DOI:
10.1039/C3RA42066C
(Paper)
RSC Adv., 2013,
3, 14571-14579
Surface functionalization of graphene quantum dots with small organic molecules from photoluminescence modulation to bioimaging applications: an experimental and theoretical investigation†
Received
26th April 2013
, Accepted 7th June 2013
First published on 7th June 2013
Abstract
Surface chemistry provides an alternative approach to modulate the emission colour and efficiency of graphene quantum dots. We systematically investigated the surface chemistry of graphene quantum dots functionalized with a series of small organic molecules combining experimental and theoretical approaches. Experimental results indicated that surface functionalization with functional groups such as alcohol, amine and thiol can effectively tune the fluorescence of graphene quantum dots, and proved that amino groups can highly elevate the quantum yields of modified graphene quantum dots. The emission efficiency of 1,2-ethylenediamine functionalized graphene quantum dots reached up to 17.6% due to specific proton transfer to the conjugated fluorophore-like structure from ammonium formed by protonation. The polyaromatic structure within the graphene quantum dots was proposed to explain the fluorescence enhancement mechanism of graphene quantum dots functionalized by diamines. The computational results suggested that not only the size of the polyaromatic structures within graphene quantum dots can change their emissions, but surface functionalization can also tune their photoluminescence through modulating their band gaps. Toxicity experiments indicated that diamine-functionalized graphene quantum dots showed low cell toxicity similar to that of pristine graphene quantum dots. Moreover, the bioimaging experiments suggested that functionalized graphene quantum dots had identical abilities to label cells at a lower concentration than pristine graphene quantum dots owing to their higher quantum yields.
1 Introduction
Graphene quantum dots (GQDs) have attracted intensive research interest for their unique properties and wide variety of promising applications1–3 since they were discovered through electrophoretic analysis of fluorescent single-walled carbon nanotube fragments.4 GQDs have shown excellent performance in photovoltaic devices,5,6 photocatalysis,7,8 and bioimaging, and their potential applications for bioimaging includes single-photon and multi-photon bioimaging, which has attracted special attention due to their outstanding luminescence, low toxicity and good resistance to photo-degradation and bleaching.9,10 Compared to traditional semiconductor inorganic dots, carbon dots have the great advantage of biocompatibility due to their low or non-toxicity to living cells.11 Hence various methods to synthesize GQDs have been developed, of which two types of approaches include top-down and bottom-up synthesis. Top-down approaches may comprise acid oxidation,12–14 electrochemical oxidation15,16 and thermal decomposition,17–19 in which GQDs are produced from a larger carbon structure. Bottom-up approaches comprise solution chemistry methods,20,21 solvothermal synthesis22,23 and microwave-assisted methods,24,25 in which GQDs are formed from molecular precursors under specific reaction conditions.
Although various synthesis methods have been proposed and GQDs have shown superior properties with regard to chemical inertness, biocompatibility and sustainability, there are still some obstacles which hinder their practical applications in bioimaging, such as relatively low luminescence quantum yields (the quantum yields of most GQDs are lower than 10%), shifting fluorescence emissions and unclear luminescence mechanisms. Improving quantum yields is an emergent and indispensable task among these limitations. Thus, a great deal of effort was exerted to increase the quantum yields of GQDs through surface chemistry. Surface modulation of GQDs can not only greatly improve their quantum yields but also effectively tune their photoluminescence, which mainly includes tuning surface oxidation,26–29 and surface functionalization with organic molecules.30–38 The studies on surface oxidation have shown that surface oxidation or reduction can be used to alter the photoluminescence properties and improve the optical performance of GQDs. However, it is not easy to control the exact oxidation state in an experiment and may damage the conjugated structures within the graphene quantum dots.
Surface functionalization or modification is an alternative promising method. Functionalized groups on the surface of the GQDs can not only tune and enhance the luminescence but also provide an important intermediate for the subsequent functionalization of GQDs and modulation of their properties. Present relevant studies are focused on the functionalization of GQDs with poly(ethylene glycol) polymers.30–34 Sun et al.30 first prepared passivated GQDs with amine-terminated PEG1500N, resulting in an increase of the quantum yield to 10%. Using the same passivating agent, Liu et al.31 adopted a silica sphere template to synthesize passivated GQDs with high luminescence efficiencies of 11–15%. Combined with a separation technique, Wang et al.32 prepared the most fluorescent PEG1500N-functionalized GQDs, and pointed out that the particle size and degree of surface passivation influences the fluorescence quantum yields of GQDs to a great extent. Later, two reports indicated that shorter PEG200N-passivated GQDs also achieved a high emission efficiency of around 10%.33,34 Although surface functionalization with such polymers is still an effective approach to enhance the luminescence of GQDs, the detailed fluorescence enhancement mechanism remains unclear and the removal of extra polymers needs complex or expensive procedures. Small organic molecules are easily removed from functionalized GQDs and are suitable for large-scale production besides their superiority in price and maintenance of GQDs' size. Recently, some small amine compounds were employed as passivation agents to modify GQDs,35–38 and the emission efficiencies of these functionalized GQDs were increased, which shows that functionalization with small organic molecules is an effective and simple method to enhance fluorescence efficiency. However, until now, the mechanism of luminescence enhancement of such functionalized GQDs remained unknown, which has hindered the development and application of GQDs. There are still some important questions to be resolved to further improve this approach: (1) Why can amino-functional groups remarkably enhance the luminescence; (2) whether other functional groups, such alcohol and thiol, apart from amino-functional groups, endow as-prepared GQDs with high luminescence performances; (3) whether the number of amino-functional groups and the size of the amine compounds obviously influence the luminescence performance of GQDs; (4) whether the toxicity and bioimaging ability of functionalized GQDs was affected by functionalized groups compared to GQDs.
