 Open Access Article
Open Access ArticleBiocomposites of 2D layered materials
Mert
Vural
and
Melik C.
Demirel
 *
*
Center for Research on Advanced Fiber Technologies (CRAFT), Materials Research Institute and Huck Institute of Life Sciences, Pennsylvania State University, University Park, Pennsylvania 16802, USA. E-mail: melik@psu.edu
First published on 7th January 2025
Abstract
Molecular composites, such as bone and nacre, are everywhere in nature and play crucial roles, ranging from self-defense to carbon sequestration. Extensive research has been conducted on constructing inorganic layered materials at an atomic level inspired by natural composites. These layered materials exfoliated to 2D crystals are an emerging family of nanomaterials with extraordinary properties. These biocomposites are great for modulating electron, photon, and phonon transport in nanoelectronics and photonic devices but are challenging to translate into bulk materials. Combining 2D crystals with biomolecules enables various 2D nanocomposites with novel characteristics. This review has provided an overview of the latest biocomposites, including their structure, composition, and characterization. Layered biocomposites have the potential to improve the performance of many devices. For example, biocomposites use macromolecules to control the organization of 2D crystals, allowing for new capabilities such as flexible electronics and energy storage. Other applications of 2D biocomposites include biomedical imaging, tissue engineering, chemical and biological sensing, gas and liquid filtration, and soft robotics. However, some fundamental questions need to be answered, such as self-assembly and kinetically limited states of organic–inorganic phases in soft matter physics.
1. Introduction
The controlled assembly of 2D crystals with biomolecules is of particular interest as it allows for precise control over the physical properties of the resulting material. To achieve this, it is crucial to understand and control the molecular interactions between 2D crystals and complementary bio-systems. Using molecular biology tools, biomolecular-based materials that can initiate multiple physical or chemical interactions with 2D crystals are suitable for constructing functional bio-inorganic composites with programmable properties. This can lead to the development of several materials and devices for electronic, terahertz, and optics applications. Examples include metallic composites and organic conductor composites for electronic applications, insulating 2D graphene oxide for actuators and bolometers in terahertz applications, and protein-based waveguides, filters, and optical switches for optics applications. The foundation of next-generation, programmable, flexible, optically superior, energy-efficient, and mechanically solid materials and devices is established by 2D-layered materials.In this review, we will thoroughly examine the interactions between biomolecules and 2D-layered composites. The paper will have three main sections: first, we will discuss the composition and structure of 2D biocomposites. Second, we will delve into various types of biomolecules used in 2D-layered composites, such as polysaccharides and polypeptides. Finally, we will summarize the unique properties that result from the interaction between these biomolecules and 2D-layered composites.
The exfoliation of layered materials into 2D crystals, starting with graphite,1 introduced a new family of nanomaterials that exhibit extraordinary physical and chemical properties.2,3 Beyond the individual qualities of these 2D materials, their proper combination with other materials projects a broad spectrum of composite materials with novel characteristics.3–7 This prospect has developed significant research focus on 2D nanocomposites, in which 2D crystals, including graphene,3,8,9 graphene oxide,10,11 transition metal dichalcogenides (TMDCs),12–15 2D metal carbides, and nitrides (MXenes), single-element 2D-Xenes,16,17 2D dielectrics,18–20 and black phosphorus21,22 are coupled with organic and inorganic materials. These supplementary materials are synthesized,23,24 assembled,11,25–27 or processed28,29 with 2D crystals to form functional composite materials. Inorganic materials are commonly synthesized or processed with 2D materials such as composites consisting of different 2D materials (heterostructures),3,24,25 and nanomaterials (nanoparticles, nanotubes, and nanowires).30,31 Inorganic 2D nanocomposites having a particular order in the nanoscale are promising for modulating electron,31–33 photon,34,35 and phonon36 transport in nanoelectronics and photonic devices. However, it isn’t easy to translate the precise structure of these nanocomposites into bulk materials and extend their utility beyond laboratory-scale device applications.37 Unlike inorganic 2D nanocomposites, composites composed of organic materials can be assembled and processed into bulk materials, hence finding a broader field of applications including flexible electronics,38–40 and energy storage,16,41,42 biomedical imaging,43,44 chemical, and biological sensing.45–48 However, organic 2D nanocomposites offer limited control over the organization of 2D crystals, mainly governed by the percolation of 2D crystals in an organic matrix.26,28,29 The inferior control over the order of 2D crystals in organic 2D nanocomposites limits the utility of these composites in device applications. In recent years, research on organic 2D nanocomposites consisting of biomaterials has demonstrated that it is possible to control the organization of 2D crystals in these composites by exploiting the assembly mechanics of biomacromolecules, biopolymers, and bio-derived polymers.10,26,40,49–51
Biomolecular building blocks of 2D biocomposites commonly consist of polysaccharides (e.g., cellulose, chitosan, alginates), polypeptides (e.g., proteins), nucleic acids, and lipids. These biomacromolecules can form strong interactions with pristine or functionalized 2D crystals.6 Hence, the initial studies concerning 2D biocomposites focus on exfoliating layered materials into 2D crystals with the help of biomacromolecules.40,52–54 As the understanding of the interactions between 2D crystals and biomacromolecules built up, researchers started to develop bioinspired nacre-like materials that are much stronger and tougher than conventional composite materials.4,49,55,56 The structural contribution of biomacromolecules to these biocomposite materials instigated more detailed research regarding the assembly mechanics of 2D crystals and biomacromolecules.10,26,40,49 These studies unveiled a new set of materials in which biomacromolecules can modulate the physical aspects of composite materials beyond mechanical properties. The templating capability of biomacromolecules can help alter electronic,26,51,57 thermal,10,58–60 and optical50,60,61 properties of composite materials via precise control over the structural organization of 2D crystals. The simplest biocomposites in which 2D crystals form a percolative network in biomacromolecule matrices can establish electronic materials that sustain a significant portion of their physical properties under mechanical deformation.40,50,59,60,62 This feature is essential for materials that are employed in flexible electronics. Biomacromolecules can more profoundly influence the structural organization of 2D crystals in composites, where intercalating biomacromolecules can alter the interlayer spacing between 2D crystals with angstrom-level precision.10,26,49 This templating capability can induce drastic changes in the isotropic and anisotropic electronic properties of biocomposites of 2D crystals. In addition, it is possible to generate electronic memory effects in these biocomposites.26 Biomacromolecules can facilitate distinct interactions with chemical and biological materials. Once coupled to 2D crystals, these specific interactions can incite a change in their electronic properties.26,40 This is the primary mechanism of detection for sensor devices based on 2D biocomposites.40,45 Beyond electronic properties, biomacromolecules can also help regulate thermal transport in composites of 2D crystals.10,58,60 Depending on the structural influence of the biomacromolecules on 2D crystals, 2D biocomposites can exhibit extraordinarily high and low (insulator) thermal conductivity.58,60 Similar to electronic transport, the isotropy of thermal transport can be modulated by combining 2D crystals with proper biomacromolecules.58,60 The structural impact of biomacromolecules on the organization of 2D crystals is significant in molding the light-matter interactions in 2D biocomposites.40,50,60,61 Biomacromolecules can help 2D crystals alter their photoconductance through structural effects and specific interactions.63 More importantly, biomacromolecules can induce a distinct separation between 2D crystals to drastically enhance their ability to trap electromagnetic waves, which results in 2D biocomposites with record-breaking electromagnetic shielding capabilities.40,50 In addition to the structural order provided by biomacromolecules in the nanoscale, 2D crystals can also be combined with biomacromolecules to form microscale structures, which is more relevant for applications in energy storage.16,64,65 The nanoscale order helps form channels for optimal ion transport, while the microscale order ensures the surface area needed to facilitate electrochemical reactions in energy storage systems.64,65 Biomacromolecules also contribute to the structural integrity of these energy storage systems.64,65 Besides energy storage, composites of biomacromolecules and 2D crystals show promise in transducing electrical, photonic, and thermal input into mechanical actuation.10,53,66 2D biocomposite actuators offer excellent alternative materials for applications in soft robotics, as they can be easily implemented into biological systems.53,66 Other vital contributions of biomacromolecules to 2D composites are biocompatibility and the capability to interact with biological systems.67–75 These features of 2D biocomposites are vitally crucial for biomedical applications such as tissue engineering,67,70,71 drug delivery,68,69,72,74,75 and antimicrobial materials.73 The most recent field of application for 2D biocomposites is gas and liquid filtration, which utilizes the nacre-like structure of 2D biocomposites to form filtration membranes and specific interactions of biomacromolecules to trap targeted ions, molecules, and particles in these membranes.76–80
2. Structure and composition of 2D biocomposites
The introduction of 2D crystals with the demonstration of graphene presented a new set of materials with extraordinary physical and chemical properties.1,81 After graphene, a plethora of 2D materials are extracted from layered materials by exfoliating single-few layered 2D crystals (graphene: 10 layers, MoS2: 6 layers) or growing 2D crystals on specific substrates.3 These 2D crystals exhibit extraordinary mechanical,82 electronic,2,3 thermal,36,83,84 and optical22,85–87 properties, significantly different from the bulk source material. However, these novel properties of 2D crystals diminish when the number of layers reaches a critical limit or individual 2D crystals interact.3,81 These phenomena necessitate proper spacing between 2D crystals in composite systems to translate a significant portion of their unique aspects into composite materials. This engenders a bottleneck; as the volumetric fraction of 2D crystals decreases in composites, their contribution to physical properties drops exponentially, which is dictated by percolation theory.29 Biomacromolecules can offer alternative assembly mechanics that differ from the composite matrix's random percolative distribution of 2D crystals.26,88,89 These bio-macromolecular templates control the assembly of 2D crystals in biocomposites by facilitating specific interactions with 2D crystals and intercalating in between 2D crystals (Fig. 1(a)-(ii)).10,26,40,49,51 Using the templating capability of natural and synthetic biomacromolecules, including polysaccharides90 and polypeptides91(Fig. 1(a)-(ii)), it is possible to reflect the unique properties of 2D crystals into bulk materials10,26,49 and nurture new properties in 2D biocomposites.10,26,49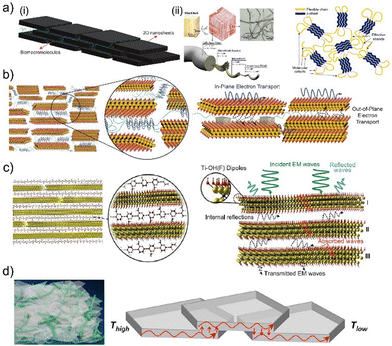 | ||
