C2Tx: a simple strategy, Anal. Bioanal. Chem., 2021, 413, 4353–4362 CrossRef CAS PubMed.
- H. Zhao and Y. Lei, 3D Nanostructures for the Next Generation of High-Performance Nanodevices for Electrochemical Energy Conversion and Storage, Adv. Energy Mater., 2020, 10(28), 2001460 CrossRef CAS.
- M. Byakodi, N. S. Shrikrishna, R. Sharma, S. Bhansali, Y. Mishra, A. Kaushik and S. Gandhi, Emerging 0D, 1D, 2D, and 3D nanostructures for efficient point-of-care biosensing, Biosens. Bioelectron. X, 2022, 12, 100284 CAS.
- E. Qiu, Z. Li and S. Han, Methods for detection of Helicobacter pylori from stool sample: current options and developments, Brazilian J. Microbiol., 2021, 52, 2057–2062 CrossRef PubMed.
- S. Su and P. M. Kang, Systemic Review of Biodegradable Nanomaterials in Nanomedicine, Nanomaterials, 2020, 10, 656 CrossRef CAS.
- R. Vainionpää and P. Leinikki, Encyclopedia of Virology, Elsevier, 2008, pp. 29–37 Search PubMed.
- C. Fernandes, M. Jathar, B. K. S. Sawant and T. Warde, Scale-Up of Nanoparticle Manufacturing Process, Pharmaceutical Process Engineering and Scale-up Principles, 2023vol. 13, pp. 173–203 Search PubMed.
- H. Zhu, H. Zhang, Y. Xu, S. Laššáková, M. Korabečná and P. Neužil, PCR past, present and future, Biotechniques, 2020, 69, 317–325 CrossRef CAS.
- E. Andresen, F. Islam, C. Prinz, P. Gehrmann, K. Licha, J. Roik, S. Recknagel and U. Resch-Genger, Assessing the reproducibility and up-scaling of the synthesis of Er,Yb-doped NaYF4-based upconverting nanoparticles and control of size, morphology, and optical properties, Sci. Rep., 2023, 13, 2288 CrossRef CAS PubMed.
- L. Gautier, Reproducibility in the lab: the proof is in the protocol, Biotechniques, 2022, 73, 71–74 CrossRef CAS.
- J. P. Gallagher, P. A. Twohig, A. Crnic and F. A. Rochling, Illicit Drug Use and Endoscopy: When Do We Say No?, Dig. Dis. Sci., 2022, 67, 5371–5381 CrossRef PubMed.
- M. Khalid and M. Abdollahi, Toxicity of inorganic nanoparticles, Compr. Anal. Chem., 2022, 99, 25–85 Search PubMed.
- G. Chakraborty, I.-H. Park, R. Medishetty and J. J. Vittal, Two-Dimensional Metal-Organic Framework Materials: Synthesis, Structures, Properties and Applications, Chem. Rev., 2021, 121, 3751–3891 CrossRef CAS.
- T. R. Kyriakides, A. Raj, T. H. Tseng, H. Xiao, R. Nguyen, F. S. Mohammed, S. Halder, M. Xu, M. J. Wu, S. Bao and W. C. Sheu, Biocompatibility of nanomaterials and their immunological properties, Biomed. Mater., 2021, 16, 042005 CrossRef CAS PubMed.
- S. S. Mirzaei, N. Mehrdadi, M. Pourmadadi, M. Ahmadi and S. Meknatkhah, Novel detection of H.pylori using ultrasensitive electrochemical aptasensor based on surface modified graphene oxide doped gold nanoparticles conjugated polythiophene, Microchem. J., 2024, 200, 110279 CrossRef.
- B. Cai, Z. Xia, J. Wang, S. Wu and X. Jin, Reduced Graphene Oxide-Based Field Effect Transistor Biosensors for High-Sensitivity miRNA21 Detection, ACS Appl. Nano Mater., 2022, 5, 12035–12044 CrossRef CAS.
- Y. Yim, H. Shin, S. M. Ahn and D.-H. Min, Graphene oxide-based fluorescent biosensors and their biomedical applications in diagnosis and drug discovery, Chem. Commun., 2021, 57, 9820–9833 RSC.
- A. M. Salama, G. Yasin, M. Zourob and J. Lu, Fluorescent Biosensors for the Detection of Viruses Using Graphene and Two-Dimensional Carbon Nanomaterials, Biosensors, 2022, 12, 460 CrossRef CAS PubMed.