Attempting to resolve the above puzzles, we prepared a series of functionalized GQDs with amines, diamines, dialcohols and dithiols, and explored and compared their photoluminescence, cell toxicity and bioimaging applications. Pristine GQDs were synthesized through an acid oxidation method and then purified by dialysis, exhibiting bright green fluorescence. Interestingly, after functionalization on the surface with 1,2-ethylenediamine, the fluorescence emission colour was tuned to blue, and the quantum yield was improved up to 17.6%. However, functionalized GQDs with glycol and dithioglycol showed only slightly enhancements of their luminescence relative to pristine GQDs. These results indicated that amino-functional groups played a significant role in the improvement of emission efficiency. Moreover, to understand the enhancement mechanism of luminescence, the photoluminescence performance of diamine-functionalized GQDs with different carbon chains was further investigated. The quantum yields of the diamine-functionalized GQDs sharply decreased with lengthening of the carbon chain of the diamines from 1,2-ethylenediamine to 1,4-butanediamine although all of them have superiority in fluorescence intensity relative to pristine GQDs. We speculated that 1,2-ethylenediamine probably acted as a special functionality to enhance photoluminescence, which was verified by further experiments and theoretical evidence. Protonated 1,2-ethylenediamine can effectively promote proton transfer from amino-functional groups to the conjugated carbon structure, which greatly enhanced luminescence emission. In addition, biological experiments demonstrated that 1,2-ethylenediamine-functionalized GQDs not only show low toxicity similar to pristine GQDs, but also performed better for in vitro bioimaging.
2 Experimental section
2.1 Synthesis of pristine GQDs
Pristine GQDs were prepared according to the method reported by Peng and co-workers.14 A typical synthesis procedure was carried out as follows: 1.0 g nano-size graphite powder was added to a mixture of H2SO4 (180 mL) and HNO3 (60 mL), and then subjected to ultrasonication for 2 h to form a homogeneous suspension. After refluxing at 120 °C for 24 h, the resulting mixture was cooled to room temperature and diluted with distilled water. The excessive acid was neutralized by NaOH and then dialyzed in a dialysis bag (remaining molecular weight: 1000 Da) 4 times.
2.2 Surface functionalization of GQDs
Several organic molecules were used for the purpose of surface functionalization. For 1,2-ethylenediamine functionalized GQDs, a mixture of as-prepared GQDs (0.1 g) and SOCl2 (20 mL) was reacted for 2 h at 80 °C, and then vacuum-distilled to remove excess SOCl2, and finally 20 mL of 1,2-ethylenediamine was added and heated at 100 °C for 4 h. The resulting mixture was subsequently vacuum-distilled to remove most of the excess 1,2-ethylenediamine and then washed with ethanol several times, and finally dried at 100 °C overnight. A similar procedure was applied for the synthesis of functionalized GQDs with 1,2-glycol, 1,2-dithioglycol, 1,3-propanediamine, 1,4-butanediamine and ethylamine. The functionalized GQDs with 1,2-glycol, 1,2-dithioglycol, 1,2-ethylenediamine, 1,3-propanediamine, 1,4-butanediamine and ethylamine were denoted G-GQDs, DTG-GQDs, EDA-GQDs, PDA-GQDs, BDA-GQDs and EA-GQDs respectively, while pristine GQDs were denoted as P-GQDs.
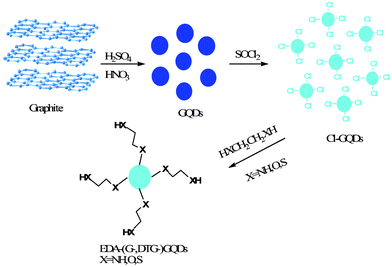 |
| | Scheme 1 A schematic route of the synthesis of functionalized GQDs with selected diamines, glycols and dithioglycols. | |
2.3 Characterization of the GQDs
Pristine or functionalized GQDs were homogenously dispersed in water with the aid of ultrasonication. The UV-Vis spectra were obtained on a PerkinElmer 950 spectrometer. The fluorescence spectra were recorded on a PerkinElmer LS-55 spectrometer, and lifetimes were determined using a FLS920 fluorescence spectrophotometer. The Fourier transform infrared spectroscopy (FTIR) was carried out on a Thermo NEXUS 670 Fourier transform infrared spectrometer, in which a KBr plate method was used. The morphologies of the GQDs were characterized by transmission electron microscopy performed on a JEOL-2100F instrument with an accelerating voltage of 200 KV. All samples were prepared by dropping aqueous suspensions of GQDs onto Cu grids coated with a holey amorphous carbon film followed by solvent evaporation under a powerful light. The X-ray photoelectron spectroscopy analyses were conducted using a Kratos Axis ULTRA X-ray photoelectron spectrometer with a 165 nm hemispherical electron energy analyzer. The incident radiation came from a monochromatic Al X-ray (1486.6 eV) at 15 kV and 3 mA. Wide survey scans were taken at an analyzer pass energy of 160 eV over a 1400–0 eV binding energy with 1.0 eV step and a dwell time of 100 ms, while narrow multiplex higher resolution scans were performed at a pass energy of 20 eV with 0.05 eV step and a dwell time of 200 ms. The pressure in the analysis chamber was less than 7.5 × 10−9 Torr during sample analysis. Atomic concentrations were calculated using Vision software.
2.4 Determination of quantum yields
Determination of the quantum yields of the GQDs was achieved by comparison of the wavelength integrated intensity of the GQDs to that of the standard quinine sulfate as described in our previous paper.39,40 The optical absorbance was kept below 0.05 to avoid inner filter effects. The quantum yields of the GQDs were calculated using the following formula:
| Φ = ΦS [(I·AS·n2)/(IS·A·ns2)] |
where Φ is the quantum yield, I is the integrated intensity, A is the optical density and n is the refractive index of the solvent. The subscript S refers to the standard reference of known quantum yield. Quinine sulfate was chosen as the standard, whose quantum yield is 0.577 and nearly constant for excitation wavelengths from 200 nm to 400 nm.
