 Open Access Article
Open Access ArticleSelf-assembled peptide-based nanofibers for cardiovascular tissue regeneration
Dhriti
Shenoy†
 a,
Sowmya
Chivukula†
a,
Sowmya
Chivukula†
 bc,
Nursu
Erdogan†
bc,
Nursu
Erdogan†
 cde,
Enrica
Chiesa
cde,
Enrica
Chiesa
 *c,
Sara
Pellegrino
*c,
Sara
Pellegrino
 *a,
Meital
Reches
*a,
Meital
Reches
 *b and
Ida
Genta
*b and
Ida
Genta
 c
c
aDISFARM, Dipartimento Di Scienze Farmaceutiche, Sezione Chimica Generale e Organica “A. Marchesini”, Università degli Studi di Milano, Milan, Italy. E-mail: sara.pellegrino@unimi.it
bInstitute of Chemistry and The Center for Nanoscience and Nanotechnology, The Hebrew University of Jerusalem, Jerusalem 9190401, Israel. E-mail: meital.reches@mail.huji.ac.il
cDipartimento di Scienze del Farmaco, Pharmaceutical Technology and Law Laboratory, Università di Pavia, Via Torquato Taramelli 12, Pavia, Italy. E-mail: enrica.chiesa@unipv.it; ida.genta@unipv.it
dPhysics of Nanostructures and Advanced Materials, Universidad del País Vasco/Euskal Herriko Unibertsitatea, Barrio Sarriena s/n, 48940 Leioa, Bizkaia, Spain
eCIC nanoGUNE BRTA, Donostia-San Sebastián, 20018, Spain
First published on 10th December 2024
Abstract
Cardiovascular diseases are the leading cause of death worldwide, claiming millions of lives every year. Cardiac tissue engineering has emerged as a versatile option for repairing cardiac tissue and helping its regeneration. The use of nanomaterials, particularly nanofiber-based scaffolds combined with biomolecular cues like peptides, has significantly improved the compatibility and efficacy of the scaffolds for cardiac tissue regeneration. By utilising the self-assembly properties of peptides to create nanofiber scaffolds, we can achieve stability that closely mimics the natural components of cardiac tissue, making them perfect for cardiac tissue regeneration. In this review, we highlighted the dynamic process of self-assembly into nanofibers and the use of various self-assembled nanofibers for cardiovascular tissue regeneration, focusing on their roles in antithrombotic, angiogenic, differentiation, proliferation, and anti-atherosclerotic interventions.
1. Introduction
Globally, cardiovascular diseases (CVDs) are the leading cause of death, accounting for 32% of all deaths in 2019 (17.9 million people) and approximately 19.1 million in 2020.1,2 In the European Union, CVDs contribute to over 1.8 million deaths annually,3 with an economic cost projected to be 210 billion euros.4 In the United States, heart diseases affected nearly 695,000 lives in 2021, with a cost larger than $239.9 billion annually between 2018 and 2019.5 Despite significant advancements in prevention and treatment, CVDs, which encompass a variety of disorders of the heart and blood circulatory system, substantially increase morbidity and mortality and place a considerable financial burden on society.6,7 There are currently a variety of clinical and surgical therapeutic options accessible, such as medication-based revascularisation, surgical procedures including heart transplantation, and installation of medical devices. Emerging regenerative medicine approaches also represent potential management options for CVD treatment.8–10 However, due to the poor ability of the heart to regenerate, injured tissues are primarily replaced by a scar that differs greatly from the original tissue in terms of biophysical characteristics. Scar tissue can cause life-threatening arrhythmias and aneurysms as well as seriously impede cardiac function, depending on the extent of the injured area.11 Furthermore, limited biological effects and resistance to conventional medications hinder the success of typical therapeutics-based treatment. To tackle these obstacles, sophisticated early disease detection and treatment methods are needed.Ischemic heart disease, particularly myocardial infarction (MI), is a major cause of death of cardiomyocytes. The limited healing capacity of cardiomyocytes and the formation of fibrotic scar tissue leads to the loss of heart function.2 Therefore, regeneration after MI aims to target the infarcted area and promote the regeneration of myocardial and vascular tissues with tissue engineering methods. Cell therapies, biomaterials or hybrid materials, and direct reprogramming of fibroblasts into cardiomyocytes are current strategies to treat infarcted areas.12 One of the solutions for MI is to develop a cellular patch of myocardial tissue that can aid in healing and regenerating the cardiac tissue. These scaffolds are designed to eventually integrate into a native myocardium tissue and can be surgically implanted in the patient's heart.13 In regenerative medicine, scaffolds are aimed to have a cellular microenvironment like the extracellular matrix (ECM) of natural tissues/organs which can support cellular adhesion, differentiation, proliferation, communication as well as assist in the transportation of nutrients and waste to/from the cells. For this purpose, it is important to select biomaterials that can direct the course of the therapeutic or diagnostic procedure through interactions with the living systems. The ideal biomaterial is the one that can mimic the corresponding extracellular matrix and thereby help in tissue regeneration.14,15 Moreover, cells and bioactive molecules enhance endogenous and exogenous therapeutic components and biomaterials can provide mechanical support for cell regeneration.16
The use of nanomaterials for tissue repair and regeneration is a very promising approach.17,18 The relative ease with which we can fine-tune their properties and combine natural and synthetic components in the scaffold material has opened innumerable possibilities in tissue regeneration. Among various nanostructure-based scaffolds, nanofiber-based scaffolds have proven to be better and more cytocompatible. Moyer et al. demonstrated that cylindrical self-assembling peptides show better affinity and selective binding to the site of injury in the arterial tissue, while spherical peptides show no binding.19 This can be attributed to the similarity in the structure of the nanofiber morphology and the ECM, a naturally occurring framework that supports the tissues. Carefully designed nanofibers, composed of biocompatible materials that can mimic the complex structure of native cardiac tissue, have been shown to promote the growth of new cells.17 The production of nanofibers can be achieved via techniques such as electrospinning, emulsion freeze-drying, and self-assembly.20
The addition of biomolecular cues, such as peptides, to the scaffolds, can enhance the regeneration process.21–23 Peptides are small protein fragments which can mimic the bioactivity of large protein molecules, thereby assisting in the targeting of protein–protein interactions in the cellular environment. The excellent bioactivity of peptides along with biocompatibility, bioresorbable nature, and commercial availability make them a suitable candidate for advanced medical applications. However, they display poor stability and undergo rapid degradation outside the cellular physiological environment.24 Peptide-based tissue scaffolds can modulate cell behaviour as they provide the biochemicals and signalling cues needed for the cell in the native tissue.25 Peptides can self-assemble into well-defined structures, such as nanofibers, nanotubes, nanospheres and hydrogels under favourable thermodynamic conditions.26 Supramolecular scaffolds formed by self-assembling peptides (SAP) hold great promise in cardiac regenerative medicine. These scaffolds mimic the structure and function of natural tissue components like the extracellular matrix, making them suitable for various tissue engineering applications. Peptide-based scaffolds can form high aspect ratio nanofibers via molecular stack motifs through non-covalent interactions. These nanofibrous scaffolds offer advantageous properties such as delayed degradation, hypo-immunogenicity, high biocompatibility, and the ability for prolonged release of specific growth factors immobilized by covalent bonding.27
Recently, a perfusable, multifunctional epicardial device consisting of a biodegradable elastic patch (BEP), permeable hierarchical microchannel networks (PHMs) and a system to enable the delivery of therapeutic agents were designed. The BEP's biomimetic elasticity and strength provide mechanical cues to infarcted myocardium while biomimetic PHMs facilitate angiogenesis, induce infiltration of reparative cells and enable to reserve therapeutic agents.28 Both SAP nanofiber and this cell-free novel device share several key features that provide an interconnected microporous environment for cells and cargo molecules as a reservoir. This novel approach emphasizes mechanical stimuli and matching besides structural enhancement. Even though SAP nanofiber may enable mechanical support they differ in their bioactivity and biocompatibility. They differ not only in biological properties but also in the way to tune and enhance the regeneration process. SAPs nanofiber scaffolds enable to have flexibility at the molecular level by different bioactive motifs such as modulating cell behaviours, cell signalling, binding, and niche properties. On the other hand, invasive surgical implantation and fixation of patches are still challenges. Patches need to be placed and fixed on damaged areas invasively whereas SAP can be delivered by local or systemic injections. Another novel cardiac patch was recruited for revascularization and magnetic accumulation of therapeutic agents for the treatment of MI. Unlike the previous design, these patches demonstrate and focus on a promising targeted delivery of therapeutic agents and cells by creating a local magnetic field to form rapid vascularization. Although this magnetic epicardial patch has similar invasive placement and needs crucial mechanical features, providing sufficient cell and therapeutic agent accumulation by magnetic force at the infarcted area and bypassing systemic delivery obstacles are exciting.29 It is believed that the integration of SAP into this design as a coating material might increase their efficiency and functionality due to their bioactive motif frame.
In this comprehensive review, we focus on the dynamic process of the general self-assembly mechanism, followed by a detailed compilation of the self-assembling peptides with different bioactivities within cardiovascular regeneration, distinct self-assembly mechanisms to elucidate their capacity to form nanofibers through non-covalent interactions and the recent studies in synthesizing self-assembling peptide nanofibers for addressing the challenge of cardiovascular tissue regeneration. This review will provide knowledge on the self-assembly of peptide nanofibers to contribute to the ongoing evolution of innovative solutions in the field of cardiovascular tissue engineering.