- T. Yang and T. V. Duncan, Challenges and potential solutions for nanosensors intended for use with foods, Nat. Nanotechnol., 2021, 16, 251–265 CrossRef CAS PubMed.
- S. Yamamoto, A. Tanaka, S. Kobayashi, Y. Oshiro, M. Ozeki, K. Maeda, K. Matsuda, K. Miyo, T. Mizoue, W. Sugiura, H. Mitsuya, H. Sugiyama and N. Ohmagari, Consistency of the results of rapid serological tests for SARS-CoV-2 among healthcare workers in a large national hospital in Tokyo, Japan, Glob. Heal. Med., 2021, 3, 90–94 CrossRef.
- O. Yilmaz, N. Sen, A. A. Küpelioğlu and I. Simşek, Detection of H. pylori infection by ELISA and western blot techniques and evaluation of anti CagA seropositivity in adult Turkish dyspeptic patients, World J. Gastroenterol., 2006, 12, 5375 CrossRef PubMed.
- K. Saxena, A. Kumar, N. Chauhan, M. Khanuja, B. D. Malhotra and U. Jain, Electrochemical Immunosensor for Detection of H. pylori Secretory Protein VacA on g-C 3 N 4/ZnO Nanocomposite-Modified Au Electrode, ACS Omega, 2022, 7, 32292–32301 CrossRef CAS.
- E. Janik-Karpinska, M. Ceremuga, M. Niemcewicz, M. Podogrocki, M. Stela, N. Cichon and M. Bijak, Immunosensors—The Future of Pathogen Real-Time Detection, Sensors, 2022, 22, 9757 CrossRef CAS.
- S. Kizhepat, A. S. Rasal, J.-Y. Chang and H.-F. Wu, Development of Two-Dimensional Functional Nanomaterials for Biosensor Applications: Opportunities, Challenges, and Future Prospects, Nanomaterial, 2023, 13, 1520 CrossRef CAS.
- I. S. Raja, M. Vedhanayagam, D. R. Preeth, C. Kim, J. H. Lee and D. W. Han, Development of Two-Dimensional Nanomaterials Based Electrochemical Biosensors on Enhancing the Analysis of Food Toxicants, Int. J. Mol. Sci., 2021, 22, 3277 CrossRef CAS PubMed.
- R. Khan, A. Radoi, S. Rashid, A. Hayat, A. Vasilescu and S. Andreescu, Two-Dimensional Nanostructures for Electrochemical Biosensor, Sensors, 2021, 21, 3369 CrossRef CAS.
- R. Hao, L. Liu, J. Yuan, L. Wu and S. Lei, Recent Advances in Field Effect Transistor Biosensors: Designing Strategies and Applications for Sensitive Assay, Biosensors, 2023, 13, 426 CrossRef CAS PubMed.
- X. Su and S. F. Li, Serological determination of Helicobacter pylori infection using sandwiched and enzymatically amplified piezoelectric biosensor, Anal. Chim. Acta, 2001, 429, 27–36 CrossRef.
- A. Kumar, H. K. Rathore, D. Sarkar and A. Shukla, Nanoarchitectured transition metal oxides and their composites for supercapacitors, Electrochem. Sci. Adv., 2022, 2, e2100187 CrossRef CAS.
- L. M. H. Al-Ameri, M. L. Israa and Saqari, The Optical Biosensor was used to Determine Antibiotic Micsagainst H. pylori, J. Res. Med. Dent. Sci., 2022, 10, 275–280 Search PubMed.
- P. M. Arun Kumar Gupta, Ashwini Yadav and Parismita Koch, Biosensors in Food Safety and Quality, 1st edn, 2022, p. 10 Search PubMed.
- B. Dick and M. Jones, The commercialization of molecular biology: Walter Gilbert and the Biogen startup, Hist. Technol., 2017, 33, 126–151 CrossRef.
- F. Gong, H. Wei, Q. Li, L. Liu and B. Li, Evaluation and Comparison of Serological Methods for COVID-19 Diagnosis, Front. Mol. Biosci., 2021, 8, 682405 CrossRef CAS.