2.5 Cellular toxicity test
Human Hela cells (105 cells mL−1) were cultivated in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum and 100 μg mL−1 penicillin/streptomycin for 12 h in an incubator (37 °C, 5% CO2). Then suspensions of GQDs with different doses were added, which maintained the final concentrations of GQDs at 100, 125, 150, 175 and 200 μg mL−1 respectively. The cells were cultivated for 24 h and then 20 μL of 5 mg mL−1 MTT solution was added to every cell well. After the cells were incubated for a further 4 h, the culture medium was removed and then 150 μL of DMSO was added. The resulting mixture was shaken for 15 min at room temperature without light. The optical density (OD) of the mixture was measured at 570 nm. The OD values were determined using a Thermo multiskan spectrum microplate spectrophotometer. The cell viability was estimated according to the following equation
| Cell viability (%) = ODTreated/ODcontrol × 100% |
where ODcontrol was obtained in the absence of GQDs, and ODTreated was obtained in the presence of GQDs.
2.6 Cellular imaging
The human Hela cells were cultivated for 12 h in culture medium containing DMEM supplemented with 10% fetal bovine serum and 100 μg mL−1 penicillin/streptomycin. Suspensions of GQDs from the stock solution were prepared with Dulbecco's phosphate buffer saline (DPBS). The suspension was added to the well of a chamber slide, and the final concentration of GQDs was 75 μg mL−1, followed by incubation at 37 °C in a 5% CO2 incubator for 24 h. Prior to inspection with a Leica laser scanning confocal microscope, the excess GQDs were removed by washing 3 times with warm DPBS. The fluorescence images were obtained at an excitation wavelength of 355 nm.
2.7 Theoretical details
A series of conjugated planar structures with different aromatic rings including 24, 42, 54, 84 and 96 sp2-carbons, and different hydroxyls and carboxyls were modeled to represent the functional structures of pristine GQDs of different sizes for photoluminescence. Similarly, for functionalized GQDs, all the carboxyls were replaced by corresponding amides. These model structures were used to evaluate the effect of size of the polyaromatic structure and functionalized groups on the band gap responsible for the photoluminescence of GQDs. Another series of conjugated planar structures with seven aromatic rings containing 24 sp2-carbons and three hydroxyls and protonated diamines with different carbon chains were constructed to explore the nature of the great fluorescence enhancement by 1,2-ethylenediamine. All the model structures were fully optimized using a DFT method at a PBE0/6-31G(d) level.41,42 The vibrational frequency was carried out to verify the stability of the structures. All the calculations were carried out with a Gaussian 09 package.43
3 Results and discussion
3.1 Characterization of GQDs
The pristine GQDs (P-GQDs) were synthesized by chemical oxidation and cutting of nano-sized graphite powder and purified by dialysis to remove most salts including Na2SO4 and NaNO3. The as-synthesized P-GQDs are highly soluble in water and other polar organic solvents. The TEM image in Fig. 1B shows that P-GQDs have a relatively narrow size distribution around 10 nm, which are close to those reported in the literature.44 P-GQDs were first reacted with SOCl2, and then with a series of small organic molecules including 1,2-ethylenediamine, 1,2-glycol, 1,2-dithioglycol, 1,3-propanediamine, 1,4-butanediamine and ethylamine to produce surface-functionalized GQDs (Scheme 1). Surface functionalization of small molecules does not change the size of the GQDs, which was verified by TEM as shown in Fig. 1A. The 1,2-ethylenediamine-functionalized GQDs (EDA-GQDs) showed a nearly identical size distribution below 10 nm, close to that for P-GQDs. The HR-TEM image in Fig. 1C showed that the EDA-GQDs have a crystalline structure consisting of parallel crystal planes.
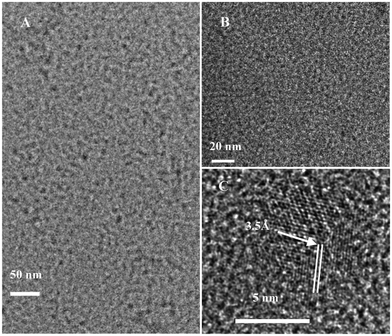 |
| | Fig. 1 TEM images of 1,2-ethylenediamine-functionalized GQDs (A) and pristine GQDs (B). HR-TEM image of 1,2-ethylenediamine-functionalized GQDs (C). | |
In order to analyze the surface state of the modified GQDs, XPS spectra of EDA-GQDs and DTG-GQDs as well as P-GQDs were recorded, which are shown in Fig. 2 and Fig. S1–S3, ESI†. All of the XPS survey spectra of pristine and modified GQDs display a predominant C1s peak at 284.8 eV and O1s peak at 532.2 eV. Since salts including Na2SO4 and possible added impurities such as Na2SiO4 were not completely removed, their XPS signals also appear in the wide spectra. The XPS spectra of EDA-GQDs show an N1s peak at 399.5 eV. The high-resolution XPS spectra of N1s for EDA-GQDs sample are given in Fig. 2d. The high resolution N1s spectrum of EDA-GQDs reveals the presence of both amide (398.4 eV) and protonated amine (400.2 eV) N atoms. The elemental analysis indicated that the composition is N 21.59%, O 25.96% and C 52.45% (Table 1). The XPS spectrum of DTG-GQDs showed an S2s peak at 227.1 eV and an S2p peak at 162.4 eV. As shown in Fig. 2c, the high-resolution XPS spectra of S2p for DTG-GQDs presents two peaks at 164.5 eV and 163.3 eV which were assigned to SH and S linked to carbonyls at the surface. The elemental analysis indicated that the composition is S 23.72%, O 26.59% and C 49.69%. These XPS results confirm the successful incorporation of N or S into the GQDs and the formation of amine-functionalized GQDs and thiol-functionalized GQDs respectively. The XPS spectrum for the G-GQDs was not recorded because the obtained sample was not in the pure solid state and thus was not accessible for XPS testing. In order to confirm the functionalization of 1,2-glycol and other diamines onto the GQDs, we recorded their FT-IR spectra as shown in Fig. S4, ESI†. The spectra for the G-GQDs showed a strong peak of a C–O stretching vibration at around 1080 cm−1, the double peaks of O–H distortion vibrations in the range 1450–1300 cm−1, and a strong and broad peak of an O–H stretching vibration at around 3400 cm−1, supporting the connecting of 1,2-glycol to the carbon dots. The characteristic C–N vibration peak at about 1100 cm−1 appeared in the FT-IR spectra of BDA-GQDs, PDA-GQDs and EA-GQDs, suggesting the successful bonding of amino groups and graphene quantum dots. In the spectra of EA-GQDs, strong peaks at 3060 and 2980 cm−1 were induced by the presence of methyl groups in ethylamine.