| Fig. 1 (a) Schematic illustration of (i) self-assembly in biocomposites, (ii) hierarchical structure of cellulose from wood to nanofibrils, (iii) microstructure of polycrystalline proteins. Adapted with permission.49,90,92 Copyright 2022, NAS, 2013, ACS, 2020, Springer Nature, respectively. (b) Schematics of the structure and electron transport in molecular composites consisting of synthetic repetitive proteins and conductive 2D crystals. Adapted with permission.26 Copyright 2020, ACS. (c) Schematic of the structure and electromagnetic wave propagation in molecular composites consisting of naturally derived polysaccharides and conductive 2D crystals. Adapted with permission.50 Copyright 2016, AAAS. (d) Schematics of the structure and thermal wave propagation in molecular composites consisting of cellulose and hexagonal boron nitride crystals. Adapted with permission.93 Copyright 2014, ACS. | ||
A recent study on 2D biocomposites composed of MXene nanosheets and recombinant proteins with repetitive amino acid sequences demonstrated that proteins consisting of 2D crystals could form a network between MXene nanosheets. This allowed MXene nanosheets to be processed using inkjet printing as responsive 2D biocomposites, which exhibit similar electronic properties as ordered MXene films.40 A follow-up study has revealed that it is possible to systematically modify the interlayer spacing between MXene nanosheets using several repeat units in proteins as tuning parameters (Fig. 1(b)).26 Systematic control over interlayer spacing between MXene nanosheets offers the ability to govern electron tunneling in these 2D biocomposites, which can be explained using a modified percolation theory.26 This theory accurately predicts in-plane and out-of-plane electronic transport in 2D biocomposites until the interlayer spacing between 2D crystals becomes large enough to accommodate 2D crystals of synthetic proteins (Fig. 1(b)).26 Once the number of tandem repeats in the protein sequence is sufficient to facilitate the formation of 2D protein crystals between MXene nanosheets, these 2D biocomposites demonstrate the electronic memory effect, which is absent in pristine MXene and protein films.26
Another 2D biocomposite of MXene nanosheets has utilized alginate to separate MXene nanosheets (Fig. 1(c)).50 The primary role of alginate is to interconnect MXene nanosheets and improve mechanical integrity. However, the detailed investigation of the electromagnetic interference (EMI) shielding capabilities of these 2D biocomposites revealed another subtle but more important role of alginate: the separation it provides between MXene nanosheets (Fig. 1(c)).50 This separation enhances the formation of internal reflections between MXene nanosheets, drastically improving the 2D biocomposite's EMI shielding efficiency. This structural contribution makes this material one of the best EMI shielding materials.50
Biocomposites of cellulose and hexagonal boron nitride (h-BN) nanosheets can be engineered to facilitate extraordinarily high93 or low94 thermal conductivity. Vacuum-assisted flocculation can help establish biocomposites of densely packed h-BN nanosheets in cellulose matrices (Fig. 1(d)).93 These composites with nanoscale order have thermal conductivities higher than films of pristine h-BN nanosheets and other composites of h-BN nanosheets (Fig. 1(d)).93,94 Cellulose acts as a binder in between h-BN nanosheets. Consequently, it enhances the interconnectivity of nanosheets and the contact area between nanosheets. This is the fundamental structural contribution of cellulose to the organization of h-BN nanosheets, which manifests itself as extraordinarily high thermal conductivity.93
In complementary to the structural templating capability of biomacromolecules, the ability to form specific interactions with 2D crystals is also critical to engineering functional materials from composites of biomaterials and 2D materials.6 Biomacromolecules, including polysaccharides, polypeptides, nucleic acids, and lipids, can establish electrostatic, hydrophobic, π–π electron interactions, covalent and hydrogen bonding with pristine or modified 2D crystals.6 These interactions can be strong enough to promote the exfoliation of 2D crystals from their bulk-layered source material (Fig. 2(a)-(i)–(iii)).52,53,95–98 After exfoliation, biomacromolecules attached to the surface of 2D crystals form inclusion complexes. These inclusion complexes can help disperse these single—or few-layered 2D crystals in aqueous or non-aqueous solvents.97 Orchestrating the intercalation of these complexes is the essence of controlling the structure and properties of resulting 2D biocomposites. In some instances, biomacromolecules interact more effectively with 2D crystals than the interactions they form with each other.89,93,99 Biomacromolecules act as molecular separators/intercalants in these composite materials (Fig. 2(b)).89,93,99 In contrast to the simple structure of these biocomposites, macromolecules can construct more complex designs since they can establish different molecular interactions with each other and 2D crystals (Fig. 2(b)-(i)–(iii)).10,26,27,49,51 Globular proteins (e.g., bovine serum albumin, BSA) can facilitate hydrophobic interactions with MoS2 and electrostatic interactions with WO3 nanosheets.27 It is possible to employ BSA to facilitate the formation of 2D heterostructures using solution-mediated self-assembly (Fig. 2(c)).27 This is an excellent alternative for building 2D heterostructures since current methods require rigorous material growth and fabrication methodologies.3 Beyond naturally derived proteins, recombinant proteins provide better control over possible interactions between proteins and 2D crystals.6,10,26,49 Synthetic proteins can be designed to exhibit specific interactions with 2D crystals while they can still initiate multiple molecular interactions among themselves. This brings out another structural contribution: the ability to control the organization of 2D crystals with Angstrom-level precision and facilitate the formation of secondary structures between 2D crystals.10,26,49 This generates unprecedented changes in the physical and chemical properties of the resulting 2D biocomposites.10,26,49
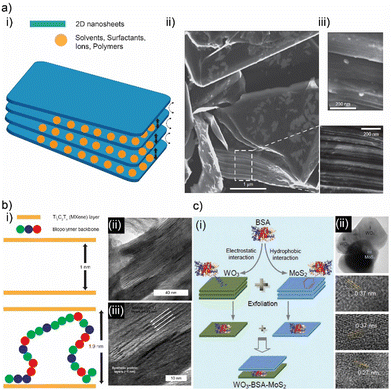 | ||
| Fig. 2 (a) (i) Schematic illustration of intercalating molecules between 2D crystals. Scanning electron microscopy images of graphite nanosheets separated by phosphoric acid particles in perspective of graphite (ii) cross-section and (iii) plane. Adapted with permission.98 Copyright 2014, Springer Nature. (b) (i) Schematic illustration of intercalating biopolymers between 2D crystals. (ii) Transmission electron microscopy images of biocomposites consisting of synthetic repetitive proteins intercalating in MXene nanocrystals. Adapted with permission.26 Copyright 2020, ACS. (c) (i) Schematic illustration of alternating stacking of various transition dichalcogenide 2D crystals facilitated using bioderived polypeptides. (ii) Transmission electron microscopy images of stacked 2D crystals and spacer polypeptides. Adapted with permission.27 Copyright 2017, Wiley-VCH. | ||
3. Family of 2D biocomposites
The advances in the spectrum of 2D materials have paved the way for new composite materials. In particular, recent research focus on composites consisting of 2D materials and macromolecules, biopolymers have led to a new set of biocomposites that can inherit the extraordinary properties of 2D materials and exhibit novel physicochemical properties originating from the diverse templating capabilities of biomaterials such as polysaccharides, polypeptides, and nucleic acids.6,100 Polysaccharides are considered the most stable matrix material for composites compared to other macromolecules since they do not chemically degrade with moderate changes in temperature.101 Besides stability, polysaccharides provide versatility in physical and chemical properties for their composites, which stem from their ability to form linear and branched biopolymers with various molecular weights.101 Polypeptides offer more diverse structural templates for 2D materials than polysaccharides due to possible amino acid sequences of natural and recombinant proteins. The amino acid sequence defines protein folding, assembly, as well as secondary and tertiary structures.100,102 Beyond individual structural characteristics of polypeptides, amino acid sequences can also be engineered to modulate the structural organization of 2D materials in a protein matrix that delineates biocomposites’ exact physical and chemical properties.10,26,49 Unlike polysaccharides that offer control over molecular weight and polymer structure, polypeptides grant additional parameters, including amino acid sequence and stereochemistry, to govern the design of biocomposites of 2D materials with Angstrom-level precision.10,26,493.1 Polysaccharide-based 2D biocomposites
Naturally abundant biomacromolecules and biopolymers of polysaccharides have recently manifested themselves as reliable alternatives to synthetic polymers as matrix material in composites. Common polysaccharide-based matrix materials include cellulose, chitosan, alginate, and pectin. These biopolymers acquired from natural resources form sustainable and biodegradable composites with 2D crystals. Biocomposites of polysaccharides and 2D crystals can exhibit extraordinary mechanical,61,103 thermal,58,93,104–106 electronic,99,107,108 electromagnetic,50,109,110 and electrochemical111–113 properties.Graphene, particularly its water-processable derivatives like graphene oxide (GO) and reduced graphene oxide (rGO), generates functional biocomposites with various forms of cellulose. Functionalized cellulose crystals can interact with oxidized graphene crystals via hydrogen bonding, ionic121 and hydrophilic interactions.78,107 In the case of bacterial cellulose, oxidized graphene crystals can initiate covalent bonding with the help of mediator molecules.122,123 Proteins and polysaccharides can also be used to connect graphene and its derivatives.97,124 The structural organization of graphene crystals is defined by the initial form of cellulose (cellulose with microfibrils, nanofibrils, or bulk wood chips with cellular structure) and the density of the biocomposites they establish with cellulose. Cellulose is an excellent template material for constructing micro and nanoporous structures, which is essential to engineering low-density biocomposite materials.101,114 Nanofibrils of cellulose can be effectively processed with aqueous solutions of GO to fabricate hydrogels. (Fig. 3(a)-(i)).76 The trapped water in these hydrogels can be removed using freeze drying to form biocomposite foams consisting of a nano fibrillar network of cellulose and GO (Fig. 3(a)-(ii)).76 GO is reduced once treated under excessive heat, and a conductive network of rGO is established (Fig. 3(a)-(ii)). Conductive nano-fibrillar networks can sustain their structure through cyclic deformations, which also helps them maintain electrical conductivity during mechanical deformation (Fig. 3(a)-(iii)).76 Besides electrical properties, nanocellulose-based foam structures can govern thermal transport in biocomposites (Fig. 3(b)-(i)).105 Biocomposite foams assembled using freeze drying of nanocellulose/GO hydrogels can also be designed to have higher porosity (Fig. 3(b)-(ii) and (iii)).105 In addition, implementing a heat gradient during hydrogel formation can help establish the formation of pores in a specific direction (Fig. 3(b)-(ii) and (iii)).105 This makes these biocomposites exhibit enhanced mechanical strength and highly anisotropic thermal conductivity (radial: 15 mW mK−1, axial: 170 mW mK−1) (Fig. 3(b)-(iv)).105 In contrast to microporous biocomposites, shrinking the pore size of biocomposites foams of cellulose and graphene derivatives to the nanoscale has a serious impact on energy storage devices relying on electrochemistry (Fig. 3(c)-(i) and (ii)).113 The high surface area of the hierarchical nanostructure established from the hydrogels of nanofibrillated cellulose and reduced graphene oxide helps realize solid-state supercapacitors with excellent specific capacitance (207 F g−1) and cyclic stability (Fig. 3(c)-(iii) and (iv)).113 Unlike low-density biocomposites, cellulose nanocrystals can govern the molecular assembly characteristics of graphene-based filler materials in compact biocomposites with higher density. Instead of building fibrillar cellulose templates that support graphene-based fillers, it is possible to utilize the specific interactions between cellulose and graphene derivatives to modulate the organization of graphene crystals in compact biocomposites.