2. Self-assembly
Self-assembly is defined as a spontaneous organization of components into ordered structures without human intervention, applicable across scales from molecular to planetary. It is a key to understanding biological processes such as cell replication, plays a crucial role in nanotechnology, improves automated systems in manufacturing and robotics, and bridges reductionist and emergent perspectives in complex systems. Processes ranging from the non-covalent association of organic molecules in solution to the growth of semiconductor quantum dots on solid substrates have been called self-assembly. Self-assembly can be classified into two main types: static and dynamic. Static self-assembly occurs at equilibrium and does not require energy dissipation, forming stable structures like molecular crystals and proteins. In contrast, dynamic self-assembly relies on continuous energy dissipation to form and maintain structures, as seen in oscillating chemical reactions and biological cells, though its study is still in its early stages.30Uncontrolled protein aggregation can lead to diseases like Alzheimer's, making it crucial to understand the self-assembly mechanism. Supramolecular nanofibrils form through a nucleation-dependent polymerization process. This involves an initial nucleation event, followed by the growth and multiplication of structures.31 Theoretical studies suggest these nuclei can form either through a direct single-step process or a two-step process involving a metastable phase, resulting in various transient intermediates due to weak noncovalent interactions and flexible conformations.32–35
Recent research by Yaun et al. showed that liquid–liquid phase separation (LLPS) into solute-rich and solvent-rich phases is the initial step before peptide self-assembly nucleation. The mechanism of LLPS depicts that the formation of peptide-rich droplets from a homogeneous solution is an entropy-driven process and is mainly attributed to solvent release from bound peptides to the bulk. The transition from these liquid droplets to nanofibrils is mediated by enthalpic interactions and structural reorganization, requiring complete depletion of the solvation shell through multiple desolvation steps.31 In contrast, traditional self-assembly involves one-step nucleation from supersaturated solutions, where peptides undergo near-complete desolvation in a single step (Fig. 1).31,36
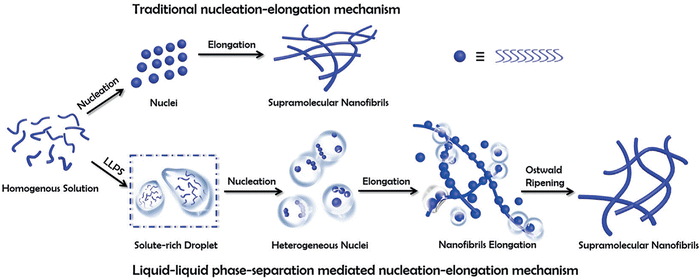 | ||
| Fig. 1 A schematic comparison of the self-assembly of supramolecular nanofibrils from amino acids or short peptides via the traditional and the liquid–liquid phase separation (LLPS) nucleation–elongation mechanism. In contrast to the traditional approach, LLPS involves the separation into solute-rich and solute-poor phases as a crucial step before nanofibril nucleation. The solute-rich droplets serve as nucleation precursors, with hydrated nanoclusters acting as nucleation sites. Subsequently, the nanofibrils elongate from metastable intermediates following Ostwald's step rule. Copyright 2019, Angewandte Chemie International Edition.31 | ||
The role of desolvation in LLPS-mediated peptide self-assembly is a multistep desolvation, it is crucial in triggering LLPS and the subsequent formation of peptide clusters (Fig. 2). The process begins with partial desolvation, followed by solvent removal from aromatic groups, leading to LLPS. Further desolvation of the LLPS-formed droplets then initiates peptide self-assembly. Unlike the one-step desolvation in classical nucleation, modulating desolvation dynamics at different stages enables control over the formation of various peptide structures, such as nanodroplets, nanobelts, nanotubes, nanoribbons, and helical nanofibers. These insights highlight a pathway to precisely control biomolecular self-assembly through desolvation modulation. Understanding the dynamics of these mechanisms is key to leveraging the unique properties of self-assembling peptide nanofibers for applications in drug delivery, tissue engineering, and other technologies.36
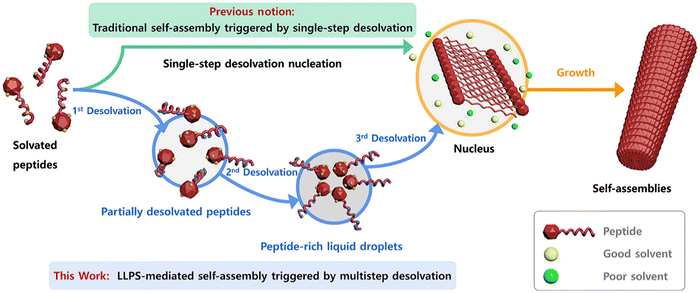 | ||
| Fig. 2 Schematic representation of traditional vs. LLPS-mediated self-assembly. Traditional self-assembly occurs via single-step desolvation. LLPS-mediated self-assembly, however, involves multistep desolvation, leading to peptide-rich droplets that form nuclei, followed by nanofibril growth. This figure has been published in CCS Chemistry 2024; multistep desolvation as a fundamental principle governing peptide self-assembly through liquid–liquid phase separation is available online at https://doi.org/10.31635/ccschem.023.202302990.36 | ||
3. Peptide-based nanofibers for cardiovascular tissue regeneration
3.1. Antithrombotic peptides
The intricate native microenvironment of the heart is essential for supporting the growth, health, and equilibrium of cardiac tissue. The cardiac environment, comprising a variety of cell types including immune cells, fibroblasts, endothelial cells, and cardiomyocytes, enables a symphony of interactions that are critical to the heart. These cells work together in harmony in the heart, carefully forming the blood channels, myocardial fibres, and extracellular matrix (ECM) framework. The extracellular matrix (ECM), a dynamic structure made up of proteoglycans, glycosaminoglycans, and many proteins such as collagen, elastin, and laminin, is fundamental to structural integrity. As a physical anchor for cells, the ECM also facilitates critical cell–matrix interactions that are essential for cellular communication and function.37In tissue engineering, integrating anti-thrombotic agents for cardiovascular regeneration is crucial to ensure the success and long-term functionality of the engineered tissue. This is particularly relevant in the context of bypass surgery, one of the most effective strategies for treating cardiovascular and peripheral vascular diseases. There are two main treatments for patients: human vascular grafts including autografts or allografts, and prosthetic vascular grafts. While autografts and allografts exhibit excellent biocompatibility, their availability is limited and they may not always be clinically feasible due to the risk of immune rejection, pre-existing medical condition of the patients or their advanced age. In contrast, prosthetic vascular grafts can be immobilized with biochemical cues to promote endothelialization and long-term functionality.38,39 However, the need to prevent the incidence of acute thrombosis at the site of the graft-blood interface remains a major challenge.
The peptide Ac-(RARADADA)2-CONH2 which is a derivative of the yeast protein zuotin repeat [Ac-(AEAEAKAK)2-CONH2], has marked a significant advancement in the field of peptide engineering.40 (RADA)4 is a 16 amino acids peptide sequence that self-assembles into stable β-sheet structures with alternating hydrophilic (R as positive residues, and D as negative residues) and hydrophobic residues (A or L residues) in aqueous conditions. As a result, nanofibers are formed through non-covalent electrostatic interactions. The charge arrangements on the hydrophilic side vary across designs, classified into four main moduli: modulus I (− + − + − + − +), modulus II (−− + + −− + +), modulus III (−−− + + +), and modulus IV (−−−− + + + +). The self-assembly is initiated by introducing electrolytes to reduce electrostatic repulsions among peptide monomers, followed by hydrophobic interactions that exclude the surrounding aqueous media between adjacent hydrophobic faces. This process leads to the formation of β-sheet bilayers, and the parallel or anti-parallel alignment of peptides results in fibrils through intra and intermolecular hydrogen bonds and electrostatic interactions.41 The mechanism of self-assembly of Ac-(RARADADA)2-CONH2 is similar to that shown for Ac-(AEAEAKAK)2-CONH2 in Fig. 3. Tissue culture studies demonstrated that (RADA)4 is compatible as a scaffold for various cell types such as myocardiocytes, neural cells, mesenchymal stem cells, chondrocytes, hepatocytes, skin epithelial, and endothelial cells.40,42
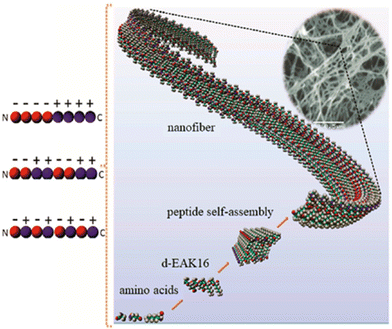 | ||
| Fig. 3 Self-assembly of the peptide Ac-(AEAEAKAK)2-CONH2 into nanofibers.43 Copyright 2018, Elsevier. | ||