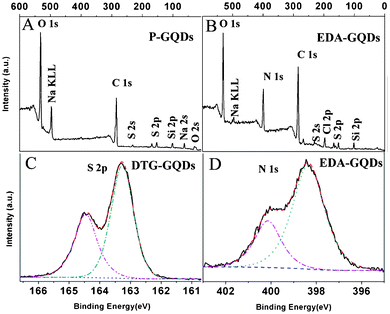 |
| | Fig. 2 XPS spectra of P-GQDs (a), EDA-GQDs (b,d) and DTG-GQDs (c). | |
Table 1 The concentrations of C, O and S or N in P-GQDs, EDA-GQDs and DTG-GQDs as determined by XPS
| Samples |
C (wt%) |
O (wt%) |
S (wt%) |
N (wt%) |
| P-GQDs |
53.43 |
46.57 |
— |
— |
| DTG-GQDs |
49.69 |
26.59 |
23.72 |
— |
| EDA-GQDs |
52.46 |
25.96 |
— |
21.59 |
3.2 Photoluminescence of GQDs
All the GQDs showed visible photoluminescence in water. P-GQDs exhibited a fluorescence band centered at 437 nm at an excitation wavelength of 322 nm. The emission peak of the DTG-GQDs was shifted to 429 nm, while the optical emission wavelengths of G-GQDs and EDA-GQDs were changed to 417 and 418 nm respectively, indicating that functionalization of GQDs with 1,2-glycol, 1,2-dithioglycol and 1,2-ethylenediamine enables the fluorescence of the GQDs to be blue-shifted(Fig. 3). This was also verified by the fact that the P-GQDs showed green fluorescence, while the EDA-GQDs emitted strong blue light under a UV lamp. We used quinine sulfate as a standard to measure the quantum yields of P-GQDs, DTG-GQDs, G-GQDs and EDA-GQDs because its quantum yield was identical (0.577) in the excitation wave length range 200–400 nm.45 As shown in Table 2, their lifetimes are between 3.56 and 7 ns, suggesting their fluorescence. However, their quantum yields are highly changeable. The quantum yield of P-GQDs is only 0.017, nevertheless, the fluorescence is enhanced after functionalization with 1,2-glycol, 1,2-dithoiglycol and 1,2-ethylenediamine. The addition of 1,2-glycol and 1,2-dithoiglycol only slightly enhanced the fluorescence of the functionalized GQDs, whereas functionalization with 1,2-ethylenediamine lead to a sharp increase in the quantum yield to 0.176. Their only difference is their surface functional group, which implies that the amino functional group has a specific function to enhance the fluorescence of GQDs. This point was consistent with previous papers, in which the quantum yields of GQDs functionalized with amino groups were remarkably elevated.24,29,35,36 Peng et al. adopted 4,7,10-trioxal-1,13-tridecanediamine (TTDDA) as a passivation agent to prepare carbon dots with a quantum yield of 0.13 through a bottom-up approach.24 Using the same passivation agent, Liu et al. obtained enhanced multicolor photoluminescent carbon dots through a bottom-up approach.35 Adopting a similar approach, Wang et al. employed 1-hexadecylamine as a passivation agent to synthesize highly fluorescent carbon dots.29 Recently, Zhu et al. conducted the surface functionalization with several alkylamines, and found that linkage of alkylamines to the surface of GQDs can modulate the fluorescence.36 In these reports, different amino groups including amines and diamines with different carbon chains were explored, however, there were no clear principles and explanation for the enhancement and modulation of the fluorescence of carbon dots. Therefore, attempting to explore these questions, we prepared additional GQDs functionalized with ethylamine (EA-GQDs), 1,3-propanediamine (PDA-GQDs) and 1,4-butanediamine (BDA-GQDs). As shown in Fig. 4, the emission peaks of the PDA-GQDs and BDA-GQDs are very similar to that of the EDA-GQDs, however, the central emission wavelength of EA-GQDs is red-shifted to 430 nm relative to those of dimaine-functionalized GQDs and P-GQDs. From the viewpoint of the influence of the length of carbon chain of the diamines on the quantum yields, one can infer that the quantum yields of GQDs functionalized with diamines are increased as the carbon chain is shortened. However, the quantum yield of the ethylamine-functionalized GQDs is only slightly elevated relative to that of P-GQDs compared with that of the EDA-GQDs. The difference between 1,2-ethylenediamine and ethylamine is the terminal amino group, which hints that the terminal amino group may play an important role to enhance the fluorescence. From their obviously different lifetimes, we can also infer that the terminal amino group has a strong ability to tune the fluorescence of carbon dots.
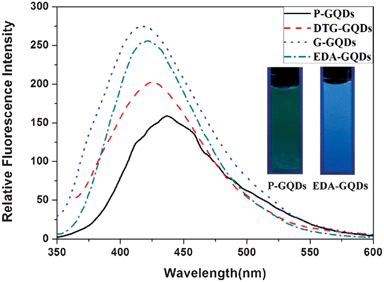 |
| | Fig. 3 The emission spectra of the P-GQDs, DTG-GQDs, G-GQDs and EDA-GQDs, and fluorescence images of P-GQDs and EDA-GQDs under a UV lamp (inset). | |
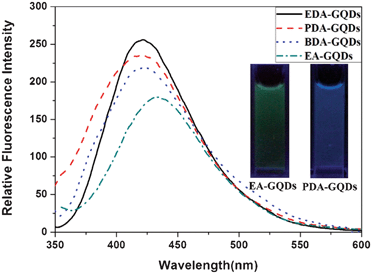 |
| | Fig. 4 The emission spectra of the EDA-GQDs, PDA-GQDs, BDA-GQDs and EA-GQDs, and fluorescence images of EA-GQDs and PDA-GQDs under a UV lamp (inset). | |
Table 2 Fluorescence parameters of all the GQDs
| Sample |
λ
ex
(nm) |
λ
em
(nm) |
Φ
f
(%) |
τ
(ns) |
|
The excitation wavelength.