- S. Malik, J. Singh, R. Goyat, Y. Saharan, V. Chaudhry, A. Umar, A. A. Ibrahim, S. Akbar, S. Ameen and S. Baskoutas, Nanomaterials-based biosensor and their applications: A review, Heliyon, 2023, 9, e19929 CrossRef CAS.
- R. Abdel-Karim, Nanotechnology-Enabled Biosensors: A Review of Fundamentals, Materials, Applications, Challenges, and Future Scope, Biomed. Mater. Dev., 2024, 2, 759–777 Search PubMed.
- A. Seitak, S. Luo, N. Cai, K. Liao, A.-M. Pappa, S. Lee and V. Chan, Emergence of MXene-based electrochemical biosensors for biomolecule and pathogen detection, Sensors and Actuators Reports, 2023, 6, 100175 CrossRef.
- D. Hristov, C. Rodriguez-Quijada, J. Gomez-Marquez and K. Hamad-Schifferli, Designing Paper-Based Immunoassays for Biomedical Applications, Sensors, 2019, 19, 554 CrossRef.
- S. A. Hashemi, S. M. Mousavi, S. Bahrani and S. Ramakrishna, Integrated polyaniline with graphene oxide-iron tungsten nitride nanoflakes as ultrasensitive electrochemical sensor for precise detection of 4-nitrophenol within aquatic media, J. Electroanal. Chem., 2020, 873, 114406 CrossRef CAS.
- L. Chen, E. Hwang and J. Zhang, Fluorescent Nanobiosensors for Sensing Glucose, Sensors, 2018, 18, 1440 CrossRef PubMed.
- S. Senapati, A. K. Mahanta, S. Kumar and P. Maiti, Controlled drug delivery vehicles for cancer treatment and their performance, Signal Transduct. Target. Ther., 2018, 3, 7 CrossRef PubMed.
- M. R. Bernsen, M. van Straten, G. Kotek, E. A. H. Warnert, J. C. Haeck, A. Ruggiero, P. A. Wielopolski and G. P. Krestin, Computed Tomography and Magnetic Resonance Imaging, Recent Results Cancer Res., 2020, 216, 31–110 CrossRef CAS PubMed.
- V. Srimaneepong, H. E. Skallevold, Z. Khurshid, M. S. Zafar, D. Rokaya and J. Sapkota, Graphene for Antimicrobial and Coating Application, Int. J. Mol. Sci., 2022, 23, 499 CrossRef CAS.
- P. J. Vikesland, Nanosensors for water quality monitoring, Nat. Nanotechnol., 2018, 13, 651–660 CrossRef CAS PubMed.
- S. Dhall, B. R. Mehta, A. K. Tyagi and K. Sood, A review on environmental gas sensors: Materials and technologies, Sensors Int., 2021, 2, 100116 CrossRef.
- Z. Wang and B. Mi, Environmental Applications of 2D Molybdenum Disulfide (MoS2) Nanosheets, Environ. Sci. Technol., 2017, 51, 8229–8244 CrossRef CAS PubMed.
- A. Aghababai Beni and H. Jabbari, Nanomaterials for Environmental Applications, Results Eng., 2022, 15, 100467 CrossRef CAS.
- J. Chen, Y. Guo, X. Zhang, J. Liu, P. Gong, Z. Su, L. Fan and G. Li, Emerging Nanoparticles in Food: Sources, Application, and Safety, J. Agric. Food Chem., 2023, 71, 3564–3582 CrossRef CAS PubMed.
- R. Biswas, M. Alam, A. Sarkar, M. I. Haque, M. M. Hasan and M. Hoque, Application of nanotechnology in food: processing, preservation, packaging and safety assessment, Heliyon, 2022, 8, e11795 CrossRef CAS.
- W. Wen, Y. Song, X. Yan, C. Zhu, D. Du, S. Wang, A. M. Asiri and Y. Lin, Recent advances in emerging 2D nanomaterials for biosensing and bioimaging applications, Mater. Today, 2018, 21, 164–177 CrossRef CAS.
- Z. Chen, T. Aziz, H. Sun, A. Ullah, A. Ali, L. Cheng, R. Ullah and F. U. Khan, Advances and Applications of Cellulose Bio-Composites in Biodegradable Materials, J. Polym. Environ., 2023, 31, 2273–2284 CrossRef CAS.