Applying this concept, Li et al. designed and synthesized the peptide (RADA)4-GGGG-U which comprises the active centre selenocysteine of glutathione peroxidase (GPx). This peptide self-assembles into nanofibers that form a hydrogel, which serves as an anticoagulant coating that promotes the antithrombotic activity of blood-contacting medical devices used in cardiovascular therapies. The anticoagulant activity of the hydrogel results from the breakdown of S-nitrosothiols (RSNOs) and its capability to stimulate in situ catalytic release of nitric oxide (NO), which can inhibit aggregation and activation of platelets. SAP nanofiber-based hydrogel scaffolds were obtained by dissolving (RADA)4 (hereinafter referred to as RADA) and (RADA)4-GGGG-U (hereinafter referred to as U) separately in water and later mixing them to obtain U&RADA peptide mixture (20% v/v). After that, the mixture was diluted with PBS and sonicated for 30 min at 25 °C to have SAP nanofiber scaffold hydrogel matrices (hereinafter referred to as U&RADA hydrogel matrices). A surface of stainless steel was coated separately with RADA, U and U&RADA hydrogel matrices. The presence of the coating was confirmed by SEM, grazing incidence reflection absorption Fourier transform infrared (GIRA-FTIR) spectroscopy and XPS. NO release was detected by the NO chemiluminescence analyser and demonstrated as tunable by U peptide solution proportion. In vitro studies with fibroblast cells showed that U&RADA nanofiber hydrogel matrices did not have a cytotoxicity effect. In addition, this hydrogel can inhibit platelet adhesion by inducing NO generation. Antithrombogenic features of U&RADA hydrogel matrices were evaluated by using an arteriovenous extracorporeal circuit with New Zealand white rabbit animal model (Fig. 4(A)). After 2 hours of blood perfusion through samples, it was noticeable that U&RADA hydrogel matrices prevented clogging significantly compared to uncoated and RADA-coated SS foils (Fig. 4(B)). Moreover, quantitative evaluations showed that U&RADA hydrogel matrices caused the lowest occlusion rate, 25.61%, where the occlusion rate of RADA coated sample was 56.39% and the uncoated sample displayed a complete occlusion (Fig. 4(C)). Moreover, the blood flow rate percentage was compared, and it was increased to 69.85% ± 7.17% by coating SS with U&RADA hydrogel matrices while flow rates were found 11.77% ± 1.79% and 42.76% ± 3.22% with uncoated and RADA coated samples respectively (Fig. 4(D)). In Fig. 4(E), thrombi formation was observed inside of uncoated SS foil and RADA coated which were 41.10 ± 6.88 mg and 13.08 ± 2.02 mg respectively although SS with U&RADA hydrogel matrices had only 4.40 ± 1.07 mg thrombosis in Fig. 4(F). Furthermore, thrombi formation was evaluated deeply by SEM. Uncoated SS foil showed abundant erythrocytes and leukocytes covering the coating of fibrin. Similarly, RADA-coated SS foil was possessed by a fibrin network enveloping activated platelets and several erythrocytes. In contrast, SS foil coated by U&RADA hydrogel matrices showed slight fibrin presence but not activated platelets Fig. 4(G).44Ex vivo investigations revealed that SAP-based nanofiber hydrogel had a remarkable antithrombotic activity. SAP, (RADA)4 and (RADA)16, have been evaluated previously for their hemostatic effect.45,46 On the other hand, this study presented that a bioactive motif can provide flexibility to have SAP nanofiber-based hydrogel scaffolds with desired features while preserving their scaffold features. It showed great promise for blood-contacting devices to overcome potential blood clogging and offered lumen patency without direct anticoagulant addition beside allowing to a tuneable NO generation consequently antithrombotic activity.44
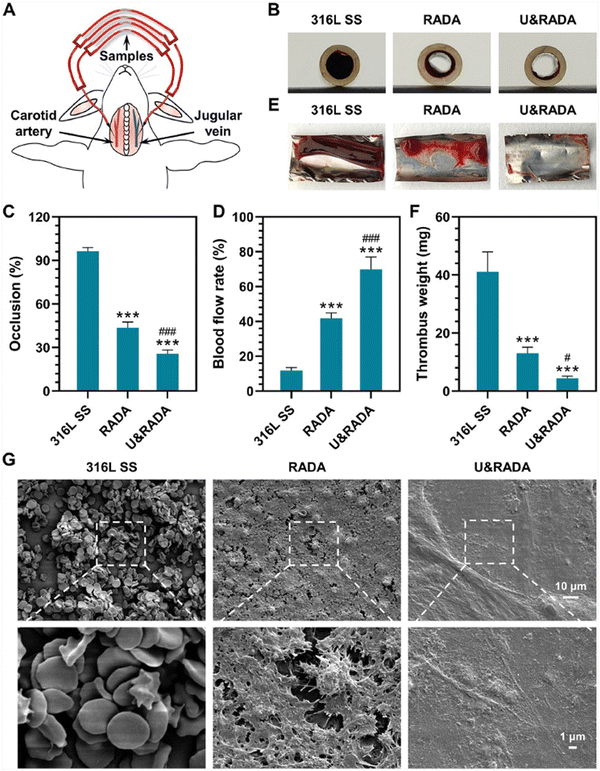 | ||
| Fig. 4 Ex vivo hemocompatibility evaluation for the hydrogel coating. (A) Representation of the rabbit arteriovenous extracorporeal circuit model. (B) Thrombi formation on the catheter inserted with samples. (C)–(F) Evaluation of occlusion rate, and blood flow rate of the catheter containing formed thrombi, and thrombus weight statistic. (G) SEM images of the thrombi on the hydrogel-coated and uncoated surfaces. Reproduced with permission.44 Copyright 2022, Elsevier. | ||
3.2. Angiogenic peptides
Angiogenesis is pivotal for cardiac regeneration, ensuring the crucial supply of nutrients and oxygen. These blood vessels not only transport oxygen and nutrients but also assist in faster immune response and disposal of metabolic waste.15 The rapid migration and proliferation of endothelial cells (EC) helps in angiogenesis. During the embryonic stage, ECs undergo rapid proliferation, however, their proliferation is extremely low in adults. This process of proliferation is mediated by interactions between the vascular growth factors and its receptors present on the cell walls.47 Pal et al. developed injectable vasculogenic hydrogels by conjugation of the peptide QK with a copolymer hydrogel of poly(N-isopropyl acrylamide) and thiol-modified gelatine. QK, a 15 amino acids long peptide (KLTWQELYQLKYKGI), is a VEGF mimicking proangiogenic sequence with better stability when compared with the VEGF molecule in solution. It exhibits better angiogenic bioactivity as it can bind to VEGF receptors (VEGFR-1 and 2).48 Unlike self-assembling peptides that form nanofibers and mimic extracellular matrix (ECM) components, the QK peptide does not self-assemble but stimulates angiogenesis through receptor interactions. Using QK in combination with self-assembling peptide-based nanofibers provides a versatile method for tissue regeneration. The nanofibers can provide structural support and a growth-friendly environment, while the QK peptide enhances angiogenesis. This combined strategy addresses both structural and functional aspects of tissue repair, potentially improving overall regenerative efficiency. Furthermore, reestablishing blood perfusion is important for preserving cardiac function after acute myocardial infarction (AMI). Activation of hepatocyte growth factor precursor (pro-HGF) has a major impact on both anti-apoptosis and angiogenesis. Ineffective pro-HGF activation is implied by the poor prognosis of AMI. Improving pro-HGF activation efficiency might have a beneficial effect on AMI therapy. A novel molecule called Nap-FFEG-IVGGYPWWMDV was produced by combining the self-assembled peptide Nap-FF with the bioactive zymogen activator peptide IVGGYPWWMDV. This compound showed a strong pro-HGF activation that encouraged pro-angiogenesis and anti-apoptosis.49 Nap-FF promotes self-assembly by largely stabilizing hydrogen bonds between the peptide moieties, which leads to an anti-parallel arrangement (Fig. 5(A)).50 The glutamic acid (E) was used for modulating the balance of hydrophilicity–hydrophobicity and glycine (G) acted as a linker to connect the two moieties, that is Nap-FFE with IVGGYPWWMDV. Resulting in Nap-FFEG-IVGGYPWWMDV that self-assembles into nanofibers with a β-sheet secondary structure (Fig. 5(B)). This structural organization through supramolecular self-assembly may address the challenges of unordered aggregation and random distribution of bioactive peptides in aqueous solutions.49 This approach improved the efficiency by utilizing self-assembled bioactive peptides to activate the endogenous growth factor instead of directly administering them, hence providing a new strategy for the therapy of AMI. It effectively reduces treatment costs and improves safety, providing excellent prospects for clinical application.49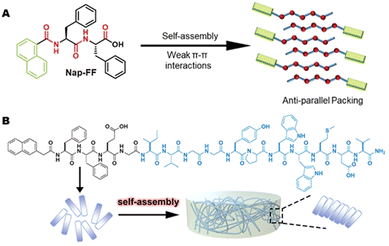 | ||
| Fig. 5 Hydrogel formation by Nap-FFEG-IVGGYPWWMDV (A) Illustration of the Nap-FF packing modes during aqueous self-assembly.50 Copyright (2021) Royal Society of Chemistry. (B) The chemical structure of FFEG-IVGGYPWWMDV. Reprinted (adapted) with permission from ref. 49, Copyright (2021) American Chemical Society. | ||
Rufaihah et al. presented a glycosaminoglycan (GAG) mimetic peptide nanofiber scaffold to enhance neovascularization after MI damage. The synthesised peptide, rich in sulfonate, hydroxyl, and carboxylic acid groups mimics heparan sulphate glycosaminoglycans without the addition of growth factors, stem cells or other biologically derived molecules. The nanofibers were formed by mixing two peptide amphiphiles (PAs) characterized by different charges particularly lauryl-VVAGEGDK (p-sulfobenzoyl)-S-Am (GAG-PA) and oppositely charged lauryl-VVAGK-Am (K-PA) (Fig. 6).27 This charge interaction neutralized the net charges, facilitating self-assembly to form higher-order nanofibers.51 This PA molecule featured a hydrophobic alkyl tail and a hydrophilic head. In an aqueous environment, these molecules are self-assembled into nanofibers with the hydrophobic tails clustered inward and the hydrophilic heads oriented outward. Lauric acid was added to the hydrophobic driving force of the self-assembly. The peptide portion has charged amino acids for solubility and four nonpolar residues that form a β-sheet structure. K-PA (lauryl-VVAGK-Am) was specifically designed with a lysine residue, which remains positively charged under acidic conditions.52 Morphological investigations showed that uniform nanofibers with approximately 7.5 nm in diameter were formed and they created an ECM-like porous structure. Activity of these nanofiber gel-based scaffolds was evaluated by injection to the infarct site in rat myocardial infarction model intramyocardially and the results indicated improved cardiac function and neovascularization with a high level of VEGF-A and Ang-1 expression. Although the treatment time considered in this work was limited, the results are noteworthy. The efficacy and safety of a new biomaterial based on SAP aimed to promote angiogenesis and cardiac regeneration after MI was successfully demonstrated in the pre-clinical small animal study, however, deep studies on the mechanism of differentiation raised by GAG mimetic nanofibers and affected signal pathways should be investigated.27
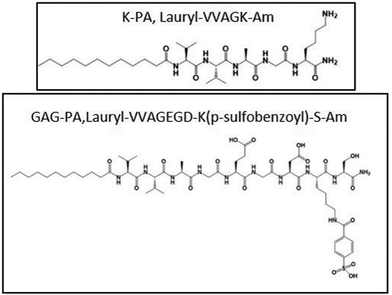 | ||
| Fig. 6 Chemical structures of the GAG peptide amphiphile.27 | ||
