Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Layer-by-layer assembly of multi-layered droplet interface bilayers (multi-DIBs)†
Matthew E.
Allen‡
 abc,
James
Albon‡
a and
Yuval
Elani
abc,
James
Albon‡
a and
Yuval
Elani
 *bc
*bc
aDepartment of Chemistry, Imperial College London, Molecular Sciences Research Hub White City, London, W12 0BZ, UK
bInstitute of Chemical Biology, Molecular Sciences Research Hub, Imperial College London, 82 Wood Lane, London, W12 0BZ, UK. E-mail: y.elani@imperial.ac.uk
cDepartment of Chemical Engineering, Imperial College London South Kensington, London, SW7 2AZ, UK
First published on 1st December 2021
Abstract
Droplet interface bilayers (DIBs) have tremendous promise as platforms for fundamental biomembrane studies and in biotechnology. Being composed of a single bilayer however limits their biomimetic potential, as many cell membrane motifs are composed of multiple aligned bilayers. We describe a technology to manufacture cell-sized multi-layered DIBs (multi-DIBs) by coating giant unilamellar vesicles with a further monolayer, and allowing such structures to make contact with themselves or a monolayer coated droplet. This easily customisable strategy will pave the way for an expanded repertoire of DIB functionality, for example by facilitating the incorporation of multiple-bilayer spanning protein complexes.
Bottom-up synthetic biology aims to replicate various aspects of living systems by building new biological structures using molecular components.1–3 Through biomimetic platforms often known as artificial cells,4 replication of diverse cellular functions, including cell–cell communication,5 conversion of light to chemical energy,6 motility,7 decision making,8 and cellular morphological remodelling9 have been achieved.
One system in particular that has found use in bottom-up synthetic biology is the droplet interface bilayer (DIB).10,11 DIBs are formed from the contact of two water-in-oil droplets which are stabilised by a lipid monolayer. Upon contact, the lipid monolayers at the water droplet interface assemble together to form a lipid bilayer, thus providing a robust chassis for a vast range of biomimetic activities. DIBs have been used as cell membrane models to probe the effects of membrane chirality,12 asymmetry,13 permeability,14,15 and embedded protein activity.16,17 Due to the potential for linking up multiple droplets in a network, they have also seen use as building blocks for artificial tissues18–20 and functional systems such as biobatteries and biomimetic sensors.21
DIBs clearly have tremendous potential as mimics of cell barriers composed of single bilayers. Many biological structures, however, consist of multiple membrane layers. These include double membranes found in gap junctions,22 mitochondria,23 Gram-negative bacterial membranes,24 nuclei25 and chloroplasts.26 Triple membrane complexes are also seen in biology, for example during pathogenesis and in bacterial secretion systems.27 As DIBs are formed by bringing together monolayer-coated droplets, they cannot be used to replicate these multi-membrane structures, which limits their utility as cell-mimetic models. Therefore, despite being effective structures for probing a wide range of protein behaviours,28 DIBs cannot be used to investigate protein structures that span over multiple bilayers (for example triple bilayer spanning type VI bacteria secretion systems,29 double bilayer spanning RND efflux pumps,30 connexin channels,22 and nuclear pore complexes31).
Herein, we describe a technology that rectifies this limitation, by making cell-sized multi-layered membrane systems using DIB analogues. Instead of using monolayer coated droplets, we use trilayer coated droplets. By bringing two such droplets together, a triple membrane (consisting of six back-to-back monolayers) is formed. Likewise, by bringing a trilayer coated droplet into contact with a monolayer coated droplet, double membrane structures can be produced. Trilayer droplets are produced by depositing a further lipid monolayer on the outer surface of a giant vesicle through a modification of an emulsion-based lipid templating approach previously used to manufacture multilamellar vesicles.32
The presence of multiple lipid layers around the droplet was confirmed through a fluorescent quenching assay before stochastic collisional based contact induced by centrifugation was achieved to produce the multi-layered DIBs, which we term multi-DIBs. The properties of the multi-DIBs were then explored through biophysical measurements and fluorescence-based assays. The platform can be scaled for custom lamellarity, and the lipid composition can be readily changed enabling access to a wide variety of DIB architectures and compositions. Thus, our system provides an excellent starting point for DIB based systems to explore and characterise multilamellar cellular systems on the cellular length scale.