- S. S. Mirzaei, M. Pourmadadi, A. Foroozandeh, A. A. Moghaddam, M. Soltani, N. Basirhaghighi and M. Ahmadi, Enhancing H. pylori detection: ultrasensitive electrochemical aptasensor with Au-doped CQDs and polythiophene conjugation, J. Appl. Electrochem., 2024, 54, 1887–1900 CrossRef CAS.
- U. Jain, S. Gupta, S. Soni, M. P. Khurana and N. Chauhan, Triple-nanostructuring-based noninvasive electro-immune sensing of CagA toxin for Helicobacter pylori detection, Helicobacter, 2020, 25, e12706 CrossRef CAS PubMed.
- M. Mathew and C. S. Rout, Electrochemical biosensors based on Ti3C2Tx MXene: future perspectives for on-site analysis, Curr. Opin. Electrochem., 2021, 30, 100782 CrossRef CAS.
- J. Chao, D. Zhu, Y. Zhang, L. Wang and C. Fan, DNA nanotechnology-enabled biosensors, Biosens. Bioelectron., 2016, 76, 68–79 CrossRef CAS.
- S. Hajihosseini, N. Nasirizadeh, M. S. Hejazi and P. Yaghmaei, A sensitive DNA biosensor fabricated from gold nanoparticles and graphene oxide on a glassy carbon electrode, Mater. Sci. Eng. C, 2016, 61, 506–515 CrossRef CAS.
- M. V. del Pozo, C. Alonso, F. Pariente and E. Lorenzo, DNA Biosensor for Detection of Helicobacter pylori Using Phen-dione as the Electrochemically Active Ligand in Osmium Complexes, Anal. Chem., 2005, 77, 2550–2557 CrossRef CAS PubMed.
- A. K. Yadav, D. Verma, S. Tabassum and P. R. Solanki, Functionalized Graphitic Carbon Nitride Based Aptasensing Platform for Electrochemical Detection of Helicobacter Pylori, IEEE Sensors Lett., 2023, 7, 1–4 Search PubMed.
- Z. Li, W. Zhang and F. Xing, Graphene Optical Biosensors, Int. J. Mol. Sci., 2019, 20, 2461 CrossRef PubMed.
- F. Ma, C. Li and C. Zhang, Development of quantum dot-based biosensors: principles and applications, J. Mater. Chem. B, 2018, 6, 6173–6190 RSC.
- M. Shahrashoob, A. Mohsenifar, M. Tabatabaei, T. Rahmani-Cherati, M. Mobaraki, A. Mota and T. R. Shojaei, Detection of Helicobacter Pylori Genome with an Optical Biosensor Based on Hybridization of Urease Gene with a Gold Nanoparticles-Labeled Probe, J. Appl. Spectrosc., 2016, 83, 322–329 CrossRef CAS.
- M. Rong, G. Kavokin and A. Sawan, Optical Biosensor Based on Porous Silicon and Tamm Plasmon Polariton for Detection of CagA Antigen of Helicobacter Pylori, Sensors, 2024, 24, 5153 CrossRef PubMed.
- J. Tan, W. Geng, J. Li, Z. Wang, S. Zhu and X. Wang, Colorimetric and Fluorescence Dual-Mode Biosensors Based on Peroxidase-Like Activity of the Co3O4 Nanosheets, Front. Chem., 2022, 10, 871013 CrossRef CAS PubMed.
- D. Zhu, B. Liu and G. Wei, Two-Dimensional Material-Based Colorimetric Biosensors: A Review, Biosensors, 2021, 11, 259 CrossRef CAS PubMed.
- G. A. R. Y. Suaifan, M. B. Shehadeh, R. M. Darwish, M. Alterify, W. Abu Jbara, F. Abu Jbara, N. Alaridah and M. Zourob, Magnetic beads-based nanozyme for portable colorimetric biosensing of Helicobacter pylori, Biosens. Bioelectron. X, 2024, 20, 100517 CAS.
- Y. Fei, R. Fang, L. Xiao, Y. Zhang, K. Fan, Y. Jiang, S. Lei, R. Xu, D. Yang, Y. Ye, S. Xiang, P. Wang, C. Zhou and T. Tang, The development of a colorimetric biosensing assay for the detection of Helicobacter pylori in feces, Anal. Biochem., 2022, 651, 114737 CrossRef CAS PubMed.