The central emission wavelength.
The fluorescence quantum yield determined with quinine sulfate (0.577) as the reference under neutral conditions.
The average lifetime.
All the parameters were determined at pH 10.
|
| P-GQDs |
322 |
437 |
1.7 |
3.56 |
| DTG-GQDs |
351 |
429 |
1.8 |
4.41 |
| G-GQDs |
335 |
418 |
2.0 |
6.26 |
| EDA-GQDs |
339 |
417 |
17.6 |
6.92 |
| PDA-GQDs |
315 |
420 |
7.8 |
8.48 |
| BDA-GQDs |
339 |
427 |
2.5 |
7.39 |
| EA-GQDs |
316 |
430 |
2.1 |
19.06 |
| EDA-GQDse |
337 |
415 |
8.9 |
7.03 |
In order to discover the underlying nature of the fluorescence enhancement by 1,2-ethylenediamine, we conducted experiments to determine the variation of fluorescence, dependent on pH. As shown in Fig. 5, the fluorescence intensity of the EDA-GQDs showed a sharp drop from pH 9.2 to pH 10.3, while its fluorescence intensity remains nearly unchanged in the ranges pH 6.5 to 9.2 and pH 10.3 to 12.8, in which the concentration of EDA-GQDs was kept identical. This phenomenon implied that pH or protons had a significant effect on the fluorescence enhancement. This was also verified by their quantum yields and lifetimes. As shown in Table 2, the lifetimes of the EDA-GQDs at pH 7 and 10 are very close, indicating that the nature of its fluorescence is not changed by the pH of the environment. However, the quantum yield at pH 10 is decreased by 0.087 relative to that at pH 7, which clearly demonstrates that the pH of the environment or presence of protons heavily impacts on the fluorescence of carbon dots. It can be deduced that the terminal amino group of EDA-GQDs is protonated below pH 9.2, and thus the possible proton transfer from the protonated amino group to conjugated carbon structure enhanced the fluorescence of the carbon dots. This is very similar to quinine: when quinine is protonated, its fluorescence efficiency can reach 0.58, but its quantum yield sharply descends when quinine is in a deprotonated state.46 It was reported that protonation and the resulting strong hydrogen bonding may make a larger contribution in the excited state than in the ground state, leading to fluorescence enhancement.47 A recent study also showed that protonation of the amino groups can effectively prevent electron transfer of the excited state and leads to recovery of the fluorescence of local emission states.48 This evidence supports the theory that protonation of amino groups neighboring fluorophores can effectively enhance fluorescence.
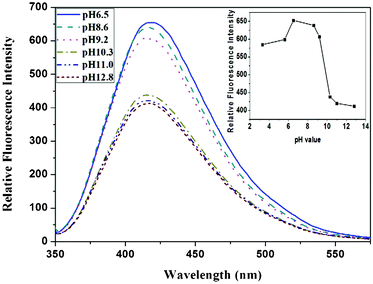 |
| | Fig. 5 The emission spectra change of EDA-GQDs at different pHs, and the intensity variation dependent on pH (inset). | |
3.3 Photoluminescence modulation mechanism
Although several speculative explanations for the strong photoluminescence have been proposed since the discovery of luminescent carbon dots, a convincing photoluminescence mechanism remains unclear. Sun et al. attributed the photoluminescence from carbon dots to the presence of surface energy traps that become emissive upon stabilization as a result of surface passivation.30 Zhou et al. developed Sun et al.'s proposal, suggesting that the mechanism is the radiative recombination of excitons from carbon dots of different sizes and a distribution of different emission trap sites.15 Pan et al. proposed that the luminescence may originate from free zigzag sites with carbene-like triplet ground states.49 Boutlinos et al. speculated that the ultrafine size of the dots combined with their disordered structure favors a high concentration of defect sites at the surface, giving rise to emission.22 Bao et al. also supported the defects mechanism that surface defects can trap excitons.50 However, Boutlinos et al. also thought that it is possible that the formation of several different polyaromatic fluorophores within the carbogenic network leads to fluorescence.22,51 Later, Li et al. provided convincing evidence for the graphite fragment structure within carbon dots, and proved that the strong emission of carbon dots comes from the quantum-sized graphite structure.7 The nature of this proposed mechanism is very similar to the explanation of polyaromatic fluorophores suggested by Boutlinos et al. Wang et al. also found that the fluorescence resembled those of band-gap transitions that may originate from similar polyaromatic clusters.32 Similar luminescence mechanisms for oxidized carbon nanotubes and graphene oxide were also proposed and investigated by our group and Shang et al., respectively.40,52 From analysis of the references above, we think that it is reasonable for polyaromatic fluorophores or quantum-sized graphite structures to explain the photoluminescence of quantum graphene dots.
Attempting to explore the photoluminescence modulation mechanism through surface functionalization with small organic molecules, we constructed a series of polyaromatic structures to model the fluorophore structure within carbon dots and diamine-functionalized carbon dots. Polyaromatic structures with 7, 13, 19, 31 and 37 aromatic cycles were constructed and optimized, and their band gaps are displayed in Fig. 6. The results showed that the band gap gradually declined with increasing size of the polyaromatic structures, and the declining trend is obvious. This result is consistent with the phenomena observed in the experiments.53 Dong et al.53 separated carbon dots into different fractions through dialysis with different membranes, and found that each fraction of similar size had different emission from the others. Their emission colours were arranged from blue to red corresponding to their sizes from small to large. This experimental evidence complements our computational results, supporting that the size of carbon dots or polyaromatic structures within the carbon dots dominated their emission. By comparing the band gaps between the original polyaromatic structures and diamine-functionalized structures, we found that the band gap can be remarkably elevated by surface functionalization with diamines, indicating that surface can also tune the fluorescence of GQDs. This is consistent with the findings by Shi et al. that both size and shape can impact the quantum mechanical and electronic properties of carbon dots.54 We optimized the structures of EDA-GQDs, PDA-GQDs, BDA-GQDs and their protonated products, and parts of them are shown in Fig. 7. We found that the structure of protonated EDA-GQDs was remarkably changed relative to those of EDA-GQDs and the other structures. When functionalized 1,2-ethylenediamine was not protonated, its carbon chain is arranged in a zigzag line, however, a cyclic structure formed through the hydrogen bonds between ammonium and the oxygen atom of the amide when it was protonated. This cyclic structure was not found in the protonated PDA-GQDs and BDA-GQDs. This cyclic structure facilitated the indirect proton transfer from ammonium to conjugated polyaromatic structure through the carboxyl groups. Thus the resulting proton transfer would have a large contribution in the excited state, and also prevent charge transfer in the excited state leading to the recovery of fluorescence.47,48 These two aspects finally enhanced the fluorescence emission. This finding rationally explain the stronger enhancement of fluorescence by 1,2-ethylenediamine than those by 1,3-propanediamine and 1,4-butanediamine.
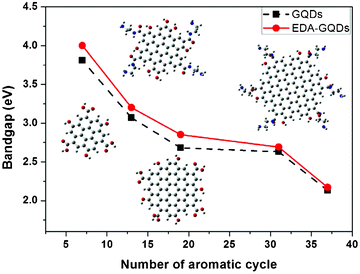 |
| | Fig. 6 The influence of the sizes of polyaromatic structures and functionalized diamines on the band gaps of modeled fluorophore structures within carbon dots. Inset: the optimized structures of different modeled structures at the PBE0/6-31G(d) level. | |
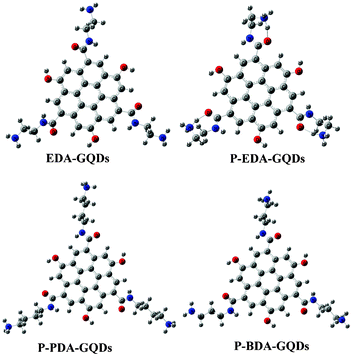 |
| | Fig. 7 The modeled structures optimized at the PBE0/6-31G(d) level. These structures represent EDA-GQDs, protonated EDA-GQDs, protonated PDA-GQDs and protonated BDA-GQDs, respectively. | |
3.4 Cellular toxicity test and bioimaging
In order to assess the possible cellular toxicity caused by the presence of functionalized diamines, the in vitro cytotoxicity of P-GQDs and EDA-GQDs was evaluated with human Hela cells by a methylthiazolydiphenyltetrazolium bromide (MTT) assay as shown in Fig. 8. The results suggested that both P-GQD and EDA-GQDs showed very low toxicity to human Hela cells with cell viabilities of higher than 80% when their concentration was below 125 μg mL−1. Their low toxicity is comparable to that of carbon dots reported in previous papers.11,17,29,33 From the results, it can be noted that EDA-GQDs showed comparably low toxicity to human Hela cells relative to P-GQDs, indicating that the functionalization with organic molecules has little influence on the cell toxicity of GQDs. We also compared their ability to label the human Hela cells, as shown in Fig. 9. The results showed that both types of GQDs easily went into cytoplasm and clearly labelled the cytoplasm. Since the emission efficiency of EDA-GQDs is higher than that of P-GQDs, smaller amounts of EDA-GQDs can be used in cell imaging to achieve an identical fluorescence performance.
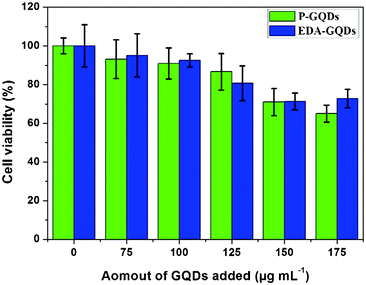 |
| | Fig. 8 Effect of P-GQDs and EDA-GQDs on human Hela cells. | |
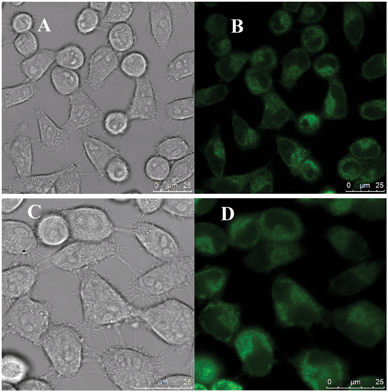 |
| | Fig. 9 Laser scanning confocal microscopy images of human Hela cells labeled by EDA-GQDs (A and B) and P-GQDs (C and D). Images for A and C were captured in bright field, and images for B and D were obtained at an excitation wavelength of 355 nm. | |
4 Conclusions
We synthesized a series of graphene quantum dots functionalized by different small organic molecules including dialcohols, diamines and dithiols. The functionalization of these functional groups on the graphene quantum dots provided an effective approach to modulate their emission colour and efficiency. Diamines showed a stronger ability to highly elevate the quantum yield of functionalized graphene quantum dots than the other functional groups due to their unique protonation mechanism. The computational results showed that 1,2-ethylenediamine functionalization on the surface of graphene quantum dots can form a specific cyclic structure which facilitates the proton transfer from the ammonium moiety to the conjugated structure, and thus lead to the largest enhancement of fluorescence. The polyaromatic structure mechanism can rationally explain the experimental results, and thus was adopted to explore the fluorescence enhancement mechanism. Computational results based on this fluorescence mechanism suggested that both size and surface functionalization can effectively modulate the photoluminescence of graphene quantum dots. Cellular toxicity tests and bioimaging experiment demonstrated that functionalized graphene quantum dots showed low toxicity, comparable to pristine graphene quantum dots, but revealed a much better bioimaging performance than pristine graphene quantum dots. This work provides a clear direction of surface functionalization with small organic molecules to improve and modulate the photoluminescence of graphene quantum dots in the future.
Acknowledgements
We are thankful for the support by the National Natural Science Foundation of China (No. 21005073 and 21275131) and Zhejiang Province (No. LY13B050001, LQ13B050002, 2011C3751 and Y201225601).
References
- S. N. Baker and G. A. Baker, Angew. Chem., Int. Ed., 2010, 49, 6726–6744 CrossRef CAS.
- J. H. Shen, Y. H. Zhu, X. L. Yang and C. Z. Li, Chem. Commun., 2012, 48, 3686–3699 RSC.
- H. T. Li, Z. K. Kang, Y. Liu and S.-T. Lee, J. Mater. Chem., 2012, 22, 24230–24253 RSC.
- X. Y. Xu, R. Ray, Y. L. Gu, H. J. Ploehn, L. Gearheart, K. Raker and W. A. Scrivens, J. Am. Chem. Soc., 2004, 126, 12736–12737 CrossRef CAS.
- V. Gupta, N. Chaudhary, R. Srivastava, G. D. Sharma, R. Bhardwaj and S. Chand, J. Am. Chem. Soc., 2011, 133, 9960–9963 CrossRef CAS.
- X. Yan, X. Cui, B. S. Li and L. S. Li, Nano Lett., 2010, 10, 1869–1873 CrossRef CAS.
- H. T. Li, X. D. He, Z. H. Kang, H. Huang, Y. Liu, J. L. Liu, S. Y. Lian, C. H. A. Tsang, X. B. Yang and S.-T. Lee, Angew. Chem., Int. Ed., 2010, 49, 4430–4434 CrossRef CAS.
- S. J. Zhuo, M. W. Shao and S.-T. Lee, ACS Nano, 2012, 6, 1059–1064 CrossRef CAS.
- L. Cao, X. Wang, M. J. Meziani, F. S. Lu, H. F. Wang, P. G. Luo, Y. Lin, B. A. Harruff, L. M. Veca, D. Murray, S.-Y. Xie and Y.-P. Sun, J. Am. Chem. Soc., 2007, 129, 11318–11319 CrossRef CAS.
- S.-T. Yang, L. Cao, P. G. Luo, F. S. Lu, X. Wang, H. F. Wang, M. J. Meziani, Y. F. Liu, G. Qi and Y.-P. Sun, J. Am. Chem. Soc., 2009, 131, 11308–11309 CrossRef CAS.
- S.-T. Yang, X. Wang, H. F. Wang, F. S. Lu, P. G. Luo, L. Cao, M. J. Meziani, J.-H. Liu, Y. F. Liu, Y. P. Huang and Y.-P. Sun, J. Phys. Chem. C, 2009, 113, 18110–18114 CAS.
- H. P. Liu, T. Ye and C. D. Mao, Angew. Chem., Int. Ed., 2007, 46, 6473–6475 CrossRef CAS.
- Z.-A. Qiao, Y. F. Wang, Y. Gao, H. W. Li, T. Y. Dai, Y. L. Liu and Q. S. Huo, Chem. Commun., 2010, 46, 8812–8814 RSC.
- J. Peng, W. Gao, B. K. Gupta, Z. Liu, R. Romero-Aburto, L. H. Ge, L. Song, L. B. Alemany, X. B. Zhan, G. H. Gao, S. A. Vithayathil, B. A. Kaipparettu, A. A. Marti, T. Hayashi, J.-J. Zhu and P. M. Ajayan, Nano Lett., 2012, 12, 844–849 CrossRef CAS.
- J. G. Zhou, C. Booker, R. Y. Li, X. T. Zhou, T.-K. Sham, X. L. Sun and Z. F. Ding, J. Am. Chem. Soc., 2007, 129, 744–745 CrossRef CAS.
- H. Ming, Z. Ma, Y. Liu, K. M. Pan, H. Yu, F. Wang and Z. H. Kang, Dalton Trans., 2012, 41, 9526–9531 RSC.
- S. J. Zhu, J. H. Zhang, C. Y. Qian, S. J. Tang, Y. F. Li, W. J. Yuan, B. Li, L. Tian, F. Liu, R. Hu, H. N. Gao, H. T. Wei, H. Zhang, H. C. Sun and B. Yang, Chem. Commun., 2011, 47, 6858–6860 RSC.
- C. Z. Zhu, J. F. Zhai and S. J. Dong, Chem. Commun., 2012, 48, 9367–9369 RSC.
- F. Yang, M. L. Zhao, B. Z. Zheng, D. Xiao, D. Wu, L. Wu and Y. Guo, J. Mater. Chem., 2012, 22, 25471–25479 RSC.
- X. Yan, X. Cui and L.-S. Li, J. Am. Chem. Soc., 2010, 132, 5944–5945 CrossRef CAS.
- R. L. Liu, D. Q. Wu, X. L. Feng and K. Mullen, J. Am. Chem. Soc., 2011, 133, 15221–15223 CrossRef CAS.
- A. B. Bourlinos, A. Stassinpoulos, D. Anglos, R. Zboril, M. Karakassides and E. P. Giannelis, Small, 2008, 4, 455–458 CrossRef CAS.
- A. B. Bourlinos, A. Stassinpoulos, D. Anglos, R. Zboril, V. Georgakilas and E. P. Giannelis, Chem. Mater., 2008, 20, 4539–4541 CrossRef CAS.
- H. Peng and J. Travas-Sejdic, Chem. Mater., 2009, 21, 5563–5565 CrossRef CAS.
- X. Y. Zhai, P. Zhang, C. J. Liu, T. Bai, W. C. Li, L. M. Dai and W. G. Liu, Chem. Commun., 2012, 48, 7955–7957 RSC.
- H. Z. Zheng, Q. L. Wang, Y. J. Long, H. J. Zhang, X. X. Huang and R. Zhu, Chem. Commun., 2011, 47, 10650–10652 RSC.
- S. J. Zhu, J. H. Zhang, X. Liu, B. Li, X. F. Wang, S. J. Tang, Q. N. Meng, Y. F. Li, C. Shi, R. Hu and B. Yang, RSC Adv., 2012, 2, 2717–2720 RSC.