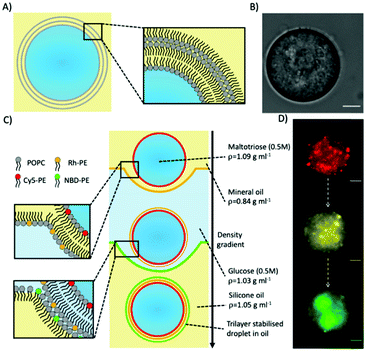
Trilayer stabilised droplets were first produced (Fig. 1A and B) by adapting the protocol used to produce giant unilamellar vesicles (GUVs) by emulsion phase transfer33 (Fig. 1C). In order to produce the trilayer droplets, the lipid POPC was dissolved in both mineral and silicone oil before a density-based oil/water/oil column was assembled with each oil/water interface being stabilised with an amphiphilic lipid monolayer (see ESI† methods for more details). An emulsion of maltotriose droplets (a trisaccharide of larger MW and density than mono/disaccharides) in lipid stabilised mineral oil was then added to the column. The density difference between the lipid-monolayer coated maltotriose droplets and the column enabled the droplets to be driven to the bottom of the column through centrifugation and through two further monolayers. Upon passing through these interfacial monolayers the maltotriose-loaded droplets were coated with another monolayer thus producing the final maltotriose droplets with a trilayer structure.
To confirm that a further lipid monolayer was deposited each time a droplet was driven through one of the two water/oil interface in the column, we used fluorescently labelled lipids. In these experiments, each lipid monolayer had 1% of a different fluorescent lipid added to it (either Cy5-PE, Rhodamine-PE, or NBD-PE). Fluorescent imaging across three different filters verified three different fluorescent signals (Fig. 1D) corresponding to the three different monolayers deposited around the maltotriose droplet.
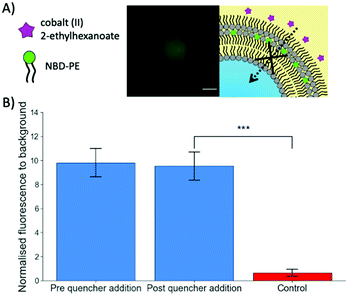
To further verify the successful coating of a trilayer around the maltotriose droplets, Cobalt(II) 2-ethylhexanoate, an oil-soluble NBD quencher,34 was added to the oil surrounding the trilayer droplets containing NBD-PE fluorophores within the central monolayer (Fig. 2). When the NBD-PE lipid was contained within the central monolayer, the addition of an external quencher had no impact upon the fluorescence. However, if the cobalt quencher was added to the same oil phase as the NBD-PE lipid, significant quenching was observed through the loss of fluorescent signal (a mean of 0.65 compared to 9.53 for the previous experiment after quencher addition). This indicates that the central monolayer had another external layer protecting it from being quenched by the cobalt(II) ions. This result indicates that multiple monolayers were present and adds credence to the previous observation that the produced droplets do contain a lipid trilayer.