- Y. Kohli, T. Kato, S. Ito, H. Miyaji, T. Azuma, H. Tsuruta, S. Matsui, K. Oka and M. Nakamura, Endoscopic Urease Sensor System for Detecting Helicobacter pylori on Gastric Wall Using a pH-sensitive Field Effect Transistor, Dig. Endosc., 1995, 7, 215–219 CrossRef.
- D. Sadighbayan, M. Hasanzadeh and E. Ghafar-Zadeh, Biosensing based on field-effect transistors (FET): Recent progress and challenges, TrAC Trends Anal. Chem., 2020, 133, 116067 CrossRef CAS PubMed.
- C.-C. Huang, Y.-H. Kuo, Y.-S. Chen, P.-C. Huang and G.-B. Lee, A miniaturized, DNA-FET biosensor-based microfluidic system for quantification of two breast cancer biomarkers, Microfluid. Nanofluidics, 2021, 25, 33 CrossRef CAS.
- L. Chao, H. Shi, K. Nie, B. Dong, J. Ding, M. Long and Z. Liu, Applications of Field Effect Transistor Biosensors Integrated in Microfluidic Chips, Nanosci. Nanotechnol. Lett., 2020, 12, 427–445 CrossRef.
- S. Lakshmi, S. Sundar, S. Menon, S. N. Kesana and M. G. Varshitha, 4th International Conference on Circuits, Control, Communication and Computing (I4C), IEEE, 2022, pp. 212–215.
- G. Dyankov, T. Eftimov, E. Hikova, H. Najdenski, V. Kussovski, P. Genova-Kalou, V. Mankov, H. Kisov, P. Veselinov, S. S. Ghaffari, M. Kovacheva-Slavova, B. Vladimirov and N. Malinowski, SPR and Double Resonance LPG Biosensors for Helicobacter pylori BabA Antigen Detection, Sensors, 2024, 24, 2118 CrossRef CAS PubMed.
- P. Kolev, E. Hikova, H. Kisov and G. Dyankov, Biosensor-based serological assay for diagnosing Helicobacter pylori infection, IOP Conf. Ser. Earth Environ. Sci., 2024, 1305, 012019 CrossRef.
- H. Kim, H. Choi, Y. Heo, C. Kim, M. Kim and K. T. Kim, Biosensors Based on Bivalent and Multivalent Recognition by Nucleic Acid Scaffolds, Appl. Sci., 2022, 12, 1717 CrossRef CAS.
- A. Sharma, K. Dulta, R. Nagraik, K. Dua, S. K. Singh, D. Kumar and D.-S. Shin, Potentialities of aptasensors in cancer diagnosis, Mater. Lett., 2022, 308, 131240 CrossRef CAS.
- M. B. Kulkarni, N. H. Ayachit and T. M. Aminabhavi, Recent Advancements in Nanobiosensors: Current Trends, Challenges, Applications, and Future Scope, Biosensors, 2022, 12, 892 CrossRef CAS.
- M. Zhang, C. Liao, C. H. Mak, P. You, C. L. Mak and F. Yan, Highly sensitive glucose sensors based on enzyme-modified whole-graphene solution-gated transistors, Sci. Rep., 2015, 5, 8311 CrossRef CAS.
- Biosensor Market Size & Share Analysis - Growth Trends & Forecasts (2024–2029) Source: https://www.mordorintelligence.com/industry-reports/biosensor-market.
- T. Beduk, D. Beduk, M. R. Hasan, E. Guler Celik, J. Kosel, J. Narang, K. N. Salama and S. Timur, Smartphone-Based Multiplexed Biosensing Tools for Health Monitoring, Biosensors, 2022, 12, 583 CrossRef PubMed.
- J. Yoon, J. Lim, M. Shin, S.-N. Lee and J.-W. Choi, Graphene/MoS2 Nanohybrid for Biosensors, Materials, 2021, 14, 518 CrossRef CAS.
- X. Sun, J. Fan, C. Fu, L. Yao, S. Zhao, J. Wang and J. Xiao, WS2 and MoS2 biosensing platforms using peptides as probe biomolecules, Sci. Rep., 2017, 7, 10290 CrossRef.
- S. Afsahi, M. B. Lerner, J. M. Goldstein, J. Lee, X. Tang, D. A. Bagarozzi, D. Pan, L. Locascio, A. Walker, F. Barron and B. R. Goldsmith, Novel graphene-based biosensor for early detection of Zika virus infection, Biosens. Bioelectron., 2018, 100, 85–88 CrossRef CAS PubMed.