Recently, Wang et al. proposed enzyme-instructed self-assembly (EISA) in situ (Fig. 7) to obtain a slow release of the basic fibroblast growth factor (bFGF) in ischemic myocardium. They exploited the increased expression of matrix metallopeptidase 9 (MMP-9) enzyme that may be present at high levels up to 15–28 days after MI. Furthermore, the typical vascular leakage following MI results in an enhanced permeability and retention (EPR) impact of ischemic myocardium, which is advantageous for nanostructure accumulations. First, they synthesized a precursor with KKPLGLAGFF (K2) for micelle formation containing a PLGLAG sequence for MMP-9 cleavage. bFGF was encapsulated into the hydrophobic core of K2 micelles successfully (bFGF@K2 micelles) to protect it from enzymatic degradation in blood circulation. The amphiphilic property of the K2 peptide causes it to form micelles in vitro, with bFGF loaded into the core. In situ when bFGF@K2 micelles are exposed to MMP-9, the K2 peptide is cleaved, enabling the micelles to change into nanofibers.53 In the presence of MMP-9, micelles underwent a shape conversion into nanofibers (bFGF@Nanofiber) and further authors investigated their potential in MI conditions. bFGF@ K2 micelles were incubated in a solution prepared by extracted serum of rats at 36 h post-myocardial ischemia-reperfusion (MI/R) injury and PBS but, the formation of nanofibers occurred after 16 h incubation. In vivo studies carried out with a rat model 24 h post-MI/R demonstrated that bFGF@K2 micelles accumulated in the infarcted area due to the EPR effect after 24 h intravenous injection and transformed to bFGF@Nanofibers which subsequently increased retention time of bFGF. Nanofiber formation at the target area was confirmed under bio-TEM and found in the ischemic myocardium of MI/R rat 24 h post bFGF@K2 micelle injection. Moreover, a higher angiogenesis effect of bFGF@micelles than free bFGF was assessed by staining vascular endothelial and vascular smooth muscle in the ischemic myocardium of the rats after MI/R. Also, they were found the most effective for inhibiting cardiomyocyte apoptosis, myocardial fibrosis and remodelling in MI/R model rats. Overall, bFGF@K2 micelle transformed into bFGF@Nanofiber on-site with the help of EPR effect of ischemic myocardium and MMP 9-based EISA strategy, then slowly releasing bFGF from nanofiber hydrogel for effective promote heart repair of MI/R rats.53 Growth factors are one of the powerful aspects of restoring infarcted areas however, their poor half-life and rapid degradation limit their use. Although the implementation of nanofibrous materials locally provides enhanced retention time of biological cues by their excellent loading capacity, it might be risky and practically not suitable. This novel on-site transformation into nanofibers might reduce systemic administration disadvantages and allow us to take advantage of and prolonged release of growth factors by fibrous features consequently improving repair.
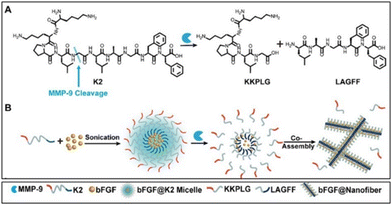 | ||
| Fig. 7 Schematic illustration of bFGF@Nanofibers formation. (A) Chemical structure of the MMP-9-responsive peptide K2. (B) Transformation of bFGF@K2 micelle to bFGF@Nanofiber upon MMP-9 cleavage. Reproduced from ref. 53. | ||
3.3. Differentiation, proliferation and cell adhesion enhancing peptides
One of the main aims of cardiovascular tissue regeneration is to develop a scaffold capable of directing the stem cells present in the scaffold to migrate to the site of injury and differentiate into cardiac cells.54 The cardiac tissue is majorly composed of cardiomyocytes supported by the fibrous cardiac skeleton of elastic fibres and other types of cells like endothelial cells, progenitor cells, smooth muscle cells, etc. The interaction of the integrins on the surface of cardiomyocytes with the extracellular matrix components like collagen, fibronectin, fibrinogen, and vitronectin have shown to modulate a wide range of cellular functions like cell differentiation, proliferation, apoptosis, migration, and adhesion. This signalling usually takes place in the form of a cascade signalling caused due to confirmational change in integrin structure on binding with intracellular molecules, i.e., protein–protein interactions. The integrin uses this type of signalling event as they do not possess enzymatic activities.55–57Table 1 lists the bioactive peptides for enhancing cell adhesion, differentiation, and proliferation.| Peptide sequence | Location of peptide | Receptor | Function | Ref. |
|---|---|---|---|---|
| RGD | Fibronectin, fibrinogen, vitronectin, collagen | α5β1, α8β1, αvβ3, αvβ5, αvβ6, αvβ8, and αIIbβ3 | Cell adhesion | 14 |
| GFOGER | Collagen | α2β1, and α11β1 | Enhances cell proliferation | 15,24 |
| YIGSR | Laminin | α3β1, α6β1, α7β1, and α6β4 | Endothelial cell-adhesive ligand | 58 |
| VGVAPG | Elastin | 67-kDa elastin-binding protein | Cell proliferation, chemotactic activity and metalloproteinase upregulation properties | 59 |
| EPLQLKM | Human bone marrow | Bone marrow-derived mesenchymal stem cells | Cell adhesion and proliferation of BMSCs | 60 |
Bioactive peptide sequences like RGD, IKVAV, etc. have shown to enhance the proliferation, differentiation, migration, and adhesion of cells. RGD sequence is found in a wide range of extracellular matrix proteins like fibronectin, fibrinogen, collagen and vitronectin has shown to aid in cell adhesion.61 It has affinity towards a wide range of integrins like α5β1, α8β1, αvβ3, αvβ5, αvβ6, αvβ8, and αIIbβ3.62 Immobilizing the RGD sequences in the tissue scaffold has shown to promote the cell–scaffold interaction as well as the biocompatibility of the scaffold.61 Clauder et al. demonstrated that simultaneously functionalizing the TiO2 surface with the peptides RGD, SIKVAV and VGVAPG significantly enhanced endothelial cell adhesion, proliferation, viability, and angiogenesis.63 Likewise, Kanda et al. developed a 3D hydrogel scaffold, namely CPCs-PRGmx containing cardiac progenitor stem cells (CPCs), PRG peptide conjugated to Puramatrix (PRGmx) and insulin-like growth factor 1 (IGF-1). PRGmx is a self-assembling peptide consisting of Puramatrix (PM) conjugated with PRGDSGYRGDS (PRG) motif. PRG motif consists of a two-unit RGD-binding peptide which is derived from collagen VI functional sequence. PM composed of alternating hydrophilic and hydrophobic amino acid residues, undergoes self-assembly into β-sheet structures, forming a stable 3D hydrogel with over 99.5% water content. This self-assembling property of PM allows it to be decorated with various bioactive motifs, enhancing its functionality for cell culturing and tissue engineering. When transplanted into cardiac tissue immediately after a myocardial infarction, the CPCs-PRGmx scaffold significantly improved vascularization, thereby demonstrating its potential to support angiogenesis.64
Angiogenesis can also be stimulated by triple-helical peptides (THPs), such as GFOGER and GLOGEN, which play a crucial role in cardiac tissue regeneration. These peptides can be used for two main functions either (1) to create anchor points for endothelial cell adhesion or (2) to instigate platelet aggregation. GFOGER, present in collagen, interacts with the α2β1 and α11β1 integrins, while GLOGEN binds to α1β1 and α10β1. By incorporating THPs, the mean cell surface area is increased, which significantly enhances the functional association of cells with collagen scaffolds. This improved integrin-THP binding promotes the projection of filopodia and lamellipodia, as well as actin polymerization, providing essential anchoring sites for endothelial cells during the process of cardiac tissue regeneration.11,12 Sun et al. demonstrated the self-assembly of GFOGER into nanofibers under specific pH conditions in the presence of lanthanide ions (La3+). They synthesized GFOGER to include negatively charged amino acids like aspartic acid/glutamic acid at both the N- and C-terminals, which facilitates radial and head-to-tail assembly into nanofibers resembling native collagen. This peptide–La3+ system mimics the pH-dependent fibrillogenesis of type I collagen, with peptide assembly driven by pH-induced changes in charged amino acid binding. At neutral pH, deprotonated amino acids strongly bind with La3+, promoting self-assembly, while acidic pH reduces this binding, disrupting the self-assembly process. The lanthanide ions also impart luminescent properties, making the nanofibers suitable for imaging and diagnostics. These sophisticated pH-responsive luminous nanofibers offer significant potential for developing functional biomaterials, especially in tissue engineering, as well as for applications in drug delivery and medical diagnostics.65
Cardiac cell therapy involves myocardial regeneration of infarcted cardiomyocytes by using cell sources such as cardiac progenitor cells, mesenchymal stem cells (MSCs), or their paracrine effect. Clinical trials on MSCs treatment for MI have been conducted. However, low engraftment, retention and viability are major problems of cardiac therapy due to insufficient nutrition in the damaged areas.66 The heart's reparative response can be influenced by communication between cell types through physical contact and paracrine signals.67 Cell spheroids offer an adaptable 3D habitat with strong cell–cell contact. 