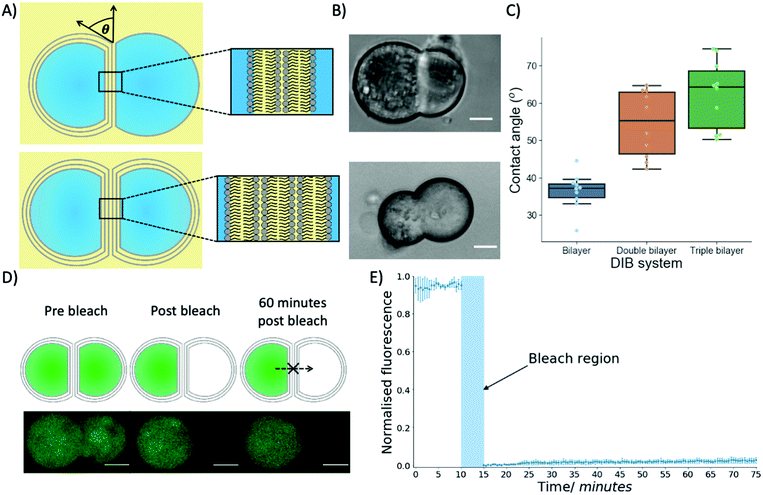
To produce multi-layered DIB structures with either double or triple bilayers (Fig. 3A and B) contact between either two separate trilayer coated droplets or a trilayer and monolayer coated droplet was achieved through stochastic collisions between the droplets.
Biophysical properties of the multi-DIBs were then characterised and compared to traditional single-bilayer DIBs formed from POPC.35,36 It was seen that the contact angle of the multi-DIBs were larger than that of the single bilayer DIBs (Fig. 3C). Furthermore, each additional bilayer increased the contact angle within the DIB. Other parameters such as the interfacial bilayer tension (Fig. S7, ESI†) and the adhesion energy (Fig. S8, ESI†) were calculated and seen to change upon the introduction of additional bilayers. The increased number of bilayers reduced the interfacial bilayer tension and increased the adhesion energy of multi-DIBs. In particular, the increase in adhesion energy (the energy saved by forming the interfacial bilayer) demonstrated that the additional bilayers increase the energy stored within the DIB complex compared to the separate droplets. It is thought that this increase in stability could be produced by the hydrogen bonding networks between the bilayer leaflets37 stabilising the interfacial membrane. As the number of bilayers present increases, the extent of the hydrogen bonding network between the bilayer leaflets increases thus increasing the adhesion energy. This is supported by our findings that the triple bilayer structures have larger adhesion energies than the double bilayer system.
In addition to demonstrating that different biophysical properties were present within multi-DIBs, the presence of a contiguous internal membrane between the trilayer droplets within a triple bilayer DIB was also shown by performing a FRAP experiment (Fig. 3D and E). This was conducted by incorporating Calcein, a dye that has very limited permeability across phospholipid membranes38 into the trilayer stabilised droplets. This meant that both compartments of the resultant multi-DIB contained Calcein dye. One compartment was then photobleached completely and the recovery of fluorescence monitored. It was seen that over an hour-long period there was negligible Calcein fluorescence recovery within the photobleached droplet, this indicates that the fluorescent Calcein was prevented from diffusing through into the other droplet by a lipidic barrier. This is again indicative of multi-DIB formation.
Our droplets systems are cell-sized (∼ pL volume; ∼15 μm diameter). While this makes them useful as artificial cell compartments, they are smaller than standard DIBs formed via manual pipetting of droplets,39 thus limiting the use of analytical techniques such as electrophysiology to reveal deeper insights into the membrane structure. Furthermore, the droplets produced through our method do not provide the same degree of precision with respect to size and positional control when compared to traditional DIBs. In future, microfluidic technologies could be developed to replicate our system through succesive layering of monolayers over droplet templates. This could help control the size and polydispersity of the trilayer droplets and could enable the production of larger multi-DIB networks.
In conclusion, we have provided a new technology to produce cell-sized DIB systems with multiple bilayers. We have confirmed that the multi-DIBs produced do contain multiple bilayers, first by showing that the constituent water in oil droplets are stabilised by a lipid trilayer. We then confirmed that the multi-DIBs have a series of contiguous bilayers that have properties distinctly different to single bilayer DIBs and characteristic of multi-layered structures. We envisage that this work will extend the operational space of DIBs to include multi-layered systems with a tuneable lipid composition and act as a chassis for the incorporation of proteins spanning multiple bilayers. This will lead to the ability to model these systems more accurately on a cellular length scale and provide previously unknown insights into how these structures function. Furthermore, the creation of multi-DIBs will provide a new building block in artificial cell design, for instance within the construction of synthetic tissues.