- R. Li, Y. Wang, J. Du, X. Wang, A. Duan, R. Gao, J. Liu and B. Li, Graphene oxide loaded with tumor-targeted peptide and anti-cancer drugs for cancer target therapy, Sci. Rep., 2021, 11, 1725 CrossRef CAS.
- P. Basu, A. Banerjee, P. D. Okoro, A. Masoumi, B. Kanjilal, M. Akbari, M. Martins-Green, D. G. Armstrong and I. Noshadi, Integration of Functional Polymers and Biosensors to Enhance Wound Healing, Adv. Healthcare Mater., 2024, 13, 2401461 CrossRef CAS PubMed.
- S. Shrestha, B. Wang and P. Dutta, Nanoparticle processing: Understanding and controlling aggregation, Adv. Colloid Interface Sci., 2020, 279, 102162 CrossRef CAS.
- H. Rasuli and R. Rasuli, Nanoparticle-decorated graphene/graphene oxide: synthesis, properties and applications, J. Mater. Sci., 2023, 58, 2971–2992 CrossRef CAS.
- J. Wu, H. Lin, D. J. Moss, K. P. Loh and B. Jia, Graphene oxide for photonics, electronics and optoelectronics, Nat. Rev. Chem., 2023, 7, 162–183 CrossRef PubMed.
- S. Zhang and C. Wang, Precise Analysis of Nanoparticle Size Distribution in TEM Image, Methods Protoc., 2023, 6, 63 CrossRef CAS.
- J. Wang, K. Fu, X. Zhang, Q. Yin, G. Wei and Z. Su, When MoS 2 meets TiO 2: facile synthesis strategies, hybrid nanostructures, synergistic properties, and photocatalytic applications, J. Mater. Chem. C, 2021, 9, 8466–8482 RSC.
- Y. Wu, J.-Y. Chen and W.-M. He, Surface-enhanced Raman spectroscopy biosensor based on silver nanoparticles@metal-organic frameworks with peroxidase-mimicking activities for ultrasensitive monitoring of blood cholesterol, Sensors Actuators B Chem., 2022, 365, 131939 CrossRef CAS.
- Y. F. Duan, Y. Ning, Y. Song and L. Deng, Fluorescent aptasensor for the determination of Salmonella typhimurium based on a graphene oxide platform, Microchim. Acta, 2014, 181, 647–653 CAS.
- Y. Xie, X. Zhi, H. Su, K. Wang, Z. Yan, N. He, J. Zhang, D. Chen and D. Cui, A Novel Electrochemical Microfluidic Chip Combined with Multiple Biomarkers for Early Diagnosis of Gastric Cancer, Nanoscale Res. Lett., 2015, 10, 477 Search PubMed.
- R. Salimian, S. Shahrokhian and S. Panahi, Enhanced Electrochemical Activity of a Hollow Carbon Sphere/Polyaniline-Based Electrochemical Biosensor for HBV DNA Marker Detection, ACS Biomater. Sci. Eng., 2019, 5, 2587–2594 CrossRef CAS PubMed.
- Z. Qian, X. Shan, L. Chai, J. Chen and H. Feng, Simultaneous Detection of Multiple DNA Targets by Integrating Dual-Color Graphene Quantum Dot Nanoprobes and Carbon Nanotubes, Chem. – A Eur. J., 2014, 20, 16065–16069 CAS.
- S. He, B. Song, D. Li, C. Zhu, W. Qi, Y. Wen, L. Wang, S. Song, H. Fang and C. Fan, A Graphene Nanoprobe for Rapid, Sensitive, and Multicolor Fluorescent DNA Analysis, Adv. Funct. Mater., 2010, 20, 453–459 CAS.
- P. Labroo and Y. Cui, Graphene nano-ink biosensor arrays on a microfluidic paper for multiplexed detection of metabolites, Anal. Chim. Acta, 2014, 813, 90–96 CAS.
- F. Mi, C. Hu, Y. Wang, L. Wang, F. Peng, P. Geng and M. Guan, Recent advancements in microfluidic chip biosensor detection of foodborne pathogenic bacteria: a review, Anal. Bioanal. Chem., 2022, 414, 2883–2902 CAS.