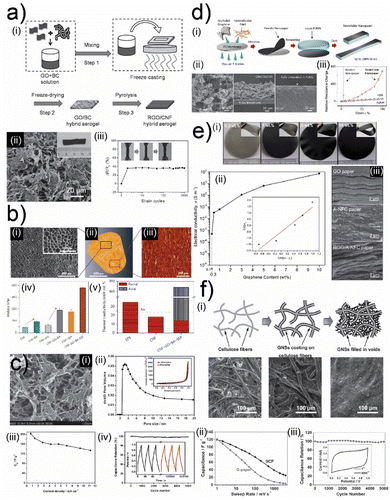 | ||
| Fig. 3 (a) (i) Schematic illustration of fabrication for hydrogels and aerogels composed of graphene oxide (GO) cellulose. (ii) Scanning electron microscopy images of GO-cellulose aerogels. (iii) Normalized resistance of GO-cellulose aerogels during cyclic deformation. Adapted with permission.76 Copyright 2017, Wiley-VCH. (b) (i) Scanning electron microscopy images of composites of nanofibrillated cellulose foams and GO crystals. (ii, iii) Reconstruction and original of X-ray microtomography images of foam composites of cellulose and GO crystals. (iv) Tensile modulus and (v) cellulose and GO crystal foam composites thermal conductivity. Adapted with permission.105 Copyright 2015, Springer Nature. (c) (i) Scanning electron microscopy and (ii) pore volume images of highly porous GO-cellulose foams. Measurements of (iii) specific capacitance and (iv) capacitance retention during charge/discharge cycles. Adapted with permission.113 Copyright 2013, RSC. (d) (i) Schematic illustration of fabrication for molecular composites of graphene crystals and fibrillar nanocellulose and their transfer on soft substrates. (ii) Scanning electron microscopy images of molecular composites transferred on soft substrates. (iii) Normalized resistance changes of molecular composites on soft substrates during continuous tensile deformation. Adapted with permission.99 Copyright 2014, Wiley-VCH. (e) (i) Images of graphene and nanofibrillar cellulose molecular composites with different graphene filler concentrations. (ii) Conductivity of graphene and nanofibrillar cellulose molecular composites with different graphene concentrations. (iii) Scanning electron microscopy images of graphene, nanofibrillar cellulose nacres, and molecular composites of graphene and nanofibrillar cellulose. Adapted with permission.125 Copyright 2011, RSC. (f) (i) Schematic illustration and SEM images of nanofibrillated cellulose, graphene-coated nanofibrillated cellulose, and composites of nanofibrillated cellulose and graphene crystals. Measurements of (ii) specific capacitance and (iii) capacitance retention during charge/discharge cycles for nanofibrillated cellulose and graphene crystal composites. Adapted with permission.111 Copyright 2011, Wiley-VCH. | ||
The earlier studies investigating amine functionalized cellulose assembly mechanics with rGO suggest that the filler and the matrix material interact through hydrogen bonding (Fig. 3(d)-(i) and (ii)).99 This strong interaction leads to the formation of biocomposites that exhibit good mechanical strength and electrical conductivity (Fig. 3(d)-(iii)).99 This robust composite can be interfaced with elastomeric substrates to construct stretchable electrodes that can maintain a significant portion of their conductivity during deformation (Fig. 3(d)-(iii)).99 Even though the fibrillar cellulose matrix did not impose a specific structural order, it acted as a bridge between conductive rGO nanoplatelets in molecular composites of cellulose and rGO (Fig. 3(e)-(i)–(iii)).125 This structural influence helped these biocomposites to exhibit a low percolation threshold (Fig. 3(e)-(ii)).125 Consequently, these biocomposites can reach high electrical conductivity values with moderate concentrations of rGO nanoplatelets (Fig. 3(e)-(ii)).125 The porous template facilitated by the cellulose matrix is an excellent host for electrochemically active high-surface-area materials like graphene to establish high-performance electrochemical energy storage systems (Fig. 3(f)-(i)).111 This cellulose-templated composite can reach promising gravimetric capacitance values of 120 F per gram of graphene (Fig. 3(f)-(ii)).111 More importantly, this robust network structure enables the fabrication of robust supercapacitor systems that can operate consistently throughout many cycles (Fig. 3(f)-(iii)).111
Besides graphene, cellulose can also form conductive biocomposites with other 2D crystals such as layered MAX (M-early transition metals, A-Group A elements, and X–C, N, and additional functional surface groups) phases, i.e., MXenes (Fig. 4(a)–(c)).16,61 Vacuum-assisted self-assembly (VASA) of cellulose nanofibrils and MXene nanosheets through solution processing leads to a nacre-like structure (Fig. 4(a)-(i), (ii) and (b)-(i), (ii)). Biocomposites composed of larger nanofibrils exhibit lower electrical conductivity (8 S cm−1) due to increased separation between MXene nanosheets (Fig. 4(a)-(iii)).61 This structure also demonstrates extraordinary EMI shielding performance as the waves get trapped in between MXene nanosheets (Fig. 4(a)-(iv)).61 Cellulose nanofibrils with smaller sizes lead to biocomposites with electrical conductivity values reaching 1000 S cm−1 (Fig. 4(b)-(iii)).118 The nacre-like structure with smaller separation in between MXene nanosheets helps these biocomposites establish extraordinary mechanical properties and specific capacitance for energy storage applications (Fig. 4(b)-(iv)).118 The extraordinary mechanical properties of these biocomposites originate from the hydrogen bonding formation between cellulose nanofibrils and MXene nanosheets (Fig. 4(a) and (b)).61,118 This significant interaction is also utilized in the 3D printing of these biocomposites (Fig. 4(c)-(i)).126 3D-printed biocomposites demonstrate the percolative distribution of MXene nanosheets in a nano fibrillar cellulose matrix (Fig. 4(c)-(i)), which offers a more random distribution than self-assembled biocomposites.126 This random distribution has minimal influence on mechanical properties (Fig. 4(c)-(ii)), but it leads to drastically low electrical conductivity (Fig. 4(c)-(iii)).126
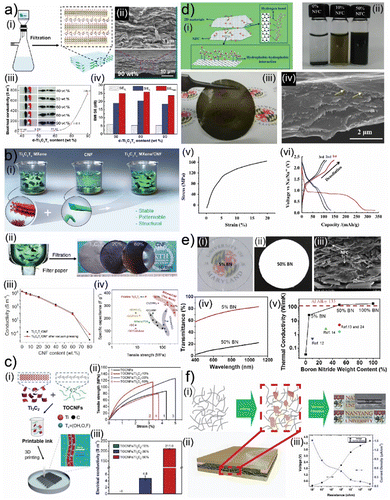 | ||
| Fig. 4 (a) (i) Schematic illustration of self-assembly for molecular composites of cellulose and MXene crystals. (ii) Scanning electron microscopy images of molecular composites of cellulose and MXene crystals. (iii) Conductivity of molecular composites of cellulose and MXene crystals with different concentrations of MXene crystals. (iv) Electromagnetic interference (EMI) shielding properties of molecular composites of cellulose and MXene crystals with different concentrations of MXene crystals. Adapted with permission.61 Copyright 2018, ACS. (b) (i) Schematic illustration and (ii) images of self-assembly for molecular composites of nanofibrillated cellulose and MXene crystals. (iii) The conductivity of molecular composites of nanofibrillated cellulose and MXene crystals with different concentrations of MXene crystals. (iv) Specific capacitance and tensile strength of molecular composites of nanofibrillated cellulose and MXene crystals. Adapted with permission.127 Copyright 2019, Wiley-VCH. (c) (i) Schematic illustration of self-assembly for molecular composite inks of cellulose and MXene crystals. (ii) Tensile strength of composites printed from inks of cellulose and MXene crystals. (iii) The conductivity of composites printed from inks of cellulose and MXene crystals. Adapted with permission.126 Copyright 2019, Wiley-VCH. (d) (i) Schematic illustration of self-assembly and exfoliation of 2D crystals through nanofibrillated cellulose. (ii) Images of 2D crystal solutions exfoliated using nanofibrillated cellulose. (iii) Images and (iv) scanning electron microscopy images of molecular composites of nanofibrillated cellulose and molybdenum disulfide crystals. (v) Stress/strain and (vi) charge/discharge profiles of molecular composites of nanofibrillated cellulose and molybdenum disulfide crystals. Adapted with permission.128 Copyright 2015, Elsevier. (e) (i, ii) Images of hexagonal boron nitride and nanofibrillated cellulose molecular composites. (iii) Scanning electron microscopy image, (iv) optical transmission spectrum, and (v) thermal conductivity values of molecular composites of hexagonal boron nitride and nanofibrillated cellulose. Adapted with permission.93 Copyright 2014, ACS. (f) Schematic illustration of (i) self-assembly for molecular composites of cellulose and black phosphorus crystals and (ii) energy generator device fabricated from these composites. (iii) Electronic properties of composites of cellulose and black phosphorus crystals. Adapted with permission.120 Copyright 2017, Wiley-VCH. | ||
Cellulose nanofibrils can establish hydrogen bonding and hydrophilic and hydrophobic interactions with other 2D crystals as well, including transition metal dichalcogenides (TMDCs), boron nitride (BN) nanosheets, and black phosphorus (bP) (Fig. 4(d)–(f)). These interactions benefit the delamination of these 2D crystals from their bulk counterparts (Fig. 4(d)-(i) and (ii)).128 The exfoliated crystals can be assembled into free-standing films using VASA techniques (Fig. 4(d)-(iii) and (iv)).128 Analogous to MXene-based biocomposites, TMDC-based biocomposites of nanofibrillated cellulose can also combine robust mechanical properties with good electrochemical properties (Fig. 4(d)-(v) and (vi)).128 These properties help these biocomposites facilitate effective charging and discharging in sodium-ion batteries while operating as a working electrode (Fig. 4(d)-(vi)).128 Cellulose nanofibrils can also help engineer thermal transport in biocomposites of 2D crystals (Fig. 4(e)).93 Cellulose nanofibrils can assemble BN nanosheets into biocomposites with excellent thermal conductivity (Fig. 4(e)-(i)–(iii)).93 Cellulose nanofibrils form bridges between BN nanosheets to keep them close and facilitate thermal transport between BN nanosheets (Fig. 4(e)-(iii)).93 This structural integrity allows these biocomposites to exhibit thermal conductivities exceeding 100 W mK−1 and the ability to combine optical transparency with moderate thermal conductivity values for biocomposites with lower BN nanosheet concentration (Fig. 4(e)-(iv) and (v)).93 In contrast to its structural contribution to graphene, MXene, TMDCs, and BN biocomposites, cellulose can act as a supporting and barrier matrix material for black phosphorus (Fig. 4(f)-(i)).120 Black phosphorus nanocrystals can be trapped inside a nano fibrillar cellulose matrix to enhance the environmental stability of bP nanocrystals (Fig. 4(f)-(ii)).120 This stable biocomposite can be used to fabricate triboelectric nanogenerator systems that utilize the extraordinary charging capability of bP nanocrystals without being concerned about the chemical degradation of bP (Fig. 4(f)-(ii) and (iii)).120
 | ||
| Fig. 5 (a) (i) Schematic illustration of self-assembly for nacre-like composites of chitosan and GO crystals. (ii) Images and (iii) scanning electron microscopy images of nacre-like composites of chitosan and GO crystals. (iv) Mechanical properties of nacre-like composites of chitosan and GO crystals. Adapted with permission.133 Copyright 2015, ACS. (b) (i, ii, iii) Scanning electron microscopy images of nacre-like composites of chitosan and conductive MXene crystals. (iv) Conductivity and (v) EMI shielding properties of nacre-like composites of chitosan and conductive MXene crystals. Adapted with permission.131 Copyright 2020, Elsevier. (c) (i) Schematic illustration of self-assembly for nacre-like composites of sodium alginate and GO crystals. (ii, iii) Scanning electron microscopy images of nacre-like composites of sodium alginate and GO crystals. (iv) Mechanical properties of nacre-like composites of sodium alginate and GO crystals. Adapted with permission.132 Copyright 2015, ACS. (d) (i) Schematic illustration of self-assembly for nacre-like composites of sodium alginate and MXene crystals. (ii) Scanning electron microscopy and (iii) transmission electron microscopy images of nacre-like composites of sodium alginate and MXene crystals. (iv) Conductivity and (v) EMI shielding properties of nacre-like composites of sodium alginate and conductive MXene crystals. Adapted with permission.50 Copyright 2016, AAAS. | ||