This work was supported by UKRI Future Leaders Fellowship grant reference MR/S031537/1 and by EPSRC grant EP/V048651/1 awarded to Y. E. For acknowledgements and author contributions, please see the ESI.†
Conflicts of interest
There are no conflicts to declare.Notes and references
- B. C. Buddingh’ and J. C. M. Van Hest, Acc. Chem. Res., 2017, 50, 769–777 CrossRef.
- K. Göpfrich, I. Platzman and J. P. Spatz, Trends Biotechnol., 2018, 36, 938–951 CrossRef PubMed.
- D. T. Gonzales, C. Zechner and T.-Y. Y. D. Tang, Curr. Opin. Syst. Biol., 2020, 24, 56–63 CrossRef.
- N. Amy Yewdall, A. F. Mason and J. C. M. Van Hest, Interface Focus, 2018, 8 Search PubMed.
- Q. Yang, Z. Guo, H. Liu, R. Peng, L. Xu, C. Bi, Y. He, Q. Liu and W. Tan, J. Am. Chem. Soc., 2021, 143, 232–240 CrossRef CAS.
- S. Berhanu, T. Ueda and Y. Kuruma, Nat. Commun., 2019, 10, 1–10 CrossRef CAS PubMed.
- S. Zhang, C. Contini, J. W. Hindley, G. Bolognesi, Y. Elani and O. Ces, Nat. Commun., 2021, 12, 1–11 CrossRef.
- K. P. Adamala, D. A. Martin-Alarcon, K. R. Guthrie-Honea and E. S. Boyden, Nat. Chem., 2017, 9, 431–439 CrossRef CAS PubMed.
- K. Y. Lee, S. J. Park, K. A. Lee, S. H. Kim, H. Kim, Y. Meroz, L. Mahadevan, K. H. Jung, T. K. Ahn, K. K. Parker and K. Shin, Nat. Biotechnol., 2018, 36, 530–535 CrossRef CAS.
- H. Bayley, B. Cronin, A. Heron, M. A. Holden, W. L. Hwang, R. Syeda, J. Thompson and M. Wallace, Mol. BioSyst., 2008, 4, 1191–1208 RSC.
- K. Funakoshi, H. Suzuki and S. Takeuchi, Anal. Chem., 2006, 78, 8169–8174 CrossRef CAS.
- J. Hu, W. G. Cochrane, A. X. Jones, D. G. Blackmond and B. M. Paegel, Nat. Chem., 2021, 13, 786–791 CrossRef CAS.
- E. B. Stephenson and K. S. Elvira, Chem. Commun., 2021, 57, 6534–6537 RSC.
- N. E. Barlow, G. Bolognesi, S. Haylock, A. J. Flemming, N. J. Brooks, L. M. C. Barter and O. Ces, Sci. Rep., 2017, 7, 1–12 CrossRef.
- J. M. Thomas, M. S. Friddin, O. Ces and Y. Elani, Chem. Commun., 2017, 53, 12282–12285 RSC.
- M. A. Elfaramawy, S. Fujii, A. Uyeda, T. Osaki, S. Takeuchi, Y. Kato, H. Watanabe and T. Matsuura, Chem. Commun., 2018, 54, 12226–12229 RSC.
- R. Strutt, J. W. Hindley, J. Gregg, P. J. Booth, J. D. Harling, R. V. Law, M. S. Friddin and O. Ces, Chem. Sci., 2021, 12, 2138–2145 RSC.
- A. Alcinesio, O. J. Meacock, R. G. Allan, C. Monico, V. Restrepo Schild, I. Cazimoglu, M. T. Cornall, R. Krishna Kumar and H. Bayley, Nat. Commun., 2020, 11, 1–13 Search PubMed.
- F. G. Downs, D. J. Lunn, M. J. Booth, J. B. Sauer, W. J. Ramsay, R. G. Klemperer, C. J. Hawker and H. Bayley, Nat. Chem., 2020, 12, 363–371 CrossRef CAS PubMed.
- M. J. Booth, I. Cazimoglu and H. Bayley, Commun. Chem., 2019, 2, 1–8 CrossRef.
- M. A. Holden, D. Needham and H. Bayley, J. Am. Chem. Soc., 2007, 129, 8650–8655 CrossRef CAS.
- S. Maeda, S. Nakagawa, M. Suga, E. Yamashita, A. Oshima, Y. Fujiyoshi and T. Tsukihara, Nature, 2009, 458, 597–602 CrossRef CAS.
- J. G. McCarron, C. Wilson, M. E. Sandison, M. L. Olson, J. M. Girkin, C. Saunter and S. Chalmers, J. Vasc. Res., 2013, 50, 357–371 CrossRef.
- T. R. D. Costa, C. Felisberto-Rodrigues, A. Meir, M. S. Prevost, A. Redzej, M. Trokter and G. Waksman, Nat. Rev. Microbiol., 2015, 13, 343–359 CrossRef CAS PubMed.
- G. Van Meer, D. R. Voelker and G. W. Feigenson, Nat. Rev. Mol. Cell Biol., 2008, 9, 112–124 CrossRef CAS.
- H. Kirchhoff, New Phytol., 2019, 223, 565–574 CrossRef PubMed.
- A. Zoued, Y. R. Brunet, E. Durand, M. S. Aschtgen, L. Logger, B. Douzi, L. Journet, C. Cambillau and E. Cascales, Biochim. Biophys. Acta, Mol. Cell Res., 2014, 1843, 1664–1673 CrossRef CAS PubMed.
- M. Allen-Benton, H. E. Findlay and P. J. Booth, Exp. Biol. Med., 2019, 244, 709–720 CrossRef CAS.
- B. T. Ho, T. G. Dong and J. J. Mekalanos, Cell Host Microbe, 2014, 15, 9–21 CrossRef CAS.
- J. M. Blair and L. J. Piddock, Curr. Opin. Microbiol., 2009, 12, 512–519 CrossRef CAS.
- F. Alber, S. Dokudovskaya, L. M. Veenhoff, W. Zhang, J. Kipper, D. Devos, A. Suprapto, O. Karni-Schmidt, R. Williams, B. T. Chait, A. Sali and M. P. Rout, Nature, 2007, 450, 695–701 CrossRef CAS PubMed.
- T. Ip, Q. Li, N. Brooks and Y. Elani, ChemBioChem, 2021, 22, 2275–2281 CrossRef CAS.
- S. Pautot, B. J. Frisken and D. A. Weitz, Langmuir, 2003, 19, 2870–2879 CrossRef CAS.
- C. M. Ajo-Franklin, C. Yoshina-Ishii and S. G. Boxer, Langmuir, 2005, 21, 4976–4983 CrossRef CAS.
- W. Helfrich, Z. Naturforsch., C: J. Biosci., 1973, 28, 693–703 CrossRef CAS PubMed.
- P. Bassereau, B. Sorre and A. Lévy, Adv. Colloid Interface Sci., 2014, 208, 47–57 CrossRef CAS PubMed.
- J. Milhaud, Biochim. Biophys. Acta, Biomembr., 2004, 1663, 19–51 CrossRef CAS.
- B. Maherani, E. Arab-Tehrany, A. Kheirolomoom, D. Geny and M. Linder, Biochimie, 2013, 95, 2018–2033 CrossRef CAS PubMed.
- M. M. Makhoul-Mansour and E. C. Freeman, Langmuir, 2021, 37, 3231–3247 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cc05155e |
| ‡ These authors contributed equally |
| This journal is © The Royal Society of Chemistry 2022 |