- M. B. Kulkarni, N. H. Ayachit and T. M. Aminabhavi, Recent Advances in Microfluidics-Based Electrochemical Sensors for Foodborne Pathogen Detection, Biosensors, 2023, 13, 246 CAS.
- S. Chen, Z. Qiao, Y. Niu, J. C. Yeo, Y. Liu, J. Qi, S. Fan, X. Liu, J. Y. Lee and C. T. Lim, Wearable flexible microfluidic sensing technologies, Nat. Rev. Bioeng., 2023, 1, 950–971 CAS.
- M. Gray, J. Meehan, C. Ward, S. P. Langdon, I. H. Kunkler, A. Murray and D. Argyle, Implantable biosensors and their contribution to the future of precision medicine, Vet. J., 2018, 239, 21–29 CAS.
- X. Mei, D. Ye, F. Zhang and C. Di, Implantable application of polymer-based biosensors, J. Polym. Sci., 2022, 60, 328–347 CrossRef CAS.
- P. Puig-Serra, M. C. Casado-Rosas, M. Martinez-Lage, B. Olalla-Sastre, A. Alonso-Yanez, R. Torres-Ruiz and S. Rodriguez-Perales, CRISPR Approaches for the Diagnosis of Human Diseases, Int. J. Mol. Sci., 2022, 23, 1757 CrossRef CAS.
- X. Zhang, H. Qiu, X. Zhong, S. Yi, Z. Jia, L. Chen and S. Hu, A CRISPR/Cas12a-assisted array for Helicobacter pylori DNA analysis in saliva, Anal. Chim. Acta, 2023, 1239, 340736 CrossRef CAS PubMed.
- E. Qiu, S. Jin, Z. Xiao, Q. Chen, Q. Wang, H. Liu, C. Xie, C. Chen, Z. Li and S. Han, CRISPR-based detection of Helicobacter pylori in stool samples, Helicobacter, 2021, 26, 12828 CrossRef.
- H. Liu, J. Wang, X. Hu, X. Tang and C. Zhang, A rapid and high-throughput Helicobacter pylori RPA-CRISPR/Cas12a-based nucleic acid detection system, Clin. Chim. Acta, 2023, 540, 117201 CrossRef CAS PubMed.
- F. Cui, Y. Yue, Y. Zhang, Z. Zhang and H. S. Zhou, Advancing Biosensors with Machine Learning, ACS Sens., 2020, 5, 3346–3364 CrossRef CAS.
- S. Kadian, P. Kumari, S. Shukla and R. Narayan, Recent advancements in machine learning enabled portable and wearable biosensors, Talanta Open, 2023, 8, 100267 CrossRef.
- P. P. Das, R. Nagraik, A. Sharma, T. K. Upadhyay, H. Lalhlenmawia, D. Balram, K.-Y. Lian, J. Singh and D. Kumar, recent update on biomimetic sensor technology for cancer diagnosis, Talanta Open, 2024, 9, 100276 CrossRef.
- R. Verma, S. K. Yadav, R. B. K. Singh, R. Verma, D. Kumar and J. Singh, Laccase-Conjugated Nanostructured ZnFe2O4/rGO-Modified Electrode-Based Interfaces for Electrochemical Impedance Monitoring of Adrenaline: A Promising Biosensor for Management of Neurodegenerative Disorders, ACS Appl. Bio Mater., 2023, 6(12), 5842–5853 CrossRef CAS PubMed.
- S. S. Chu, H. A. Nguyen, J. Zhang, S. Tabassum and H. Cao, Towards Multiplexed and Multimodal Biosensor Platforms in Real-Time Monitoring of Metabolic Disorders, Sensors, 2022, 14, 5200 CrossRef PubMed.
- R. Tomichan, A. Sharma, K. Akash, A. A. Siddiqui, A. Dubey, T. K. Upadhyay, D. Kumar, S. Pandey and R. Nagraik, Insight of smart biosensors for COVID-19: A review, Luminescence, 2023, 38, 1102 CrossRef PubMed.
- B. Gil Rosa, O. E. Akingbade, X. Guo, L. Gonzalez-Macia, M. A. Crone, L. P. Cameron, P. Freemont, K.-L. Choy, F. Güder, E. Yeatman, D. J. Sharp and B. Li, Multiplexed immunosensors for point-of-care diagnostic applications, Biosens. Bioelectron., 2022, 203, 114050 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2025 |