3.2 Polypeptide-based 2D biocomposites
The success of polysaccharides as matrix materials in 2D composites has initiated research on composite materials based on other macromolecules, particularly polypeptides.6,51,67,134 Polypeptides are produced from natural resources or biomanufacturing methods like fermentation.102,135 Biomanufacturing enables large-scale production and, more importantly, engineering opportunities on the amino acid sequence of the polypeptides. The recent advances in synthetic biology can offer polypeptide designs with immense diversity, which renders polypeptides a better alternative for governing the structural organization of 2D crystal fillers in biocomposites.6,135 The initial studies demonstrated that polypeptides acquired from natural resources could facilitate strong and specific interactions with various 2D materials, including graphene, GO, h-BN, MXenes, and TMDCs.6 These studies have provided basic information concerning interactions between 2D crystals and polypeptides and paved the way for new studies based on engineered proteins.6 Recent studies on composites of 2D crystals and recombinant proteins acquired from fermentation processes have revealed the true potential of polypeptides’ templating capability in 2D biocomposites.10,26,49 These works have demonstrated that it is possible to systematically alter composite materials’ physical properties, including thermal, electronic, and mechanical properties, without being bound by the rules of percolation, which was unprecedented in composite materials.10,26,49 Incorporating chitosan-based materials with 2D layered materials like molybdenum disulfide (MoS2),136 hexagonal boron nitride (h-BN),137 and black phosphorus138 indicates promising uses across different domains. Specifically, MoS2 holds promise for electronic devices,136 h-BN acts as a catalyst,137 and black phosphorus is valuable in electrochemistry.138We recently studied atomically thin inorganic layers with tandem repeat proteins.10,26,49 By controlling their molecular weight, we could fine-tune their mechanical properties, which exceeded those of state-of-the-art composites.49 However, an existing theoretical model was unable to explain this phenomenon. Our findings suggest that the failure mechanism for composites depends more on interfacial rather than bulk properties.49 Our inspiration for this study came from squid ring teeth, where recombinant proteins were found to adhere to inorganic sheets through secondary structures such as β-sheets and α-helices. This bonding mechanism resulted in high stretchability (59 ± 1% fracture strain) and toughness (54.8 ± 2 MJ m−3).49 We also discovered that the mechanical properties can be optimized by adjusting the protein molecular weight and tandem repetition. The exceptional mechanical responses we observed greatly exceeded the current state-of-the-art stretchability for layered composites by over a factor of three. This demonstrates the potential of engineering materials with reconfigurable physical properties.49
Naturally derived polypeptides have demonstrated intriguing assembly characteristics when matched with graphene and its derivatives,45,67,71,140 which incited many recent studies on polypeptide-based 2D biocomposites.26,49,51 In particular, silk fibroin can form various structures, from nano micelles to single molecule protofibrils on the surface of graphene derivatives, depending on the fibroin concentration, pH, humidity, and other physical conditions.45,67,71,140 BSA is another naturally derived polypeptide that can facilitate strong interactions with graphene,139 which is also commonly employed to exfoliate layered bulk materials to generate 2D crystals.52,141 These fundamental studies have helped develop an understanding of the physical properties of biocomposites fabricated from 2D crystals and naturally derived polypeptides. Silk and amyloid fibrils have formed functional composite materials with graphene derivatives (Fig. 6(a) and (b)).45,134 Silk has proven to be an effective matrix material for constructing mechanically tough and strong 2D biocomposites (Fig. 6(a)-(i) and (ii)).134 On the other hand, amyloid fibrils facilitated the specific assembly of 2D crystals, which resulted in enhanced interconnectivity of conductive graphene oxide fillers and, consequently, higher electrical conductivity (Fig. 6(b)-(i) and (ii)).45 The assembly kinetics of amyloid fibrils and graphene oxide nanosheets have also led to anisotropic mechanical properties (Fig. 6(b)-(iii)).45 The interactions between graphene derivatives are also used to construct hydrogel structures, wherein filler materials are employed to cross-link naturally derived gelatin matrix.67,71 Gelatin can also be processed with graphene derivatives and h-BN nanosheets to form nacre-like structures, which can be used to construct mechanically robust biomaterials.70 Beyond graphene derivatives, other conductive 2D filler materials like MXenes can create specific assemblies with silk fibroin (Fig. 6(c)). However, unlike amyloid fibrils, these assemblies did not incite controllable changes in the electrical properties of the resultant 2D biocomposite.51 Silk fibroin instead acts as a mechanical support and protective enclosure against oxidation of MXenes (Fig. 6(c)), which tends to degrade their electronic and optical properties.51
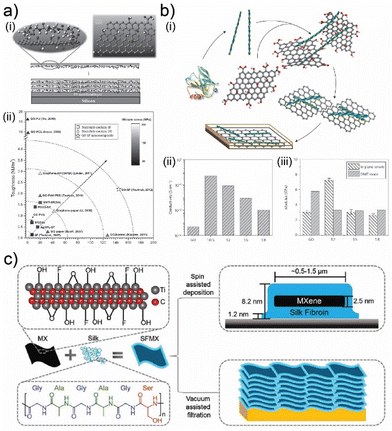 | ||
| Fig. 6 (a) (i) Schematic illustration of self-assembly for nacre-like composites of silk fibroin and GO crystals. (ii) Mechanical properties of nacre-like composites of nacre-like composites of silk fibroin and GO crystals. Adapted with permission.134 Copyright 2013, Wiley-VCH. (b) (i) Schematic illustration of self-assembly for nacre-like composites of amyloid fibrils and GO crystals. (ii) Conductivity of nacre-like composites of amyloid fibrils and GO crystals. (iii) Tensile modulus of nacre-like composites of amyloid fibrils and GO crystals. Adapted with permission.45 Copyright 2015, Springer Nature. (c) Schematic illustration of self-assembly for nacre-like composites of silk and MXene crystals. Adapted with permission.51 Copyright 2020, Wiley-VCH. | ||
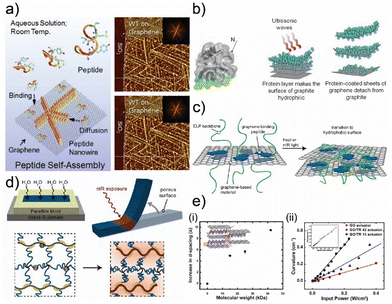 | ||
| Fig. 7 (a) Schematic illustration and atomic force microscopy images of self-assembly of recombinant proteins and GO crystals. Adapted with permission.142 Copyright 2016, Springer Nature. (b) Schematic illustration of recombinant proteins and their assembly mechanics on graphene crystals. Adapted with permission.97 Copyright 2010, Wiley-VCH. (c) Schematic illustration of synthetic proteins derived from elastin-like proteins and their response to physical probing. Adapted with permission.143 Copyright 2014, ACS. (d) Schematic illustration of graphene–gelatin composites and their response to infrared radiation. Adapted with permission.144 Copyright 2013, ACS. (e) (i) The interlayer distance between GO crystals as a function of the molecular weight of recombinant repetitive proteins. (ii) The influence of structural order introduced by synthetic proteins on the actuation performance of their composites with GO crystals. Adapted with permission.10 Copyright 2017, Elsevier. | ||
The recombinant polypeptides can also form functional assemblies with a new family of 2D crystals like MXenes.16 MXene nanocrystals can interact with naturally derived and synthetic polypeptides based on silk and squid ring teeth, commonly through hydrogen bonding and charge interactions.10,26,49,51,146 These interactions can be programmed to define the overall organization of graphene oxide and MXene nanocrystals.10,26 In a more complex system consisting of more than 2D crystal, synthetic proteins elect to organize a single 2D crystal while the secondary 2D crystal acts as a percolative filler material (Fig. 8(a)-(i)).49 This is reflected in the assembly characteristics of the 2D crystals that are systematically separated by synthetic proteins (Fig. 8(a)-(ii)).49 It is possible to explain this assembly characteristic by implementing interfacial interactions between 2D crystals and repetitive proteins.49
 | ||
| Fig. 8 (a) (i) Schematic illustration of self-assembly for molecular composites of synthetic repetitive proteins and 2D crystals (GO and MXene). (ii) The interlayer distance between 2D crystals as a function of the number of repeat units of repetitive proteins in these molecular composites. (b) (i) The influence of the number of repeat units on the mechanical toughness of these molecular composites. (ii) The landscape of mechanical properties of existing 2D composites and their comparison with these molecular composites. Adapted with permission.49 Copyright 2022, NAS. (c) (i) The interlayer distance between MXene crystals as a function of the number of repeat units of repetitive proteins for molecular composites of repetitive proteins and MXene crystals. (ii) The experimental and theoretical in-plane conductivity of these molecular composites as a function of the interlayer separation between MXene crystals. (d) (i) The experimental and theoretical out-of-plane conductivity of these molecular composites as a function of the interlayer separation between MXene crystals. (ii) I/V curve of molecular composites composed of synthetic repetitive proteins with 11 repeat units exhibiting resistive memory effect. Adapted with permission.26 Copyright 2020, ACS. | ||
This physical model reveals that the structural organization of these two distinct 2D crystals orchestrated by synthetic proteins and interfacial interactions can engender structural material systems that exhibit extraordinary toughness (Fig. 8(b)-(i)).49 Unlike existing structural materials engineered using synthetic chemistry, these structural materials programmed using synthetic biology employ enhancements in the extent of deformation instead of improving material strength (Fig. 8(b)-(ii)).49 This unorthodox approach helps these novel materials match and overcome the toughness of the strongest composites based on 2D crystals (Fig. 8(b)-(ii)).49 Another important aspect of this approach is the ability to modify this enhancement systematically using the amino acid sequence of these repetitive proteins.49 In a more straightforward material system composed of MXene nanocrystals and these repetitive proteins, the organization of conductive MXene crystals is altered with precision to govern electronic transport in a composite material (Fig. 8(c)-(i)).26 Besides percolative distribution simply relying on the volumetric concentration of the conductive filler material, synthetic repetitive proteins offer a more refined control over the tunneling distance between MXene nanocrystals among a bulk freestanding composite (Fig. 8(c)-(ii)).26 The influence of this structural control over tunneling distance can be easily modeled and confirmed using experimental conductivity measurements, which confirms the overall organization of MXene crystals is indeed governed by self-assembly kinetics defined by the amino acid sequence of these repetitive proteins (Fig. 8(c)-(ii)).26 This model is also indicative of a topological order in these conductive composite materials, which is documented by the distinct difference between in-plane and out-of-plane electron transport among these bulk composites (Fig. 8(c)-(ii) and d-(i)). The physical model and experimental investigation of out-of-plane electronic conductivity indicated a critical dimension for a transition from electron-dominated transport to ion-dominated transport in between MXene nanocrystals (Fig. 8(d)-(ii)).26 Once the separation between MXene nanocrystals becomes large enough to accommodate the formation of beta-sheet crystals, these composites exhibit an electronic memory behavior (Fig. 8(d)-(ii)).26 Globular and structural proteins are known to exhibit memory effects in more delicate device geometries based on lithographic fabrication methods. However, this is one of the first demonstrations of memristive behavior in bulk materials, particularly based on biocomposites.57,147,148