3D cell spheroids can boost the secretion of bioactive factors needed for cell survival, angiogenesis, and immunomodulation. This effect is enhanced by the diverse distribution of oxygen, nutrients, and signalling molecules within cell aggregates.68 Recently, Fan et al. utilized SAP nanofibers to induce mesenchymal stem cell spheroids formation. The final aim was to exploit the paracrine effect of MSC spheroids on MI-affected tissues. They designed a phosphatase-responsive peptide (Nap-pD-E7) with MSCs anchoring motif which self-assembled into a fibrillar matrix, providing a dynamic microenvironment where MSCs can form 3D spheroids. As shown in Fig. 8(A), the Nap-pD-E7 consists of three main components: (i) a well-established self-assembling backbone (Nap-DFDF), (ii) an enzymatic trigger with phosphatase responsiveness (phosphotyrosine, DpY), (iii) an anchoring motif with MSC surface targeting attribute (EPLQLKM, E7). In the presence of phosphatases, the phosphopeptide underwent a partial dephosphorylation at the intercellular space. This enzymatic trigger created a dynamic microenvironment conducive to the formation of a fibrillar matrix through instructed intercellular self-assembly. The fibrillar matrix modulated by cell–cell adhesive forces, prompted MSCs to aggregate into spheroids. This process mimics the natural cell-mediated unfolding of fibronectin, leading to the formation of 3D spheroids. The incorporation of the MSC-anchoring motif E7 ensured robust adhesion and interaction with MSCs, distinguishing the synthetic peptide's adhesive properties from those of endogenous ligands such as RGD or YIGSR. The Nap-pD-E7 peptide successfully formed a hydrogel in the presence of alkaline phosphatase (ALP) and exhibited similar mechanical stiffness to two other control hydrogel groups lacking either the phosphate group or the E7 motif. After gelation, self-assembled fibrillary structures were observed from Nap-pD-E7 peptide hydrogel by TEM. Then, MSC spheroids formation was evaluated in vitro and after treatment with Nap-pD-E7 for 24 h, MSC aggregated from a 2D cell sheet to form 3D cell spheroids while MSC in the standard medium without Nap-pD-E7 remained as a 2D sheet. Resulted cell spheroids mimic the dynamic of ECM remodelling as shown in Fig. 8(B). The gene expression levels of associated angiogenic growth factors, including VEGF, IGF, and HGF, were assessed using real-time PCR to investigate the paracrine action of Nap-pD-E7-induced MSC spheroids. They concluded that their peptide assembly could improve mRNA levels of angiogenic factors. Furthermore, in vivo studies showed that Nap-pD-E7 hydrogel-encapsulated MSC spheroids could improve cell retention, promote tissue angiogenesis, and attenuate ventricular remodelling.68 In tissue engineering, it is aimed to mimic native structures and benefit their unique features as much as possible. MSCs bring great promise to regenerative medicine especially on ischemic diseases due to their self-renewal capacity and paracrine effect. However, cell senescence, loss of stemness, limited resources and reduced paracrine ability resulting from MSCs expansion in vitro require preserving them and promoting their function. Here, SAP showed excellent potential to induce MSCs spheroids by forming a fibrillar matrix at intracellular and consequently reveal their paracrine and recovery features which are already in MSCs nature.
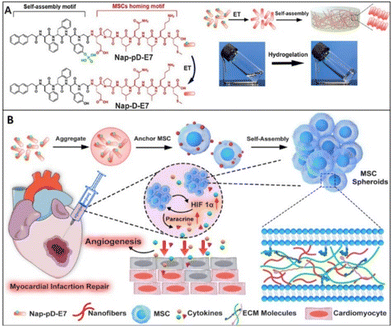 | ||
| Fig. 8 Schematic demonstration of enzymatic self-assembly nanofibers to enhance paracrine function for MI treatment (A) structural formula of Nap-pD-E7 to Nap-D-E7 by enzymatic transformation and images of the solution and hydrogel (B) illustration of the procedures of Nap-pD-E7 to induce MSC spheroids and transplantation of MSC spheroids carried by Nap-pD-E7 hydrogel. Reproduced with permission.68 Copyright 2022, Elsevier. | ||
Moreover, self-assembled peptides are utilized to functionalize electrospun nanofibers and develop a biomimetic hybrid nano matrix. The combination of electrospinning with SAPs results in unique polymer structures. Electrospinning is simple, inexpensive, and reliable involving high voltage, a syringe pump, a nozzle, and a collector. Here, a polymer solution or melt droplet is subjected to electrostatic forces that form a conical Taylor cone. After that, a jet is ejected from the Taylor cone when accumulated charges overcome surface charge toward the collector and solidification occurs by rapid solvent evaporation.69,70 The viscosity of the polymer solution is crucial; too high a viscosity hinders extrusion, while too low a viscosity disrupts filament stability.71 As the viscosity of the polymer solution is related to molecular weight, higher molecular weight is desired because longer polymer chains entangle better and facilitate nanofiber production.69 This cutting-edge technology has been used to manufacture nanofibers with biomedical purposes to utilize these fibres’ high surface area to volume ratio which remarkably affects cell behaviours.72
For instance, Andukuri et al. designed a hybrid scaffold using electrospun PCL (ePCL) nanofibers and PAs, including C16-GTAGLIGQ-YIGSR (PA-YIGSR) and C16-GTAGLIGQ-KKKKK (PA-KKKKK), which were mixed in a 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio to form PA-YK. The PAs include matrix metalloprotease-2 (MMP-2) degradable sites GTAGLIGQ. PAs also contain cell-adhesive sequences such as YIGSR, promoting endothelial cell adhesion and spreading and the polylysine (KKKKK) sequence acts as an NO donor, which is crucial for reducing smooth muscle cell proliferation and platelet adhesion. The PA-YIGSR and PA-KKKKK were then reacted with nitric oxide (NO) to produce PA-YK-NO. The PA-YK-NO was self-assembled onto ePCL nanofibers at pH 7 to create a hybrid nano matrix, ePCL-PA-YK-NO. The self-assembly of these SAPs into nanofibers is driven by their amphiphilic nature as mentioned above. The ePCL scaffolds were coated with PAs and shaken for 24 hours to allow them to self-assemble onto ePCL nanofibers through solvent evaporation and drying in a chemical hood. The results showed that hybrid scaffolds promoted endothelialization and on the contrary, limited platelet adhesion and smooth muscle cell proliferation. This combination of SAPs and nanofibers could be promising for cardiovascular use such as heart valves and grafts.73
10 ratio to form PA-YK. The PAs include matrix metalloprotease-2 (MMP-2) degradable sites GTAGLIGQ. PAs also contain cell-adhesive sequences such as YIGSR, promoting endothelial cell adhesion and spreading and the polylysine (KKKKK) sequence acts as an NO donor, which is crucial for reducing smooth muscle cell proliferation and platelet adhesion. The PA-YIGSR and PA-KKKKK were then reacted with nitric oxide (NO) to produce PA-YK-NO. The PA-YK-NO was self-assembled onto ePCL nanofibers at pH 7 to create a hybrid nano matrix, ePCL-PA-YK-NO. The self-assembly of these SAPs into nanofibers is driven by their amphiphilic nature as mentioned above. The ePCL scaffolds were coated with PAs and shaken for 24 hours to allow them to self-assemble onto ePCL nanofibers through solvent evaporation and drying in a chemical hood. The results showed that hybrid scaffolds promoted endothelialization and on the contrary, limited platelet adhesion and smooth muscle cell proliferation. This combination of SAPs and nanofibers could be promising for cardiovascular use such as heart valves and grafts.73
3.4. Anti-atherosclerotic peptides
Stem/progenitor cells, while holding promise for tissue revascularization, also contribute to atherosclerosis progression, plaque destabilization, and vessel remodelling. This impedes cardiovascular regeneration through chronic inflammation, endothelial dysfunction, and tissue damage, hampering natural healing processes in affected arteries.74 Another key player in atherosclerotic plaque development is the PCSK9 enzyme. The proprotein convertase subtilisin/kexin type 9 (PCSK9) enzyme plays a crucial role in regulating hepatic low-density lipoprotein (LDL) receptors. By promoting the degradation of these receptors, PCSK9 effectively reduces their availability on hepatocyte surfaces, resulting in elevated serum levels of low-density lipoprotein cholesterol (LDL-C). This dysregulation of LDL-C is a key pathogenic factor in the development and progression of atherosclerosis. Harbour et al. proposed a short self-assembling peptide, EPep2-8 comprising of E2 domain and Pep2-8, as a possible PCSK9 inhibitor. EPep2-8, a multidomain peptide, consists of a bioactive domain (Pep2-8) attached to the C-terminus of the canonical self-assembling E2 domain. The E2 domain facilitates the self-assembly of EPep2-8 into long, nanofibrous polymers characterised by a supramolecular β-sheet secondary structure, while peptide Pep2-8, comprising of 13-amino-acids, binds to PCSK9 and inhibits its interaction with LDL receptors (Fig. 9). The E2 sequence (EESLSLSLSLSLSLEE) contains terminal glutamic acid residues (E) which sandwich amino acid repeats of hydrophobic (L) and hydrophilic (S) residues that eventually undergo neutralization in the presence of charge-shielding ions such as Ca2+, allowing nanofibers to self-assemble. Upon the addition of calcium chloride, EPep2-8 changes into a viscous hydrogel made of nanofibers. The self-assembly process is predominantly propelled by hydrophobic residues (L), forming a compact structure that avoids water, while the hydrophilic residues (S) are exposed to the aqueous environment in an entropically favourable manner. The polymer elongates and adopts an intermolecular β-sheet secondary structure as the packing expands. This innovative approach holds promise in addressing the challenge of high LDL cholesterol levels and provides a potential avenue for mitigating the risk of cardiovascular diseases.75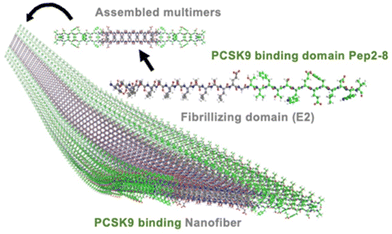 | ||
| Fig. 9 3D illustration of the nanofiber self-assembled by the peptide EPep2-8. EPep2-8 comprises the PCSK9 binding domain, Pep2-8 (shown in green carbons) and the fibrillating domain E2 (shown in grey carbons). These multimers combine to form extended nanofibers, which in turn entangle to create hydrogels. Reprinted (adapted) with permission from Copyright (2020) American Chemical Society.75 | ||
Another study recruited by Mansukhani et al. shows us the versatility of SAP-based nanofibers, for atherosclerosis treatment. Apolipoproteins, the key component in high-density lipoprotein (HDL), are naturally capable of promoting cholesterol efflux from atherosclerotic plaques. However, their hydrophobicity and size limit their use as therapeutic agents. Here, they developed Apo-1 mimicking peptide 4F (Ac-D-W-F-K-A-F-Y-D-K-V-A-E-K-F-K-E-A-F-NH2), which has a higher affinity towards the oxidized lipid than the ApoA1 protein. Additionally, liver X receptor agonists (LXR) can reduce atherosclerosis by promoting cholesterol efflux. However, liver toxicity can occur by systemic administration of LXR. Therefore, in this study, Mansukhani et al. used a peptide amphiphile covalently linked 4F peptide (ApoA1 PAs) to target an atherosclerosis plaque and aimed LXR agonist delivery to reduce its toxicity. These SAPs nanofibers acted as a nanocarrier (ApoA1-LXR PAs).76 The ApoA1 peptide amphiphile (PA) was meticulously designed with a C16 aliphatic tail, a β-sheet-forming sequence (V2A2), two negatively charged glutamic acid residues, a glycine spacer, and the 4F ApoA1 mimetic sequence (DWFKAFYDKVAEKFKEAF-NH2). The addition of the glutamic acid residues imparts a negative charge to the PA, enhancing its solubility and uptake by macrophages in physiological conditions. Given the correlation between oxidized LDL and atherosclerosis severity, the ApoA1 PA targets atherosclerotic plaques through its high-affinity binding to oxidized lipids. Hence, we developed the ApoA1 PA as a platform to use oxidized-lipid binding to selectively target atherosclerotic plaques. The ApoA1 PA formed aggregated mesh-like networks instead of nanofibers. By successfully co-assembling with a diluent PA (C16V2A2E2-NH2), ApoA1 PA naturally forms long nanofibers. This co-assembly process involved dissolving the PAs in HFIP, a solvent that disaggregates peptides, followed by solvent evaporation to achieve the desired nanofibers.77 Nanofiber formation was confirmed by conventional and cryogenic transmission electron microscopy except for 4F and 4F-LXR groups as expected. The median nanofiber length for ApoA1, ApoA1-LXR, scrambled, and diluent PAs was found 616, 712, 258, and 2541 nm, respectively. In vivo, results showed that nanofibers were accumulated at the targeted area in 24 h, maintained a high concentration for two days and eliminated 10–14 days post-injection. It was found that ApoA1-LXR PAs and ApoA1 PAs did not cause liver toxicity in male mice on the contrast LXR and scrambled PA. In the scope of plaque reduction, their outcomes depended on gender and control groups. They reported that ApoA1-LXR PAs have a significant effect on plaque reduction in comparison to the scrambled PAs in male mice, but not in females while they showed a similar reduction size in comparison to LXR in both genders. As a result, this study demonstrated the safety and effective potential of SAP-based nanofibers as a nanocarrier. However, their drug release mechanism and circumstances within blood should be investigated in detail.76
4. Conclusion
The past decades have seen growing research in the area of tissue regeneration at both clinical and preclinical levels. Specifically, studies on cardiovascular regeneration are desperately needed, as cardiovascular disorders account for the majority of fatalities in humans. Biomimetic scaffolds promise great opportunities to enhance cell proliferation, differentiation, etc., by triggering cell signalling, thereby promoting tissue regeneration.Peptide-based tissue scaffolds are a promising candidate as they are an efficient method to enhance the functional properties of inert and synthetic materials. The knowledge of the assembling mechanism of the peptides can be used to create new scaffolds by self-assembly of bioactive peptides or a combination of self-assembling peptides with the bioactive peptide. SAP-based nanofibers are favourable scaffolds as their morphological features are similar to ECM and versatile. They are capable of influencing and customising for specific assignments including signalling pathways, angiogenesis, cell proliferation as well as cargo other biomolecules. Their utilities are valued to develop scaffolds and may meet with requirements of cardiovascular tissue regeneration.
5. Future perspectives
Research on SAPs materials is rapidly increasing, and it has shown the potential of SAPs in medicine. SAPs are defined as the spontaneous formation of well-ordered and fairly stable structures under specific conditions, and studying this phenomenon is crucial for understanding molecular interactions within biological systems.78 Although self-assembly is prevalently involved in various biological functions, this field is still developing. SAP nanofibers are biocompatible and biodegradable but often lack mechanical strength. This limitation can be addressed by blending or conjugating peptides or by encapsulating peptides with nanofibers made of synthetic polymers. Improving the stability in physiological environments such as in vivo studies, and understanding the underlying mechanisms are vital for advancing their biomedical applications. Introducing inorganic molecules may enhance both stability and mechanical strength by stable interactions between the peptide and the inorganic ion. Also combining peptides with other biomolecules could further increase their stability and target selectivity. However, without a clear understanding of the fundamental molecular events driving self-assembly, these techniques have limited applications and hinder the rational design of new peptides with desired properties. Computational tools such as molecular dynamic simulations can help predict peptide behaviour, and mechanism of assembly and guide the design for more stable sequences, ultimately improving nanofibers for biomedical applications, especially in cardiovascular diseases. However, their conceptualization of cardiovascular regeneration needs to be deeply evaluated. The aforementioned studies highlight the promising role of SAPs in cardiovascular regeneration, emphasizing the need for safety and scalability to facilitate their transition into further preclinical and clinical applications. While most of the SAP-based scaffolds discussed in this review are primarily studied at the laboratory level, however, some SAPs such as RADA16, have shown significant potential in clinical trials and have been successfully commercialized.45 Despite the various peptide synthesis methods available—such as fermentation, chemical synthesis, and enzymatic synthesis—there is a pressing need for greener, faster, and optimized synthesis techniques. Innovations in these synthesis methods could greatly enhance the development of peptide materials, improving their efficacy and applicability in clinical settings. By advancing these methodologies, we can better harness the regenerative capabilities of SAPs and move toward more effective therapeutic solutions.Additionally, the use of external forces for the formation of peptide-based nanofibers is particularly promising. The electrospinning technique can be used to enhance or induce assembly in peptides that do not naturally self-assemble into fibres.79,80 SAPs can be directly electrospun due to their high anisotropic nature80 or mixed with polymers before electrospinning to improve the mechanical and thermal stability of the resulting functionalized nanofiber scaffolds.58,81–84 Electrospun peptide scaffolds are promising in tissue engineering due to their biocompatibility; however, an effective strategy needs to be developed to overcome challenges like low viscosity, low solubility, and lack of molecular entanglement which can impede fibre formation.85 Yet, the biggest challenge in peptide electrospinning is the need for a highly viscous solution (often 1 to 10 Pa s), which can only be reached by high concentrations of 10–50 wt%. In addition to being prone to affect by environmental factors such as temperature and humidity, suitable solvents are restricted.79
Although the electrospinning of peptides has been widely studied and explored,80,86–88 further research is needed to integrate SAPs with electrospinning techniques and to evaluate their potential in cardiovascular applications more deeply.
Author contributions
All authors conceptualized the article. Graphical abstract was created by N. E., S. C. and article was structured by S.C. Writing – original draft: D. S., S. C. and N. E. Supervision, writing – review & editing: E. C., S. P., M. R. and I. G.Data availability
All data is available upon request for the corresponding authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project has received funding from the European Union's research and innovation programme under the Marie Skłodowska-Curie grant agreement no. 101072645.References
- World Health Organization, Cardiovascular diseases (CVDs)-Key facts, https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds), (accessed January 1, 2024).
- C. W. Tsao, A. W. Aday, Z. I. Almarzooq, A. Alonso, A. Z. Beaton, M. S. Bittencourt, A. K. Boehme, A. E. Buxton, A. P. Carson, Y. Commodore-Mensah, M. S. V. Elkind, K. R. Evenson, C. Eze-Nliam, J. F. Ferguson, G. Generoso, J. E. Ho, R. Kalani, S. S. Khan, B. M. Kissela, K. L. Knutson, D. A. Levine, T. T. Lewis, J. Liu, M. S. Loop, J. Ma, M. E. Mussolino, S. D. Navaneethan, A. M. Perak, R. Poudel, M. Rezk-Hanna, G. A. Roth, E. B. Schroeder, S. H. Shah, E. L. Thacker, L. B. VanWagner, S. S. Virani, J. H. Voecks, N.-Y. Wang, K. Yaffe, S. S. Martin and on behalf of the American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee, Circulation, 2022, 145(8), e153–e639, DOI:10.1161/CIR.0000000000001052
.
- P. Piko, Z. Kosa, J. Sandor and R. Adany, Sci. Rep., 2021, 11, 3085 CrossRef CAS
.
- N. K. Movsisyan, M. Vinciguerra, J. R. Medina-Inojosa and F. Lopez-Jimenez, Ann. Glob. Health, 2020, 86, 21 CrossRef PubMed
.
- Center for Disease Control and Prevention, Heart Disease Facts|cdc.gov, https://www.cdc.gov/heartdisease/facts.htm, (accessed January 1, 2024).
-
M. Thiriet, in Vasculopathies: Behavioral, Chemical, Environmental, and Genetic Factors, ed. M. Thiriet, Springer International Publishing, Cham, 2018, pp. 1–90 Search PubMed
.
- S. U. Khan, S. U. Khan, M. Suleman, M. U. Khan, M. S. Khan, F. M. Arbi, T. Hussain, A. Mohammed Alsuhaibani and M. S. Refat, Curr. Probl. Cardiol., 2024, 49, 102084 CrossRef PubMed
.
- M. I. Nasser, X. Qi, S. Zhu, Y. He, M. Zhao, H. Guo and P. Zhu, Biomed. Pharmacother., 2020, 132, 110813 CrossRef CAS
.
- T. A. McDonagh, M. Metra, M. Adamo, R. S. Gardner, A. Baumbach, M. Böhm, H. Burri, J. Butler, J. Čelutkienė, O. Chioncel, J. G. F. Cleland, A. J. S. Coats, M. G. Crespo-Leiro, D. Farmakis, M. Gilard, S. Heymans, A. W. Hoes, T. Jaarsma, E. A. Jankowska, M. Lainscak, C. S. P. Lam, A. R. Lyon, J. J. V. McMurray, A. Mebazaa, R. Mindham, C. Muneretto, M. Francesco Piepoli, S. Price, G. M. C. Rosano, F. Ruschitzka, A. Kathrine Skibelund and ESC Scientific Document Group, Eur. J. Heart Failure, 2022, 24, 4–131 CrossRef
.
- B. Arjmand, M. Abedi, M. Arabi, S. Alavi-Moghadam, M. Rezaei-Tavirani, M. Hadavandkhani, A. Tayanloo-Beik, R. Kordi, P. P. Roudsari and B. Larijani, Front. Cell Dev. Biol., 2021, 9, 704903 CrossRef
.
- P. Decuzzi and J. P. Cooke, Methodist Debakey Cardiovasc J., 2013, 9, 186 CrossRef PubMed
.
- G. Basara, G. Bahcecioglu, S. G. Ozcebe, B. W. Ellis, G. Ronan and P. Zorlutuna, Biophys. Rev., 2022, 3, 031305 CrossRef CAS
.
- P. R. Schmitt, K. D. Dwyer and K. L. K. Coulombe, ACS Appl. Bio Mater., 2022, 5, 2461–2480 CrossRef CAS
.
- X. Sun, W. Altalhi and S. S. Nunes, Adv. Drug Delivery Rev., 2016, 96, 183–194 CrossRef CAS
.
- O. J. Mezu-Ndubuisi and A. Maheshwari, Pediatr. Res., 2021, 89, 1619–1626 CrossRef
.
-
K. Maeda, E. J. Suuronen and M. Ruel, in Biology and Engineering of Stem Cell Niches, ed. A. Vishwakarma and J. M. Karp, Academic Press, Boston, 2017, pp. 459–478 Search PubMed
.
- L. Shariati, Y. Esmaeili, I. Rahimmanesh, S. Babolmorad, G. Ziaei, A. Hasan, M. Boshtam and P. Makvandi, Environ. Res., 2023, 238, 116933 CrossRef CAS
.
- Y. Wu, L. Wang, B. Guo and P. X. Ma, ACS Nano, 2017, 11, 5646–5659 CrossRef CAS PubMed
.
- T. J. Moyer, H. A. Kassam, E. S. M. Bahnson, C. E. Morgan, F. Tantakitti, T. L. Chew, M. R. Kibbe and S. I. Stupp, Small, 2015, 11, 2750–2755 CrossRef CAS
.
- P.-H. Kim and J.-Y. Cho, BMB Rep., 2016, 49, 26–36 CrossRef CAS
.
- H. Shin, S. Jo and A. G. Mikos, Biomaterials, 2003, 24, 4353–4364 CrossRef CAS PubMed
.
- K. Klimek and G. Ginalska, Polymers, 2020, 12, 844 CrossRef CAS PubMed
.
- M. Shachar, O. Tsur-Gang, T. Dvir, J. Leor and S. Cohen, Acta Biomater., 2011, 7, 152–162 CrossRef CAS PubMed
.
- E. I. Biru, M. I. Necolau, A. Zainea and H. Iovu, Polymers, 2022, 14, 1032 CrossRef CAS PubMed
.
- G. Onak, O. Gökmen, Z. B. Yaralı and O. Karaman, J. Tissue Eng. Regener. Med., 2020, term.3095 CrossRef
.
- S. Lee, T. H. T. Trinh, M. Yoo, J. Shin, H. Lee, J. Kim, E. Hwang, Y. Lim and C. Ryou, Int. J. Mol. Sci., 2019, 20, 5850 CrossRef CAS PubMed
.
- A. J. Rufaihah, I. C. Yasa, V. S. Ramanujam, S. C. Arularasu, T. Kofidis, M. O. Guler and A. B. Tekinay, Acta Biomater., 2017, 58, 102–112 CrossRef CAS PubMed
.
- S. Huang, D. Lei, Q. Yang, Y. Yang, C. Jiang, H. Shi, B. Qian, Q. Long, W. Chen, Y. Chen, L. Zhu, W. Yang, L. Wang, W. Hai, Q. Zhao, Z. You and X. Ye, Nat. Med., 2021, 27, 480–490 CrossRef CAS
.
- B. Qian, A. Shen, S. Huang, H. Shi, Q. Long, Y. Zhong, Z. Qi, X. He, Y. Zhang, W. Hai, X. Wang, Y. Cui, Z. Chen, H. Xuan, Q. Zhao, Z. You and X. Ye, Adv. Sci., 2023, 10, 2303033 CrossRef CAS
.
- G. M. Whitesides and B. Grzybowski, Science, 2002, 295, 2418–2421 CrossRef CAS PubMed
.
- C. Yuan, A. Levin, W. Chen, R. Xing, Q. Zou, T. W. Herling, P. K. Challa, T. P. J. Knowles and X. Yan, Angew. Chem., Int. Ed., 2019, 58, 18116–18123 CrossRef CAS PubMed
.
- T. P. J. Knowles, C. A. Waudby, G. L. Devlin, S. I. A. Cohen, A. Aguzzi, M. Vendruscolo, E. M. Terentjev, M. E. Welland and C. M. Dobson, Science, 2009, 326, 1533–1537 CrossRef CAS
.
- S. I. A. Cohen, M. Vendruscolo, M. E. Welland, C. M. Dobson, E. M. Terentjev and T. P. J. Knowles, J. Chem. Phys., 2011, 135(6), 065105 CrossRef PubMed
.
- M.-C. Hsieh, D. G. Lynn and M. A. Grover, J. Phys. Chem. B, 2017, 121, 7401–7411 CrossRef CAS PubMed
.
- K. Tao, A. Levin, L. Adler-Abramovich and E. Gazit, Chem. Soc. Rev., 2016, 45, 3935–3953 RSC
.
- C. Yuan, R. Xing, J. Cui, W. Fan, J. Li and X. Yan, CCS Chem., 2024, 6, 255–265 CrossRef CAS
.
- A. Khan, P. Kumari, N. Kumari, U. Shaikh, C. Ekhator, R. Halappa Nagaraj, V. Yadav, A. W. Khan, S. Lazarevic, B. Bharati, G. Lakshmipriya Vetrivendan, A. Mulmi, H. Mohamed, A. Ullah, B. Kadel, S. B. Bellegarde and A. Rehman, Cureus, 2023, 15(8), e43431 Search PubMed
.
- L. Wang, X. Xin, P. Li, J. Dou, X. Han, J. Shen and J. Yuan, Colloids Surf., B, 2021, 205, 111855 CrossRef CAS
.
- V. S. Chernonosova, A. A. Gostev, Y. A. Chesalov, A. A. Karpenko, A. M. Karaskov and P. P. Laktionov, Int. J. Polym. Mater. Polym. Biomater., 2019, 68, 34–43 CrossRef CAS
.
- S. Koutsopoulos, J. Biomed. Mater. Res., Part A, 2016, 104, 1002–1016 CrossRef CAS
.
- J. Chen and X. Zou, Bioact. Mater., 2019, 4, 120–131 Search PubMed
.
-
A. L. Boyle, in Peptide Applications in Biomedicine, Biotechnology and Bioengineering, ed. S. Koutsopoulos, Woodhead Publishing, 2018, pp. 51–86 Search PubMed
.
- B. He, J. Zhao, Y. Ou and D. Jiang, Mater. Sci. Eng., C, 2018, 90, 728–738 CrossRef CAS
.
- H. Li, Q. Guo, Q. Tu, K. Xiong, W. Wang, L. Lu, W. Zhang, N. Huang and Z. Yang, J. Mater. Sci. Technol., 2022, 131, 106–114 CrossRef CAS
.
- S. Sankar, K. O’Neill, M. Bagot D’Arc, F. Rebeca, M. Buffier, E. Aleksi, M. Fan, N. Matsuda, E. S. Gil and L. Spirio, Front. Bioeng. Biotechnol., 2021, 9, 679525 CrossRef
.
- A. Saini, K. Serrano, K. Koss and L. D. Unsworth, Acta Biomater., 2016, 31, 71–79 CrossRef CAS
.
- L. D. D’Andrea, G. Iaccarino, R. Fattorusso, D. Sorriento, C. Carannante, D. Capasso, B. Trimarco and C. Pedone, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 14215–14220 CrossRef PubMed
.
- A. Pal, C. I. Smith, J. Palade, S. Nagaraju, B. A. Alarcon-Benedetto, J. Kilbourne, A. Rawls, J. Wilson-Rawls, B. L. Vernon and M. Nikkhah, Acta Biomater., 2020, 107, 138–151 CrossRef CAS
.
- W. Guo, W. Feng, J. Huang, J. Zhang, X. Fan, S. Ma, M. Li, J. Zhan, Y. Cai and M. Chen, ACS Appl. Mater. Interfaces, 2021, 13, 22131–22141 CrossRef CAS PubMed
.
- G. Ghosh, K. K. Kartha and G. Fernández, Chem. Commun., 2021, 57, 1603–1606 RSC
.
- R. Mammadov, B. Mammadov, M. O. Guler and A. B. Tekinay, Biomacromolecules, 2012, 13, 3311–3319 CrossRef CAS
.
- S. Toksoz, R. Mammadov, A. B. Tekinay and M. O. Guler, J. Colloid Interface Sci., 2011, 356, 131–137 CrossRef CAS PubMed
.
- Y. Wang, D. Wang, C. Wu, B. Wang, S. He, H. Wang, G. Liang and Y. Zhang, Theranostics, 2022, 12, 7237–7249 CrossRef CAS
.
- Z.-S. Razavi, M. Soltani, G. Mahmoudvand, S. Farokhi, A. Karimi-Rouzbahani, B. Farasati-Far, S. Tahmasebi-Ghorabi, H. Pazoki-Toroudi and H. Afkhami, Front. Bioeng. Biotechnol., 2024, 12, 1385124 CrossRef PubMed
.
- I. Valiente-Alandi, A. E. Schafer and B. C. Blaxall, J. Mol. Cell. Cardiol., 2016, 91, 228–237 CrossRef CAS
.
- X. Pang, X. He, Z. Qiu, H. Zhang, R. Xie, Z. Liu, Y. Gu, N. Zhao, Q. Xiang and Y. Cui, Signal Transduction Targeted Ther., 2023, 8, 1–42 CrossRef CAS
.
- P. Dhavalikar, A. Robinson, Z. Lan, D. Jenkins, M. Chwatko, K. Salhadar, A. Jose, R. Kar, E. Shoga, A. Kannapiran and E. Cosgriff-Hernandez, Adv. Healthcare Mater., 2020, 9, 2000795 CrossRef CAS
.
- A. Tambralli, B. Blakeney, J. Anderson, M. Kushwaha, A. Andukuri, D. Dean and H.-W. Jun, Biofabrication, 2009, 1, 025001 CrossRef PubMed
.
- K. A. Szychowski and J. Gmiński, Sci. Rep., 2019, 9, 20165 CrossRef CAS PubMed
.
- Q. Li, D. Xing, L. Ma and C. Gao, Mater. Sci. Eng., C, 2017, 73, 562–568 CrossRef CAS PubMed
.
- S. Vigneswari, J. M. Chai, K. H. Kamarudin, A.-A. A. Amirul, M. L. Focarete and S. Ramakrishna, Front. Bioeng. Biotechnol., 2020, 8, 567693 CrossRef
.
- M. Barczyk, S. Carracedo and D. Gullberg, Cell Tissue Res., 2010, 339, 269–280 CrossRef CAS
.
- F. Clauder, A. S. Czerniak, S. Friebe, S. G. Mayr, D. Scheinert and A. G. Beck-Sickinger, Bioconjugate Chem., 2019, 30, 2664–2674 CrossRef CAS PubMed
.
- M. Kanda, T. Nagai, N. Kondo, K. Matsuura, H. Akazawa, I. Komuro and Y. Kobayashi, Cell Transplant., 2023, 32, 096368972311740 Search PubMed
.
- X. Sun, M. He, L. Wang, L. Luo, J. Wang and J. Xiao, ACS Omega, 2019, 4, 16270–16279 CrossRef CAS PubMed
.
- S. Firoozi, S. Pahlavan, M.-H. Ghanian, S. Rabbani, S. Tavakol, M. Barekat, S. Yakhkeshi, E. Mahmoudi, M. Soleymani and H. Baharvand, Biomolecules, 2020, 10, 205 CrossRef CAS
.
- R. Alonaizan and C. Carr, Biochem. Soc. Trans., 2022, 50, 269–281 CrossRef CAS PubMed
.
- X. Fan, J. Zhan, X. Pan, X. Liao, W. Guo, P. Chen, H. Li, W. Feng, Y. Cai and M. Chen, Chem. Eng. J., 2022, 436, 135224 CrossRef CAS
.
- L. É. Uhljar and R. Ambrus, Pharmaceutics, 2023, 15, 417 CrossRef CAS
.
- A. Keirouz, Z. Wang, V. S. Reddy, Z. K. Nagy, P. Vass, M. Buzgo, S. Ramakrishna and N. Radacsi, Adv. Mater. Technol., 2023, 8, 2201723 CrossRef CAS
.
- N. Amariei, L. R. Manea, A. P. Bertea, A. Bertea and A. Popa, IOP Conf. Ser.: Mater. Sci. Eng., 2017, 209, 012092 Search PubMed
.
- B. Yan, Y. Zhang, Z. Li, P. Zhou and Y. Mao, SN Appl. Sci., 2022, 4, 172 CrossRef CAS
.
- A. Andukuri, M. Kushwaha, A. Tambralli, J. M. Anderson, D. R. Dean, J. L. Berry, Y. D. Sohn, Y.-S. Yoon, B. C. Brott and H.-W. Jun, Acta Biomater., 2011, 7, 225–233 CrossRef CAS PubMed
.
- O. Dotsenko, Open Cardiovasc. Med. J., 2010, 4, 97–104 CAS
.
- V. Harbour, C. Casillas, Z. Siddiqui, B. Sarkar, S. Sanyal, P. Nguyen, K. K. Kim, A. Roy, P. Iglesias-Montoro, S. Patel, F. Podlaski, P. Tolias, W. Windsor and V. Kumar, ACS Appl. Bio Mater., 2020, 3, 8978–8988 CrossRef CAS PubMed
.
- N. A. Mansukhani, E. B. Peters, M. M. So, M. S. Albaghdadi, Z. Wang, M. R. Karver, T. D. Clemons, J. P. Laux, N. D. Tsihlis, S. I. Stupp and M. R. Kibbe, Macromol. Biosci., 2019, 19, 1900066 CrossRef
.
- M. M. So, N. A. Mansukhani, E. B. Peters, M. S. Albaghdadi, Z. Wang, C. M. Rubert Pérez, M. R. Kibbe and S. I. Stupp, Adv. Biosys., 2018, 2, 1700123 CrossRef
.
- T. Li, X.-M. Lu, M.-R. Zhang, K. Hu and Z. Li, Bioact. Mater., 2022, 11, 268–282 CAS
.
- R. Bucci, E. Georgilis, A. M. Bittner, M. L. Gelmi and F. Clerici, Nanomaterials, 2021, 11, 1262 CrossRef CAS PubMed
.
- W. Nuansing, D. Frauchiger, F. Huth, A. Rebollo, R. Hillenbrand and A. M. Bittner, Faraday Discuss., 2013, 166, 209 RSC
.
- M. S. Liberato, S. Kogikoski, E. R. Da Silva, D. R. De Araujo, S. Guha and W. A. Alves, J. Mater. Chem. B, 2016, 4, 1405–1413 RSC
.
- R. Gharaei, G. Tronci, R. P. W. Davies, C. Gough, R. Alazragi, P. Goswami and S. J. Russell, J. Mater. Chem. B, 2016, 4, 5475–5485 RSC
.
- P. Brun, F. Ghezzo, M. Roso, R. Danesin, G. Palù, A. Bagno, M. Modesti, I. Castagliuolo and M. Dettin, Acta Biomater., 2011, 7, 2526–2532 CrossRef CAS PubMed
.
- M. Nune, U. M. Krishnan and S. Sethuraman, Mater. Sci. Eng., C, 2016, 62, 329–337 CrossRef CAS
.
-
S. L. Levit, R. C. Walker, A. L. Pham and C. Tang, in Green Electrospinning, ed. N. Horzum, M. M. Demir, R. Muñoz-Espí and D. Crespy, De Gruyter, 2019, pp. 41–68 Search PubMed
.
- S. Locarno, A. Eleta-Lopez, M. G. Lupo, M. L. Gelmi, F. Clerici and A. M. Bittner, RSC Adv., 2019, 9, 20565–20572 RSC
.
- M. Maleki, A. Natalello, R. Pugliese and F. Gelain, Acta Biomater., 2017, 51, 268–278 CrossRef CAS
.
- Y. Hamedani, P. Macha, E. L. Evangelista, V. R. Sammeta, V. Chalivendra, S. Rasapalli and M. C. Vasudev, J. Biomed. Mater. Res., 2020, 108, 829–838 CrossRef CAS PubMed
.
Footnote |
| † All the authors contributed equally to the review. |
| This journal is © The Royal Society of Chemistry 2025 |