- Y.-M. Long, C.-H. Zhou, Z.-L. Zhang, Z.-Q. Tian, L. Bao, Y. Lin and D.-W. Pang, J. Mater. Chem., 2012, 22, 5917–5920 RSC.
- S. J. Zhu, J. H. Zhang, S. J. Tang, C. Y. Qian, L. Wang, H. Y. Wang, X. Liu, B. Li, W. L. Yu, X. F. Wang, H. C. Sun and B. Yang, Adv. Funct. Mater., 2012, 22, 4732–4740 CrossRef CAS.
- Y.-P. Sun, B. Zhou, Y. Lin, W. Wang, K. A. S. Fernando, P. Pathak, M. J. Meziani, B. A. Harruff, X. Wang, H. F. Wang, P. G. Luo, H. Yang, M. E. Kose, B. L. Chen, L. M. Veca and S.-Y. Xie, J. Am. Chem. Soc., 2006, 128, 7756–7757 CrossRef CAS.
- R. L. Liu, D. Q. Wu, S. H. Liu, K. Koynov, W. Knoll and Q. Li, Angew. Chem., Int. Ed., 2009, 48, 4598–4601 CrossRef CAS.
- X. Wang, L. Cao, S.-T. Yang, F. S. Lu, M. J. Meziani, L. L. Tian, K. W. Su, M. A. Bloodgood and Y.-P. Sun, Angew. Chem., Int. Ed., 2010, 49, 5310–5314 CrossRef CAS.
- A. Jaiswal, S. S. Ghosh and A. Chattopadhyay, Chem. Commun., 2012, 48, 407–409 RSC.
- S.-L. Hu, K.-Y. Niu, J. Sun, J. Yang, N.-Q. Zhao and X.-W. Du, J. Mater. Chem., 2009, 19, 484–488 RSC.
- F. Wang, S. P. Pang, L. Wang, Q. Li, M. Kreiter and C.-Y. Liu, Chem. Mater., 2010, 22, 4528–4530 CrossRef CAS.
- C. J. Liu, P. Zhang, F. Tian, W. C. Li, F. Li and W. G. Liu, J. Mater. Chem., 2011, 21, 13163–13167 RSC.
- Y. H. Yang, J. H. Cui, M. T. Zheng, C. F. Hu, S. Z. Tan, Y. Xiao, Q. Yang and Y. L. Liu, Chem. Commun., 2012, 48, 380–382 RSC.
- S. Chandra, S. H. Pathan, S. Mitra, B. H. Modha, A. Goswami and P. Pramanik, RSC Adv., 2012, 2, 3602–3606 RSC.
- J. Zhou, C. Wang, Z. S. Qian, C. C. Chen, J. J. Ma, G. H. Du, J. R. Chen and H. Feng, J. Mater. Chem., 2012, 22, 11912–11914 RSC.
- Z. S. Qian, J. Zhou, J. J. Ma, X. Y. Shan, C. C. Chen, J. R. Chen and H. Feng, J. Mater. Chem. C, 2013, 1, 307–314 RSC.
- V. Karunakaran, K. Kleinermanns, R. Improta and S. A. Kovalenko, J. Am. Chem. Soc., 2009, 131, 5839–5850 CrossRef CAS.
- F. Santora, V. Barone and R. Improta, J. Am. Chem. Soc., 2009, 131, 15232–15245 CrossRef.
-
M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Bobb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, O. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09, Revision A.02, Gaussian Inc., Wallingford CT, 2009 Search PubMed.
- S. Kim, S. W. Hwang, M. Kim, D. Y. Shin, D. S. Shin, C. O. Kim, S. B. Yang, J. H. Park, E. Hwang, S. Choi, G. Ko, S. Sim, C. Sone, H. J. Choi, S. Bae and B. H. Hong, ACS Nano, 2012, 6, 8203–8208 CrossRef CAS.
- J. N. Demas and G. A. Crosby, J. Phys. Chem., 1971, 75, 991–1024 CrossRef.
- J. N. Moorthy, T. Shevchenko, A. Magon and C. Bohne, J. Photochem. Photobiol., A, 1998, 113, 189–195 CrossRef CAS.
- B. L. Van Duuren, Chem. Rev., 1963, 63, 325–354 CrossRef.
- J. H. Qian and A. M. Brouwer, Phys. Chem. Chem. Phys., 2010, 12, 12562–12569 RSC.
- D. Y. Pan, J. C. Zhang, Z. Li and M. H. Wu, Adv. Mater., 2010, 22, 734–738 CrossRef CAS.
- L. Bao, Z. L. Zhang, Z. Q. Tian, L. Zhang, C. Liu, Y. Lin, B. P. Qi and D. W. Pang, Adv. Mater., 2011, 23, 5801–5806 CrossRef CAS.
- A. B. Bourlinos, A. Stassinopoulos, D. Anglos, R. Zboril, V. Georgakilas and E. P. Giannelis, Chem. Mater., 2008, 20, 4539–4541 CrossRef CAS.
- J. Z. Shang, L. Ma, J. W. Li, W. Ai, T. Yu and G. G. Gurzadyan, Sci. Rep., 2012, 2, 792 CrossRef.
- Y. Q. Dong, N. N. Zhou, X. M. Lin, J. P. Lin, Y. W. Chi and G. N. Chen, Chem. Mater., 2010, 22, 5895–5899 CrossRef CAS.
- H. Shi, A. S. Barnard and I. K. Snook, Nanoscale, 2012, 4, 6761–6767 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available: XPS and fluorescence spectra and experimental details. See DOI: 10.1039/c3ra42066c |
| ‡ Abbreviations: GQDs, graphene quantum dots; EDA, 1,2-ethylenediamine; PEA, 1,3-propanediamine; BEA, 1,4-butanediamine; G, glycol; DTG, dithioglycol; EA, ethylamine. |
|
| This journal is © The Royal Society of Chemistry 2013 |
Click here to see how this site uses Cookies. View our privacy policy here. 