3.3 Nucleic acid-based 2D biocomposites
Nucleic acids, such as DNA and RNA, have been increasingly explored as functional components in developing innovative 2D layered composites.6,100 These composites, often consisting of graphene or graphene oxide, offer exceptional mechanical, thermal, and electrical properties have been obtained. By integrating nucleic acids into these 2D layered structures, hybrid materials with enhanced functionality, such as biosensing, drug delivery, and tissue engineering capabilities. For example, it has been shown that the construction of nucleic acids on the surface of graphene has a strong sensitivity to metal ions,149 and can also enhance its electrochemical performance.1503.4 Summary of materials properties of 2D biocomposites
To summarize the available 2D biocomposites, we created a table outlining composite materials consisting of different 2D crystals and biopolymers. These materials can form a hierarchical order that translates to novel physical properties, as shown in Table 1. In the current state of composite theory, 2D biocomposites predominantly rely on the filler fraction as a primary parameter to modify the physical characteristics of the resultant composites. The potential of biopolymers is reflected in their ability to attain a wide range of properties, which can be adjusted across multiple orders of magnitude. This extensive range is difficult to achieve using filler fraction alone, as it is limited by the rules of percolation. Therefore, biopolymer assembly characteristics, including hydrogen bonding networks, charge, and hydrophobic interactions, are utilized to create 2D biocomposites with novel material properties. In recent years, there has been a push to separate the structural influence from the filler fraction by introducing a secondary parameter that defines this influence. To achieve this, researchers have developed bioengineered proteins with a distinct number of identical repetition units among their amino acid sequences.151 The number of repetition units helps define the interlayer separation between 2D crystals with Angstrom-level precision, hence introducing a definite model for electrical conductivity,26 mechanical deformation,49 and electromagnetic shielding.40 This secondary parameter has extended the range of physical aspects to be engineered across three orders of magnitude and even enabled the conversion of physical behavior like switching from conductive to memristive (i.e., observed in thin dielectrics) state.26 Other researchers follow these works, employing similar block copolymer-like protein sequences, such as silk, to establish functional 2D biocomposites.51 We believe the advances in synthetic biology tools will drive this approach further as new protein designs become available to match the surface chemistry of an ever-growing family of 2D crystals. This way, the fundamentals of composite research will be redefined as novel matrix materials developed by protein researchers will introduce a new set of parameters for engineering composite materials.| Biopolymer | 2D crystal | Physical properties | Control parameter | Range | |
|---|---|---|---|---|---|
| Ref. 103 | Nanocellulose | Graphene | Tensile strength | Filler fraction | 170–351 MPa |
| Tensile modulus | 8–16.9 MPa | ||||
| Elongation at break | 9–12% | ||||
| Toughness | 11–22 MJ m−3 | ||||
| Ref. 61 | Nanocellulose | MXene | Tensile strength | Filler fraction | 42–135.4 MPa |
| Tensile modulus | 1–3.8 GPa | ||||
| Elongation at break | 3–16.7% | ||||
| Toughness | 1.2–15 MJ m−3 | ||||
| Electrical conductivity | 0.62–739 S cm−1 | ||||
| EMI shielding | 1200–2647 dB cm2 g−1 | ||||
| Ref. 93 | Nanocellulose | Hexagonal boron nitride | Thermal conductivity | Filler fraction | 0.04–145 W m−1 K−1 |
| Ref. 106 | Nanocellulose | Hexagonal boron nitride | Thermal conductivity | Filler fraction | 0.184–3.13 W m−1 K−1 |
| Ref. 105 | Nanocellulose | Graphene oxide | Thermal conductivity | Filler fraction | 15–170 mW m−1 K−1 |
| Ref. 113 | Nanocellulose | Graphene | Specific capacitance | Filler fraction | 138–207 F g−1 |
| Ref. 99 | Nanocellulose | Graphene | Gauge factor | Filler fraction | 7.1–2427 |
| Elongation at break | 6–100% | ||||
| Ref. 111 | Cellulose | Graphene | Electrical resistance | Filler fraction | 1 MΩ–1 kΩ |
| Elongation at break | 0.7–3.2% | ||||
| Ref. 119 | Bacterial cellulose | MXene | Capacitance | Filler fraction | 550–3400 mF cm−2 |
| Ref. 118 | Nanocellulose | MXene | Tensile strength | Filler fraction | 25.8–271.5 MPa |
| Tensile modulus | 8.25–42 MPa | ||||
| Electrical conductivity | 1–1850 S cm−1 | ||||
| Specific capacitance | 240–348 F g−1 | ||||
| Ref. 126 | Nanocellulose | MXene | Tensile strength | Filler fraction | 75.6–136.5 MPa |
| Tensile modulus | 4.5–9.3 GPa | ||||
| Ref. 128 | Nanocellulose | Hexagonal boron Nntride | Tensile strength | Filler fraction | 80–182 MPa |
| Ref. 120 | Nanocellulose | Phosphorene | Optical transparency | Filler fraction | 60–90% |
| Power density | 0.2–9.36 μW cm−2 | ||||
| Ref. 129 | Chitosan | Graphene | Tensile strength | Filler fraction | 100–527 MPa |
| Tensile modulus | 2.4–19 GPa | ||||
| Elongation at break | 1–10% | ||||
| Toughness | 1.6–17.7 MJ m−3 | ||||
| Electrical conductivity | 34.6–155.3 S cm−1 | ||||
| Ref. 131 | Chitosan | MXene | Electrical conductivity | Filler fraction | 92–1402 S cm−1 |
| Effective EMI shielding | 26–34.7 dB | ||||
| Ref. 132 | Alginate | Graphene | Tensile strength | Filler fraction | 75–272 MPa |
| Tensile modulus | 4.5–9.7 GPa | ||||
| Elongation at break | 1.6–7.9% | ||||
| Toughness | 2–18.1 MJ m−3 | ||||
| Ref. 50 | Alginate | MXene | Electrical conductivity | Filler fraction | 0.5–4665 S cm−1 |
| Effective EMI shielding | 15–58 dB | ||||
| Ref.134 | Silk fibroin | Graphene | Tensile strength | Filler fraction | 75–300 MPa |
| Tensile modulus | 9.5–145 GPa | ||||
| Toughness | 0.7–2.4 MJ m−3 | ||||
| Ref. 51 | Silk fibroin | MXene | Electrical conductivity | Filler fraction | 0.05–1400 S cm−1 |
| Ref. 71 | Gelatin | Graphene | Tensile strength | Filler fraction | 10–120 kPa |
| Tensile modulus | 2–7 kPa | ||||
| Ref. 140 | Gelatin | Graphene | Electrical conductivity | Filler fraction | 1 μ–0.1 S cm−1 |
| Ref. 45 | Amyloid fibrils | Graphene | Tensile strength | Filler fraction | 6–16.5 MPa |
| Tensile modulus | 2.8–7.2 GPa | ||||
| Electrical conductivity | 50 μ–0.5 S cm−1 | ||||
| Ref. 70 | Gelatin | Hexagonal boron nitride | Tensile strength | Filler fraction | 96.4–148.7 MPa |
| Tensile modulus | 11.8–31 GPa | ||||
| Ref. 64 | Recombinant viral protein | Graphene | Specific capacity | Filler fraction | 128–181 mA h g−1 |
| Ref. 142 | Recombinant proteins | Graphene | Transconductance | Filler fraction | 0.6–1.4 mS |
| Ref. 10 | Recombinant tandem repeat proteins | Graphene | Actuation curvature | Molecular weight | 0.12–1.2 cm−1 |
| Actuation power density | (number of repeat units in amino acid sequence) | 0.2–1.2 W cm−2 | |||
| Ref. 26 | Recombinant tandem repeat proteins | MXene | Electrical conductivity | Molecular weight (number of repeat units in amino acid sequence) | 30–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 S cm−1 000 S cm−1 |
| Ref. 49 | Recombinant tandem repeat proteins | Graphene/MXene | Tensile strength | Molecular weight (number of repeat units in amino acid sequence) | 100–150 MPa |
| Elongation at break | 6–58% | ||||
| Toughness | 4–55 MJ m−3 |
4. Conclusions and future prospects of 2D biocomposites
The molecular architecture of layered composites was inspired by exoskeletons and endoskeletons, which rely heavily on polypeptides and polysaccharides, such as collagen or chitin. The evolutionary paths of exoskeletons and endoskeletons are intertwined. Early life forms probably lacked either type of structure, but support systems emerged as creatures became more complex. They became internalized and bony, providing strong support for muscles and organs. Meanwhile, exoskeletons took various forms like shells or scales, offering protection but limiting growth potential. The future of layered composites will go beyond imitating nature, as sustainable materials and high-performance biocomposites make them more competitive with established materials.Layered composites hold great promise for the future, as they allow the properties of different materials to be combined. An important area for development will be the customization of interfaces between layers to optimize these properties. The ability to control interfacial chemistry, electrical contact resistance, and thermal boundary resistance using biomolecular interfaces in contact with 2D materials is essential to the flexible 2D devices made from these materials. Additionally, layered 2D systems provide an exciting platform to study the control of optical, electrical, thermal, and mechanical properties in a molecular composite by finely tuning the interlayer distance based on the molecular weight of the biomolecules. By creating programmable 2D materials, we can explore new directions in equilibrium and non-equilibrium materials by studying their phase transition and response to perturbations such as magnetic susceptibility, conductivity, optical reflectivity, and nonlinearity. Moreover, the practical requirements for various functionalities—like achieving waterproof, anti-fouling, conductive, and biocompatible properties—along with intelligent responsiveness, which includes automatically adapting to external factors such as temperature, pH, humidity, and other conditions, are crucial for designing 2D layered materials.
Exploiting the asymmetric transport properties, such as thermal or electrical conductivity, of 2D layered materials requires innovative design rules, which can open up new ways to control emergent physical properties. However, many fundamental challenges still need to be addressed to fully comprehend the capabilities of layered structures in material science and their potential applications in manipulating transport properties. One of the main challenges is the gap between computationally designed and experimentally manufactured materials, which can result in imperfections due to manufacturing and synthesis. To overcome this issue, possible solutions include the directed evolution of biomanufactured materials to enhance robustness or the selection of 2D materials that meet the physical requirements.
In the upcoming years, there will be a notable surge in the exploration of sustainable materials, including a deeper understanding of biomolecules and biodegradable 2D-layered composites. As the production of these materials scales up, biocomposites are expected to become more affordable, accelerating their adoption across various industries, such as automobile, aerospace, and consumer goods. This will not only promote the use of eco-friendly materials but also positively impact the environment.
Author contributions
M. V. and M. C. D. wrote the manuscript. All authors participated in revisions, discussions, and interpretation of the data.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
The authors declare that the research was conducted without any commercial or financial relationships that could potentially create a conflict of interest.Acknowledgements
M. V. and M. C. D. were partially supported by DARPA (D19AC00016), Airforce Office of Sponsored Research (FA9550-18-1-0235), Army Research Office (W911NF-16-1-0019), and Huck Endowment of Pennsylvania State University.References
- K. S. Novoselov, D. Jiang, F. Schedin, T. J. Booth, V. V. Khotkevich, S. V. Morozov and A. K. Geim, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 10451–10453 CrossRef CAS PubMed.
- K. S. Novoselov, A. K. Geim, S. V. Morozov, D. Jiang, Y. Zhang, S. V. Dubonos, I. V. Grigorieva and A. A. Firsov, Science, 2004, 306, 666–670 CrossRef CAS PubMed.
- K. S. Novoselov, A. Mishchenko, A. Carvalho and A. H. Castro Neto, Science, 2016, 353, aac9439 CrossRef CAS PubMed.
- R. J. Young, I. A. Kinloch, L. Gong and K. S. Novoselov, Compos. Sci. Technol., 2012, 72, 1459–1476 CrossRef CAS.
- S. Stankovich, D. A. Dikin, G. H. B. Dommett, K. M. Kohlhaas, E. J. Zimney, E. A. Stach, R. D. Piner, S. T. Nguyen and R. S. Ruoff, Nature, 2006, 442, 282–286 CrossRef CAS PubMed.
- M. C. Demirel, M. Vural and M. Terrones, Adv. Funct. Mater., 2018, 28, 1704990 CrossRef.
- Z. Ling, C. E. Ren, M.-Q. Zhao, J. Yang, J. M. Giammarco, J. Qiu, M. W. Barsoum and Y. Gogotsi, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 16676–16681 CrossRef CAS PubMed.
- Y. Zhang, S. Gong, Q. Zhang, P. Ming, S. Wan, J. Peng, L. Jiang and Q. Cheng, Chem. Soc. Rev., 2016, 45, 2378–2395 RSC.
- P. Samorì, I. A. Kinloch, X. Feng and V. Palermo, 2D Mater., 2015, 2, 030205 CrossRef.
- M. Vural, Y. Lei, A. Pena-Francesch, H. Jung, B. Allen, M. Terrones and M. C. Demirel, Carbon, 2017, 118, 404–412 CrossRef CAS.
- K. W. Putz, O. C. Compton, C. Segar, Z. An, S. T. Nguyen and L. C. Brinson, ACS Nano, 2011, 5, 6601–6609 CrossRef CAS PubMed.
- A. K. Geim and I. V. Grigorieva, Nature, 2013, 499, 419–425 CrossRef CAS PubMed.
- D. Jariwala, T. J. Marks and M. C. Hersam, Nat. Mater., 2016, 16, 170–181 CrossRef PubMed.
- K. Kalantar-zadeh, J. Z. Ou, T. Daeneke, M. S. Strano, M. Pumera and S. L. Gras, Adv. Funct. Mater., 2015, 25, 5086–5099 CrossRef CAS.
- C. Tan and H. Zhang, Chem. Soc. Rev., 2015, 44, 2713–2731 RSC.
- B. Anasori, M. R. Lukatskaya and Y. Gogotsi, Nat. Rev. Mater., 2017, 2, 16098 CrossRef CAS.
- A. VahidMohammadi, J. Rosen and Y. Gogotsi, Science, 2021, 372, eabf1581 CrossRef CAS PubMed.
- M. G. Rasul, A. Kiziltas, B. Arfaei and R. Shahbazian-Yassar, npj 2D Mater. Appl., 2021, 5, 56 CrossRef CAS.
- C. R. Dean, A. F. Young, I. Meric, C. Lee, L. Wang, S. Sorgenfrei, K. Watanabe, T. Taniguchi, P. Kim, K. L. Shepard and J. Hone, Nat. Nanotechnol., 2010, 5, 722–726 CrossRef CAS PubMed.
- D. Lee, S. H. Song, J. Hwang, S. H. Jin, K. H. Park, B. H. Kim, S. H. Hong and S. Jeon, Small, 2013, 9, 2602–2610 CrossRef CAS PubMed.
- H. Mu, S. Lin, Z. Wang, S. Xiao, P. Li, Y. Chen, H. Zhang, H. Bao, S. P. Lau, C. Pan, D. Fan and Q. Bao, Adv. Opt. Mater., 2015, 3, 1447–1453 CrossRef CAS.
- G. Hu, T. Albrow-Owen, X. Jin, A. Ali, Y. Hu, R. C. T. Howe, K. Shehzad, Z. Yang, X. Zhu, R. I. Woodward, T.-C. Wu, H. Jussila, J.-B. Wu, P. Peng, P.-H. Tan, Z. Sun, E. J. R. Kelleher, M. Zhang, Y. Xu and T. Hasan, Nat. Commun., 2017, 8, 278 CrossRef PubMed.
- Z. Zhang, P. Chen, X. Duan, K. Zang, J. Luo and X. Duan, Science, 2017, 357, 788–792 CrossRef CAS PubMed.
- K. S. Kim, D. Lee, C. S. Chang, S. Seo, Y. Hu, S. Cha, H. Kim, J. Shin, J.-H. Lee, S. Lee, J. S. Kim, K. H. Kim, J. M. Suh, Y. Meng, B.-I. Park, J.-H. Lee, H.-S. Park, H. S. Kum, M.-H. Jo, G. Y. Yeom, K. Cho, J.-H. Park, S.-H. Bae and J. Kim, Nature, 2023, 614, 88–94 CrossRef CAS PubMed.
- B.-I. Park, J. Kim, K. Lu, X. Zhang, S. Lee, J. M. Suh, D.-H. Kim, H. Kim and J. Kim, Nano Lett., 2024, 24, 2939–2952 CrossRef CAS PubMed.
- M. Vural, H. Zhu, A. Pena-Francesch, H. Jung, B. D. Allen and M. C. Demirel, ACS Nano, 2020, 14, 6956–6967 CrossRef CAS PubMed.
- G. Guan, J. Xia, S. Liu, Y. Cheng, S. Bai, S. Y. Tee, Y.-W. Zhang and M.-Y. Han, Adv. Mater., 2017, 29(32), 1700326 CrossRef PubMed.
- M. Shtein, R. Nadiv, M. Buzaglo, K. Kahil and O. Regev, Chem. Mater., 2015, 27, 2100–2106 CrossRef CAS.
- J. Li and J.-K. Kim, Compos. Sci. Technol., 2007, 67, 2114–2120 CrossRef CAS.
- I. A. Kinloch, J. Suhr, J. Lou, R. J. Young and P. M. Ajayan, Science, 2018, 362, 547–553 CrossRef CAS PubMed.
- D. Jariwala, V. K. Sangwan, C.-C. Wu, P. L. Prabhumirashi, M. L. Geier, T. J. Marks, L. J. Lauhon and M. C. Hersam, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 18076–18080 CrossRef CAS PubMed.
- L. Britnell, R. V. Gorbachev, R. Jalil, B. D. Belle, F. Schedin, A. Mishchenko, T. Georgiou, M. I. Katsnelson, L. Eaves, S. V. Morozov, N. M. R. Peres, J. Leist, A. K. Geim, K. S. Novoselov and L. A. Ponomarenko, Science, 2012, 335, 947–950 CrossRef CAS PubMed.
- T. Georgiou, R. Jalil, B. D. Belle, L. Britnell, R. V. Gorbachev, S. V. Morozov, Y.-J. Kim, A. Gholinia, S. J. Haigh, O. Makarovsky, L. Eaves, L. A. Ponomarenko, A. K. Geim, K. S. Novoselov and A. Mishchenko, Nat. Nanotechnol., 2012, 8, 100–103 CrossRef PubMed.
- L. Britnell, R. M. Ribeiro, A. Eckmann, R. Jalil, B. D. Belle, A. Mishchenko, Y. Kim, R. V. Gorbachev, T. Georgiou, S. V. Morozov, A. N. Grigorenko, A. K. Geim, C. Casiraghi, A. H. C. Neto and K. S. Novoselov, Science, 2013, 340, 1311–1314 CrossRef CAS PubMed.
- F. Withers, O. Del Pozo-Zamudio, A. Mishchenko, A. P. Rooney, A. Gholinia, K. Watanabe, T. Taniguchi, S. J. Haigh, A. K. Geim, A. I. Tartakovskii and K. S. Novoselov, Nat. Mater., 2015, 14, 301–306 CrossRef CAS PubMed.
- S. E. Kim, F. Mujid, A. Rai, F. Eriksson, J. Suh, P. Poddar, A. Ray, C. Park, E. Fransson, Y. Zhong, D. A. Muller, P. Erhart, D. G. Cahill and J. Park, Nature, 2021, 597, 660–665 CrossRef CAS PubMed.
- H. Kim, Y. Liu, K. Lu, C. S. Chang, D. Sung, M. Akl, K. Qiao, K. S. Kim, B.-I. Park, M. Zhu, J. M. Suh, J. Kim, J. Jeong, Y. Baek, Y. J. Ji, S. Kang, S. Lee, N. M. Han, C. Kim, C. Choi, X. Zhang, H.-K. Choi, Y. Zhang, H. Wang, L. Kong, N. N. Afeefah, M. N. M. Ansari, J. Park, K. Lee, G. Y. Yeom, S. Kim, J. Hwang, J. Kong, S.-H. Bae, Y. Shi, S. Hong, W. Kong and J. Kim, Nat. Nanotechnol., 2023, 18, 464–470 CrossRef CAS PubMed.
- S. J. Kim, K. Choi, B. Lee, Y. Kim and B. H. Hong, Annu. Rev. Mater. Res., 2015, 45, 63–84 CrossRef CAS.
- H. Jang, Y. J. Park, X. Chen, T. Das, M.-S. Kim and J.-H. Ahn, Adv. Mater., 2016, 28, 4184–4202 CrossRef CAS PubMed.
- M. Vural, A. Pena-Francesch, J. Bars-Pomes, H. Jung, H. Gudapati, C. B. Hatter, B. Allen, B. Anasori, I. T. Ozbolat, Y. Gogotsi and M. C. Demirel, Adv. Funct. Mater., 2018, 28, 1801972 CrossRef.
- X. Zhang, L. Hou, A. Ciesielski and P. Samorì, Adv. Energy Mater., 2016, 6, 1600671 CrossRef.
- F. Bonaccorso, L. Colombo, G. Yu, M. Stoller, V. Tozzini, C. Ferrari, R. S. Ruoff and V. Pellegrini, Science, 2015, 347, 1246501 CrossRef PubMed.
- Q. Xue, H. Zhang, M. Zhu, Z. Pei, H. Li, Z. Wang, Y. Huang, Y. Huang, Q. Deng, J. Zhou, S. Du, Q. Huang and C. Zhi, Adv. Mater., 2017, 29, 1604847 CrossRef PubMed.
- J. Kuchlyan, N. Kundu, D. Banik, A. Roy and N. Sarkar, Langmuir, 2015, 31, 13793–13801 CrossRef CAS PubMed.
- C. Li, J. Adamcik and R. Mezzenga, Nat. Nanotechnol., 2012, 7, 421–427 CrossRef CAS PubMed.
- J. Lee, P. Dak, Y. Lee, H. Park, W. Choi, M. A. Alam and S. Kim, Sci. Rep., 2015, 4, 7352 CrossRef PubMed.
- L. Wang, Y. Wang, J. I. Wong, T. Palacios, J. Kong and H. Y. Yang, Small, 2014, 10, 1101–1105 CrossRef CAS PubMed.
- K. Kalantar-zadeh and J. Z. Ou, ACS Sens., 2016, 1, 5–16 CrossRef CAS.
- M. Vural, T. Mazeed, D. Li, O. Colak, R. F. Hamilton, H. Gao and M. C. Demirel, Proc. Natl. Acad. Sci. U. S. A., 2022, 119(31), e2120021119 CrossRef CAS PubMed.
- F. Shahzad, M. Alhabeb, C. B. Hatter, B. Anasori, S. Man Hong, C. M. Koo and Y. Gogotsi, Science, 2016, 353, 1137–1140 CrossRef CAS PubMed.
- M. C. Krecker, D. Bukharina, C. B. Hatter, Y. Gogotsi and V. V. Tsukruk, Adv. Funct. Mater., 2020, 30, 2004554 CrossRef CAS.
- G. Guan, S. Zhang, S. Liu, Y. Cai, M. Low, C. P. Teng, I. Y. Phang, Y. Cheng, K. L. Duei, B. M. Srinivasan, Y. Zheng, Y. W. Zhang and M. Y. Han, J. Am. Chem. Soc., 2015, 137, 6152–6155 CrossRef CAS PubMed.
- L. Zong, M. Li and C. Li, Adv. Mater., 2017, 29, 1604691 CrossRef PubMed.
- H. B. Sim, J. Y. Lee, B. Park, S. J. Kim, S. Kang, W. H. Ryu and S. C. Jun, Nano Res., 2016, 9, 1709–1722 CrossRef CAS.
- D. Akinwande, C. J. Brennan, J. S. Bunch, P. Egberts, J. R. Felts, H. Gao, R. Huang, J.-S. Kim, T. Li, Y. Li, K. M. Liechti, N. Lu, H. S. Park, E. J. Reed, P. Wang, B. I. Yakobson, T. Zhang, Y.-W. Zhang, Y. Zhou and Y. Zhu, Extreme Mech. Lett., 2017, 13, 42–77 CrossRef.
- Y. Cheng, L. D. Koh, D. Li, B. Ji, Y. Y. W. Zhang, J. Yeo, G. Guan, M. Y. Han and Y. Y. W. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 21787–21796 CrossRef CAS PubMed.
- A. Moudgil, N. Kalyani, G. Sinsinbar, S. Das and P. Mishra, ACS Appl. Mater. Interfaces, 2018, 10, 4866–4873 CrossRef CAS PubMed.
- N. Song, D. Jiao, S. Cui, X. Hou, P. Ding and L. Shi, ACS Appl. Mater. Interfaces, 2017, 9, 2924–2932 CrossRef CAS PubMed.
- G. Song, R. Kang, L. Guo, Z. Ali, X. Chen, Z. Zhang, C. Yan, C.-T. Lin, N. Jiang and J. Yu, New J. Chem., 2020, 44, 7186–7193 RSC.
- W. Yang, Z. Zhao, K. Wu, R. Huang, T. Liu, H. Jiang, F. Chen and Q. Fu, J. Mater. Chem. C, 2017, 5, 3748–3756 RSC.
- W.-T. Cao, F.-F. Chen, Y.-J. Zhu, Y.-G. Zhang, Y.-Y. Jiang, M.-G. Ma and F. Chen, ACS Nano, 2018, 12, 4583–4593 CrossRef CAS PubMed.
- B. Liang, L. Fang, Y. Hu, G. Yang, Q. Zhu and X. Ye, Nanoscale, 2014, 6, 4264–4274 RSC.
- M. Qi, Y. Zhou, Y. Huang, L. Zhu, X. Xu, Z. Ren and J. Bai, Nanoscale, 2017, 637–646 RSC.
- D. Oh, X. Dang, H. Yi, M. A. Allen, K. Xu, Y. J. Lee and A. M. Belcher, Small, 2012, 8, 1006–1011 CrossRef CAS PubMed.
- S. Wan, J. Peng, L. Jiang and Q. Cheng, Adv. Mater., 2016, 28, 7862–7898 CrossRef CAS PubMed.
- T. Benselfelt, J. Shakya, P. Rothemund, S. B. Lindström, A. Piper, T. E. Winkler, A. Hajian, L. Wågberg, C. Keplinger and M. M. Hamedi, Adv. Mater., 2023, 35, 2303255 CrossRef CAS PubMed.
- N. Annabi, S. R. Shin, A. Tamayol, M. Miscuglio, M. A. Bakooshli, A. Assmann, P. Mostafalu, J. Y. Sun, S. Mithieux, L. Cheung, X. Tang, A. S. Weiss and A. Khademhosseini, Adv. Mater., 2016, 28, 40–49 CrossRef CAS PubMed.
- S.-H. H. Hu, R.-H. H. Fang, Y.-W. W. Chen, B.-J. J. Liao, I.-W. W. Chen and S.-Y. Y. Chen, Adv. Funct. Mater., 2014, 24, 4144–4155 CrossRef CAS.
- R. Deng, H. Yi, F. Fan, L. Fu, Y. Zeng, Y. Wang, Y. Li, Y. Liu, S. Ji and Y. Su, RSC Adv., 2016, 6, 77083–77092 RSC.
- S. C. Yoo, Y. K. Park, C. Park, H. Ryu and S. H. Hong, Adv. Funct. Mater., 2018, 28, 1805948 CrossRef.
- C. Cha, S. R. Shin, X. Gao, N. Annabi, M. R. Dokmeci, X. (Shirley) Tang and A. Khademhosseini, Small, 2014, 10, 514–523 CrossRef CAS PubMed.
- C. Ye, Z. A. Combs, R. Calabrese, H. Dai, D. L. Kaplan and V. V. Tsukruk, Small, 2014, 10, 5087–5097 CrossRef CAS PubMed.
- E. A. Mayerberger, R. M. Street, R. M. McDaniel, M. W. Barsoum and C. L. Schauer, RSC Adv., 2018, 8, 35386–35394 RSC.
- W. Yin, L. Yan, J. Yu, G. Tian, L. Zhou, X. Zheng, X. Zhang, Y. Yong, J. Li, Z. Gu and Y. Zhao, ACS Nano, 2014, 8, 6922–6933 CrossRef CAS PubMed.
- H. Bao, Y. Pan, Y. Ping, N. G. Sahoo, T. Wu, L. Li, J. Li and L. H. Gan, Small, 2011, 7, 1569–1578 CrossRef CAS PubMed.
- C. Li, Z.-Y. Wu, H.-W. Liang, J.-F. Chen and S.-H. Yu, Small, 2017, 13, 1700453 CrossRef PubMed.
- C. Zhu, P. Liu and A. P. Mathew, ACS Appl. Mater. Interfaces, 2017, 9, 21048–21058 CrossRef CAS PubMed.
- C. Zhu, S. Monti and A. P. Mathew, ACS Nano, 2018, 12, 7028–7038 CrossRef CAS PubMed.
- M. Abolhassani, C. S. Griggs, L. A. Gurtowski, J. A. Mattei-Sosa, M. Nevins, V. F. Medina, T. A. Morgan and L. F. Greenlee, ACS Omega, 2017, 2, 8751–8759 CrossRef CAS PubMed.
- Y. Wang, A. G. El-Deen, P. Li, B. H. L. Oh, Z. Guo, M. M. Khin, Y. S. Vikhe, J. Wang, R. G. Hu, R. M. Boom, K. A. Kline, D. L. Becker, H. Duan and M. B. Chan-Park, ACS Nano, 2015, 9(10), 10142–10157 CAS.
- A. K. K. Geim and K. S. S. Novoselov, Nat. Mater., 2007, 6, 183–191 CrossRef CAS PubMed.
- D. A. Dikin, S. Stankovich, E. J. Zimney, R. D. Piner, G. H. Dommett, G. Evmenenko, S. T. Nguyen and R. S. Ruoff, Nature, 2007, 448, 457–460 CrossRef CAS PubMed.
- Y. Fu, J. Hansson, Y. Liu, S. Chen, A. Zehri, M. K. Samani, N. Wang, Y. Ni, Y. Zhang, Z.-B. Zhang, Q. Wang, M. Li, H. Lu, M. Sledzinska, C. M. S. Torres, S. Volz, A. A. Balandin, X. Xu and J. Liu, 2D Mater., 2019, 7, 12001 CrossRef.
- H. Jang, J. D. Wood, C. R. Ryder, M. C. Hersam and D. G. Cahill, Adv. Mater., 2015, 27, 8017–8022 CrossRef CAS PubMed.
- S. B. Jo, H. H. Kim, H. Lee, B. Kang, S. Lee, M. Sim, M. Kim, W. H. Lee and K. Cho, ACS Nano, 2015, 9, 8206–8219 CrossRef CAS PubMed.
- G. Cassabois, P. Valvin and B. Gil, Nat. Photonics, 2015, 10, 262–266 CrossRef.
- J. Simon, C. Fruhling, H. Kim, Y. Gogotsi and A. Boltasseva, Opt. Photonics News, 2023, 34, 42–49 CrossRef.
- C. R. So, Y. Hayamizu, H. Yazici, C. Gresswell, D. Khatayevich, C. Tamerler and M. Sarikaya, ACS Nano, 2012, 6, 1648–1656 CrossRef CAS PubMed.
- A. J. Patil, J. L. Vickery, T. B. Scott and S. Mann, Adv. Mater., 2009, 21, 3159–3164 CrossRef CAS.
- H. Zhu, Z. Jia, Y. Chen, N. Weadock, J. Wan, O. Vaaland, X. Han, T. Li and L. Hu, Nano Lett., 2013, 13, 3093–3100 CrossRef CAS PubMed.
- A. Pena-Francesch, H. Jung, M. C. Demirel and M. Sitti, Nat. Mater., 2020, 19, 1230–1235 CrossRef CAS PubMed.
- A. Pena-Francesch, H. Jung, M. C. Demirel and M. Sitti, Nat. Mater., 2020, 19, 1230–1235 CrossRef CAS PubMed.
- H. Zhu, Y. Li, Z. Fang, J. Xu, F. Cao, J. Wan, C. Preston, B. Yang and L. Hu, ACS Nano, 2014, 8, 3606–3613 CrossRef CAS PubMed.
- K. Wu, J. Fang, J. Ma, R. Huang, S. Chai, F. Chen and Q. Fu, ACS Appl. Mater. Interfaces, 2017, 9, 30035–30045 CrossRef CAS PubMed.
- V. Nicolosi, M. Chhowalla, M. G. Kanatzidis, M. S. Strano and J. N. Coleman, Science, 2013, 340, 1226419 CrossRef.
- J. N. Coleman, M. Lotya, A. O’Neill, S. D. Bergin, P. J. King, U. Khan, K. Young, A. Gaucher, S. De, R. J. Smith, I. V. Shvets, S. K. Arora, G. Stanton, H.-Y. Kim, K. Lee, G. T. Kim, G. S. Duesberg, T. Hallam, J. J. Boland, J. J. Wang, J. F. Donegan, J. C. Grunlan, G. Moriarty, A. Shmeliov, R. J. Nicholls, J. M. Perkins, E. M. Grieveson, K. Theuwissen, D. W. McComb, P. D. Nellist and V. Nicolosi, Science, 2011, 331, 568–571 CrossRef CAS PubMed.
- P. Laaksonen, M. Kainlauri, T. Laaksonen, A. Shchepetov, H. Jiang, J. Ahopelto and M. B. Linder, Angew. Chem., Int. Ed., 2010, 49, 4946–4949 CrossRef CAS PubMed.
- N. I. Kovtyukhova, Y. Wang, A. Berkdemir, R. Cruz-Silva, M. Terrones, V. H. Crespi and T. E. Mallouk, Nat. Chem., 2014, 6, 957–963 CrossRef CAS PubMed.
- C. Yan, J. Wang, W. Kang, M. Cui, X. Wang, C. Y. Foo, K. J. Chee and P. S. Lee, Adv. Mater., 2014, 26, 2022–2027 CrossRef CAS PubMed.
- U. G. K. Wegst, H. Bai, E. Saiz, A. P. Tomsia and R. O. Ritchie, Nat. Mater., 2015, 14, 23–36 CrossRef CAS PubMed.
- H. Zhu, W. Luo, P. N. Ciesielski, Z. Fang, J. Y. Zhu, G. Henriksson, M. E. Himmel and L. Hu, Chem. Rev., 2016, 116, 9305–9374 CrossRef CAS PubMed.
- A. Pena-Francesch, N. E. Domeradzka, H. Jung, B. Barbu, M. Vural, Y. Kikuchi, B. D. Allen and M. C. Demirel, APL Mater., 2018, 6, 10701 CrossRef.
- J.-M. Malho, P. Laaksonen, A. Walther, O. Ikkala and M. B. Linder, Biomacromolecules, 2012, 13, 1093–1099 CrossRef CAS PubMed.
- N. Song, X. Hou, L. Chen, S. Cui, L. Shi and P. Ding, ACS Appl. Mater. Interfaces, 2017, 9, 17914–17922 CrossRef CAS PubMed.
- B. Wicklein, A. Kocjan, G. Salazar-Alvarez, F. Carosio, G. Camino, M. Antonietti and L. Bergström, Nat. Nanotechnol., 2015, 10, 277–283 CrossRef CAS PubMed.
- J. Chen, X. Huang, Y. Zhu and P. Jiang, Adv. Funct. Mater., 2017, 27, 1604754 CrossRef.
- S.-S. Kim, J.-H. Jeon, H.-I. Kim, C. D. Kee and I.-K. Oh, Adv. Funct. Mater., 2015, 25, 3560–3570 CrossRef CAS.
- Y. Li, H. Zhu, F. Shen, J. Wan, X. Han, J. Dai, H. Dai and L. Hu, Adv. Funct. Mater., 2014, 24, 7366–7372 CrossRef CAS.
- Z. Zhou, J. Liu, X. Zhang, D. Tian, Z. Zhan and C. Lu, Adv. Mater. Interfaces, 2019, 6, 1802040 CrossRef.
- A. C. E. Camilo, A. J. de Menezes, M. A. Pereira-da-Silva, F. E. G. Guimarães and R. H. Longaresi, Cellulose, 2020, 27, 713–728 CrossRef CAS.
- Z. Weng, Y. Su, D.-W. Wang, F. Li, J. Du and H.-M. Cheng, Adv. Energy Mater., 2011, 1, 917–922 CrossRef CAS.
- L. Liu, Z. Niu, L. Zhang, W. Zhou, X. Chen and S. Xie, Adv. Mater., 2014, 26, 4855–4862 CrossRef CAS PubMed.
- K. Gao, Z. Shao, J. Li, X. Wang, X. Peng, W. Wang and F. Wang, J. Mater. Chem. C, 2013, 1, 63–67 CAS.
- Y. Habibi, L. A. Lucia and O. J. Rojas, Chem. Rev., 2010, 110, 3479–3500 CrossRef CAS PubMed.
- W. S. Lee and J. Choi, ACS Appl. Mater. Interfaces, 2019, 11, 19363–19371 CrossRef CAS PubMed.
- C. Yoo, M. G. Kaium, L. Hurtado, H. Li, S. Rassay, J. Ma, T.-J. Ko, S. S. Han, M. S. Shawkat, K. H. Oh, H.-S. Chung and Y. Jung, ACS Appl. Mater. Interfaces, 2020, 12, 25200–25210 CrossRef CAS PubMed.
- F. Lai, D. Yong, X. Ning, B. Pan, Y.-E. Miao and T. Liu, Small, 2017, 13, 1602866 CrossRef PubMed.
- W. Tian, A. VahidMohammadi, M. S. Reid, Z. Wang, L. Ouyang, J. Erlandsson, T. Pettersson, L. Wågberg, M. Beidaghi and M. M. Hamedi, Adv. Mater., 2019, 31, 1902977 CrossRef PubMed.
- Y. Wang, X. Wang, X. Li, Y. Bai, H. Xiao, Y. Liu, R. Liu and G. Yuan, Adv. Funct. Mater., 2019, 29, 1900326 CrossRef.
- P. Cui, K. Parida, M.-F. Lin, J. Xiong, G. Cai and P. S. Lee, Adv. Mater. Interfaces, 2017, 4, 1700651 CrossRef.
- R. Xiong, K. Hu, A. M. Grant, R. Ma, W. Xu, C. Lu, X. Zhang and V. V. Tsukruk, Adv. Mater., 2016, 28, 1501–1509 CrossRef CAS PubMed.
- Y. Liu, J. Zhou, E. Zhu, J. Tang, X. Liu and W. Tang, J. Mater. Chem. C, 2015, 3, 1011–1017 RSC.
- Y. Liu, J. Zhou, J. Tang and W. Tang, Chem. Mater., 2015, 27, 7034–7041 CrossRef CAS.
- P. Laaksonen, A. Walther, J. M. Malho, M. Kainlauri, O. Ikkala and M. B. Linder, Angew. Chem., Int. Ed., 2011, 50, 8688–8691 CrossRef CAS PubMed.
- N. D. Luong, N. Pahimanolis, U. Hippi, J. T. Korhonen, J. Ruokolainen, L.-S. Johansson, J.-D. Nam and J. Seppälä, J. Mater. Chem., 2011, 21, 13991–13998 RSC.
- W.-T. Cao, C. Ma, D.-S. Mao, J. Zhang, M.-G. Ma and F. Chen, Adv. Funct. Mater., 2019, 29, 1905898 CrossRef CAS.
- W. Tian, A. VahidMohammadi, M. S. Reid, Z. Wang, L. Ouyang, J. Erlandsson, T. Pettersson, L. Wågberg, M. Beidaghi and M. M. Hamedi, Adv. Mater., 2019, 31, 1902977 CrossRef PubMed.
- Y. Li, H. Zhu, F. Shen, J. Wan, S. Lacey, Z. Fang, H. Dai and L. Hu, Nano Energy, 2015, 13, 346–354 CrossRef CAS.
- S. Wan, J. Peng, Y. Li, H. Hu, L. Jiang and Q. Cheng, ACS Nano, 2015, 9, 9830–9836 CrossRef CAS PubMed.
- M. Naguib, ACS Cent. Sci., 2020, 6, 344–346 CrossRef CAS PubMed.
- F. Liu, Y. Li, S. Hao, Y. Cheng, Y. Zhan, C. Zhang, Y. Meng, Q. Xie and H. Xia, Carbohydr. Polym., 2020, 243, 116467 CrossRef CAS PubMed.
- K. Chen, B. Shi, Y. Yue, J. Qi and L. Guo, ACS Nano, 2015, 9, 8165–8175 CrossRef CAS PubMed.
- S. Wan, J. Peng, Y. Li, H. Hu, L. Jiang and Q. Cheng, ACS Nano, 2015, 9, 9830–9836 CrossRef CAS PubMed.
- K. Hu, M. K. Gupta, D. D. Kulkarni and V. V. Tsukruk, Adv. Mater., 2013, 25, 2301–2307 CrossRef CAS PubMed.
- A. P. Liu, E. A. Appel, P. D. Ashby, B. M. Baker, E. Franco, L. Gu, K. Haynes, N. S. Joshi, A. M. Kloxin, P. H. J. Kouwer, J. Mittal, L. Morsut, V. Noireaux, S. Parekh, R. Schulman, S. K. Y. Tang, M. T. Valentine, S. L. Vega, W. Weber, N. Stephanopoulos and O. Chaudhuri, Nat. Mater., 2022, 21, 390–397 CrossRef CAS PubMed.
- M. Ding, D. Ji and W. Hu, ACS Appl. Electron. Mater., 2024, 6(12), 8655–8670 CrossRef CAS.
- S. Yang, Y. Zhu, J. Liu, X. Zheng, X. Zhang and P. Liu, Int. J. Hydrogen Energy, 2023, 48(49), 18708–18718 CrossRef CAS.
- K. W. Liu, Y. B. Ma, Y. Guo, H. Wang, Y. N. Xu, X. D. Zhang, X. Zhang, X. Z. Sun, K. Wang, L. Yu and Y. W. Ma, Adv. Funct. Mater., 2024, 34, 2410451 CrossRef CAS.
- J. Liu, S. Fu, B. Yuan, Y. Li and Z. Deng, J. Am. Chem. Soc., 2010, 132, 7279–7281 CrossRef CAS PubMed.
- H. Nassira, A. Sánchez-Ferrer, J. Adamcik, S. Handschin, H. Mahdavi, N. Taheri Qazvini and R. Mezzenga, Adv. Mater., 2016, 6914–6920 CrossRef CAS PubMed.
- L. Liu, Z. Liu, P. Huang, Z. Wu and S. Jiang, RSC Adv., 2016, 6, 113315 RSC.
- Y. Hayamizu, C. R. So, S. Dag, T. S. Page, D. Starkebaum and M. Sarikaya, Sci. Rep., 2016, 6, 33778 CrossRef CAS PubMed.
- E. Wang, M. S. Desai, K. Heo and S. W. Lee, Langmuir, 2014, 30, 2223–2229 CrossRef CAS PubMed.
- E. Wang, M. S. Desai and S. W. Lee, Nano Lett., 2013, 13, 2826–2830 CrossRef CAS PubMed.
- T. H. Han, W. J. Lee, D. H. Lee, J. E. Kim, E. Y. Choi and S. O. Kim, Adv. Mater., 2010, 22, 2060–2064 CrossRef CAS PubMed.
- Z. Wang, S. Kang, S. Cao, M. Krecker, V. V. Tsukruk and S. Singamaneni, MRS Bull., 2020, 45, 1017–1026 CrossRef.
- F. Meng, B. Sana, Y. Li, Y. Liu, S. Lim and X. Chen, Small, 2014, 10, 277–283 CrossRef CAS PubMed.
- H. Wang, B. Zhu, H. Wang, X. Ma, Y. Hao and X. Chen, Small, 2016, 12, 3360–3365 CrossRef CAS PubMed.
- M. Wang, S. Zhang, Z. Ye, L. He, F. Yan, Y. Yang, H. Zhang and Z. Zhang, Microchim. Acta, 2015, 182, 2251–2258 CrossRef CAS.
- X. He, H. Han, W. Shi, J. Dong, X. Lu, W. Yanga and X. Lu, Anal. Methods, 2020, 12, 3462–3469 RSC.
- H. Jung, A. Pena-Francesch, A. Saadat, A. Sebastian, D. H. Kim, R. F. Hamilton, I. Albert, B. D. Allen and M. C. Demirel, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 6478–6483 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |
