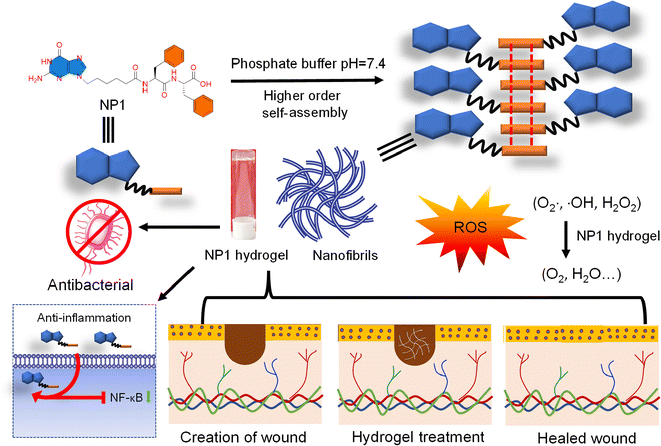
Design and synthesis of a nucleobase functionalized peptide hydrogel: in vitro assessment of anti-inflammatory and wound healing effects†
Sourav
Bhowmik
a,
Budhadev
Baral
b,
Tanmay
Rit
a,
Hem Chandra
Jha
 b and
Apurba K.
Das
b and
Apurba K.
Das
 *a
*a
aDepartment of Chemistry, Indian Institute of Technology Indore, Indore 453552, India. E-mail: apurba.das@iiti.ac.in
bDepartment of Biosciences and Biomedical Engineering, Indian Institute of Technology Indore, Simrol, Khandwa Road, Indore 453552, India
First published on 13th June 2024
Abstract
Over the past several years, a significant increase in the expanding field of biomaterial sciences has been observed due to the development of biocompatible materials based on peptide derivatives that have intrinsic therapeutic potential. In this report, we synthesized nucleobase functionalized peptide derivatives (NPs). Hydrogelation in the synthesized NPs was induced by increasing their hydrophobicity with an aromatic moiety. The aggregation behavior of the NPs was analyzed by performing molecular dynamics simulations and DOSY NMR experiments. We performed circular dichroism (CD), thioflavin-T binding and PXRD to characterize the supramolecular aggregation in the NP1 hydrogel. The mechanical strength of the NP1 hydrogel was tested by performing rheological experiments. TEM and SEM experiments were performed to investigate the morphology of the NP1 hydrogel. The biocompatibility of the newly synthesized NP1 hydrogel was investigated using McCoy and A549 cell lines. The hemolytic activity of the NP1 hydrogel was examined in human blood cells. The stability of the newly formed NP1 hydrogel was examined using proteinase K and α-chymotrypsin. The NP1 hydrogel was used for in vitro wound healing. Western blotting, qRT-PCR and DCFDA assay were performed to determine the anti-inflammatory activity of the NP1 hydrogel. The synthesized NP1 hydrogel also exhibits antibacterial efficacy.
Introduction
The skin, being the largest organ and the body's first line of defense, is crucial for shielding the body from germs and environmental damage. However, the skin is easily damaged, and once it has significant flaws, it takes a long time to heal.1 Millions of people experience agony, suffering, and in extreme instances, disability due to wounds from burns, trauma, surgery, and chronic diseases. The global demand for wound dressing material is on the rise. Consequently, the financial burden of medical treatment is also increasing.2 As a result, basic and efficient wound care remains a challenging issue that presents significant obstacles for the healthcare system. Wound healing is a crucial aspect of healthcare, aiming to restore normal cell and tissue function to protect against external threats, chemical and mechanical damage, and regulate body temperature.3–5 However, wound repair encounters numerous obstacles, including bacterial infections and immune and inflammatory processes involved in the restoration of healthy cells and tissues.6–10 The skin is recognized as the biggest and most rapidly developing organ in an animal's body, functioning as a regenerative and self-restoring barrier between the body and its extrinsic surroundings. Therefore, a potential hydrogel for wound healing should possess antimicrobial properties, facilitate gas exchange, and maintain a moist environment to support cell proliferation and cell migration. Wound healing is a complex process that involves several stages, including (a) inflammation, (b) coagulation, (c) mesenchymal cell proliferation, differentiation and migration, (d) re-epithelization and (e) angiogenesis.11,12 The initial and preliminary stages of the wound healing process involve coagulation or hemostasis, which is achieved through fibrin formation, platelet aggregation, and vascular constriction to staunch haemorrhage. The wound tissues and hemostatic clots secrete proadhesive molecules, proinflammatory cytokines and several growth factors such as platelet-based growth factor, epidermal growth factor, and fibroblast growth factor.13 The proinflammatory cytokines initiate inflammation by sequentially attracting inflammatory cells, such as neutrophils, macrophages and lymphocytes at the injury site. Inflammatory cells are responsible for removing invading microorganisms, cellular debris and apoptotic cells from the wound site.13,14 However, if inflammation is excessive and prolonged, it can trigger a cycle of pro-inflammatory signals that delay wound healing.15 Therefore, controlling these proinflammatory signals is also important for the wound healing process. It is widely recognized that moist environments play a key role in accelerating wound healing, while dry surroundings can cause cell death, resulting in delayed healing.16 However, excessive moisture near the wound can lead to microbial infection, resulting in the wound becoming a non-healing inflammatory condition.17 Intrinsic antibacterial hydrogels, which eliminate bio-contaminants and maintain a moist environment, are an alternate kind of biomaterial for the wound healing process.18 Furthermore, it is crucial that the optimal biomaterials for wound healing do not elicit any unwanted inflammatory or allergic reactions.19 Recently, peptide-based soft materials have become significant.20,21 As peptides have unique biological roles, activity, and specificity and are broadly distributed in the human body, both naturally occurring and chemically synthesized peptide molecules display a wide spectrum of biological effects.22–26 Interestingly, peptides can form supramolecular architectures through various non-covalent weak interactions such as H-bonding and π–π stacking.27–30 Numerous studies in the literature report self-assembling peptide-based biomaterials that are efficient at repairing wound tissue.31–34 These well-organized peptides possess a wide range of properties, including stimulating cellular and humoral immunity, mimicking the extracellular matrix, and delivering drug molecules. The presence of nanofiber in the self-assembled peptide nanostructure resembles the composition and structure of fibrin in the extracellular matrix, which aids in the healing of injured tissues and facilitates the recovery of their cellular activities.35–37 Moreover, low-molecular-weight peptide functionalized hydrogelators have better biocompatibility and biodegradability. Therefore, these peptide functionalized hydrogelators are suitable for most tissue engineering applications.38 Additionally, self-assembling peptide functionalized molecules can adapt to changes in complex wound healing processes and have shown significant promise for multimodal therapy.39 Consequently, hydrogels based on self-assembling peptides have several uses in biomedicine and nanotechnology, such as topical medications, immunological adjuvant therapy, tumor treatment, 3D tissue cell culture, tissue healing, and tissue regeneration.40–42 Despite the progress in peptide-based self-assembling materials, they still lack certain properties such as anti-inflammation. Nucleoside analogs are known to have anti-inflammatory effects.43,44 In this study, we functionalized a self-assembling peptide with the nucleobase guanine to form a nucleopeptide hydrogel. Guanine, which has a number of H-bonding donor–acceptor sites, can form self-assembling structures. Hence, this study focuses on the development and production of nucleopeptide-based bioactive biomaterials, which have the ability to heal wounds. The key objectives of this project include: (a) designing and synthesizing guanine functionalized peptide molecules, (b) forming self-assembling biomaterials from the newly synthesized nucleopeptides, (c) testing the self-assembly propensity through molecular dynamics simulations, (d) conducting in vitro cytotoxicity assays, (e) performing in vitro wound healing assays, (f) assessing the hemolytic activity of the synthesized hydrogel, (g) evaluating the anti-inflammatory activity of the NP1 hydrogel and (h) testing the antibacterial activity of the NP1 hydrogel.Experimental section
Nucleopeptide synthesis
Nucleopeptide derivatives were prepared using the solution phase synthetic process. Purine base guanine was conjugated with the dipeptide moieties to synthesize nucleobase functionalized peptide derivatives.Gelation method
The newly developed 30 mM nucleopeptide molecules were placed in a clean 5 mL vial. Then, freshly prepared phosphate buffer (100 mM, pH = 7.4) was added in the vial. Then the resulting solution was vortexed for 5 min to homogeneously mix the solution. Finally, the solution was sonicated for 10 min and kept inside the incubator. After 30 min, NP1 formed the hydrogel and the rest of the synthesized NPs (NP2 and NP3) formed the precipitate.Isolation of blood
A vacuum tube with the anticoagulant, sodium citrate solution (3.0 w/v%), was used to collect the blood sample. Notably, the blood sample was provided by one of the ambulance drivers as part of a standard physical examination at a medical facility. Blood plasma was then extracted, and the residual cells were used for tests.Separation of WBCs and erythrocytes
By centrifuging blood at 2000 rpm for five minutes under room temperature, erythrocytes were separated from the blood. With the use of a sterile syringe, WBCs were removed from the buffy coat, and the remaining cells were then washed with 10 mM phosphate buffered saline (PBS) of pH 7.4.Hemolytic activity
A 1% erythrocyte suspension in PBS was employed to assess hemolytic activity. To evaluate the NP1 hydrogel for hemolysis, the erythrocytes were immediately mixed with different concentrations of the hydrogel and left to incubate for 1 h. PBS was utilized as a negative control and 1% (w/v) Triton X was used as a positive control. After that, the mixtures were incubated for 1 h at 37 °C. After the compilation of the incubation period, centrifugation was carried out at 1000 rpm for 10 min at 4 °C.Antibacterial properties of the hydrogel
The effectiveness of the NP1 hydrogel against B. subtilis and E. coli bacteria was examined in vitro. The optical density (OD625) technique was used to examine the hydrogels’ antibacterial properties. The NP1 hydrogel with varied concentrations (2 to 0.031 mM) was used for the antibacterial study. The control was a bacterial solution in nutrient broth devoid of hydrogels. The absorbance level of the test solution and the control were estimated at 625 nm.Animal cell culture
Mouse fibroblast cells (McCoy) and human lung epithelial cells (A549) were procured from NCCS Pune and cultured as per standard protocol. Briefly, the cells were grown in Dulbecco's modified Eagle's medium (Himedia, Mumbai, India) with the standard amount of FBS (Gibco, New York, USA) and penicillin/streptomycin (Himedia, Mumbai, India). The culture was maintained in a CO2 incubator (Forma, Steri-cycle i160, Thermo Scientific, Waltham, USA).Cytotoxicity of the hydrogel
The cytotoxicity was determined by the MTT assay. For these, an approximate 103 cells were placed in each well of a 96-well plate. The plates are incubated over night (12 h). The hydrogel was treated for 24 h with concentrations ranging from 2 mM to 0.03 mM. After completion of the incubation period the medium was aspirated and 50 μl of 0.5 mg ml−1 MTT solution was added to each well of the 96-well plate, followed by incubation at 37 °C for 3 h. Post incubation the solution was aspirated and DMSO was added to dissolve the formazan and kept on an orbital shaker for 1.5 h. The reading was taken using a microplate reader (Biotek Synergy H1) at 570 and 590 nm wavelengths. The IC50 of the hydrogel could not be determined due to minimal toxicity of the hydrogel. For all further experiments involving animal cell, 0.1 mM concentration of hydrogel was used.ROS estimation by DCFDA assay
To understand the ROS scavenging activity of the hydrogel, cells were seeded in a 12 well cell culture plate and kept under incubation overnight. Then, the cells were treated with 100 μM H2O2 for 1 h followed by treatment of the hydrogel for 12 h after completion of the treatment period. The DCFDA staining, imaging and data analysis were performed as described earlier.45RNA isolation and qRT PCR
To determine the transcript level of inflammatory markers, 1 million cells were seeded in a 60 mm disk and allowed to settle, and were further treated with 100 ng μL−1 of lipopolysaccharides (Cat# L8880 ANJ Biologicals, NJ, USA) for 12 h followed by treatment of the hydrogel for 12 h. For the positive control an equivalent amount of LPS was treated and untreated cells were also kept as the negative control. After completion of the incubation period the cells were harvested by scraping and then centrifuged at 2500 rpm for 5 min. The pellets were washed with PBS, and a RNA extraction reagent (Himedia, Mumbai, India) was added and RNA was isolated as mentioned previously.46 The isolated RNA was quantified using Nanodrop and cDNA was prepared from 2 μg of RNA by using the cDNA synthesis kit. qRT PCR of IL6, TNFα and NFκB was conducted using gene specific primers. For McCoy cells, mouse specific primers were used.Western blot
To understand the expression of specific markers at protein levels western blotting was performed as mentioned in our earlier work.47 Briefly post completion of the treatment time (LPS + hydrogel treatment) cells were harvested and protein was isolated by RIPA buffer and an equal amount of protein was loaded into each well of a running gel. The separated proteins were then transferred to a 0.45 μM pore size nitrocellulose membrane and after blocking, these were incubated with a primary antibody followed by incubation with secondary antibody and visualization. Analysis and quantification of blots were performed using Image J software (National Institutes of Health, USA). Antibodies for NF-κB p65 (#8242, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) were from Cell Signaling Technology, Danvers, USA. GAPDH (#MA5-15738, 1
1000) were from Cell Signaling Technology, Danvers, USA. GAPDH (#MA5-15738, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) was from Invitrogen, Waltham, USA.
2000) was from Invitrogen, Waltham, USA.
Results and discussion
The nucleobase, guanine, was conjugated with a peptide moiety to synthesize a new kind of nucleopeptide (NP) moiety. The nucleopeptide was synthesized using a solution phase synthetic procedure. In a previous report, we enhanced the gelation properties of the synthesized NPs by increasing their hydrophobicity by introducing a long fatty acid chain.48 In this report, we aim to demonstrate the gelation properties of the synthesized nucleopeptide by increasing its hydrophobicity through the introduction of an aromatic amino acid into it. We have designed three different nucleopeptides (NP1, NP2, and NP3) and synthesized them according to the reported solution phase synthetic procedure.49,50 To form a self-assembled hydrogel, 30 mM synthesized NP1 was taken in a vial and then 500 μl of phosphate buffer with a pH of 7.4 was added. After the buffer was added, the solution was ultrasonicated vigorously for 10 min. The solution was then left at room temperature for 15 min. The formation of the hydrogel was verified by inverting the vial. The minimum gelation concentration was 30 mM. In a similar manner, we attempted to prepare the hydrogel with the other two synthesized NPs. A viscous solution appeared for the remaining two NPs (Fig. S1†). Therefore, only NP1 formed a stable, self-supported NP1 hydrogel in phosphate buffer (Scheme 1).The formation of a stable hydrogel by NP1 can be attributed to the proper balance of hydrophobicity and hydrophilicity. In a previous report, we demonstrated that increasing hydrophobicity through the introduction of variable chain lengths of the fatty acid led to gelation in the synthesized nucleopeptides.48 In this instance, the increase in hydrophobicity due to the aromatic amino acid plays a crucial role in inducing gelation in the synthesized NPs. The CLogP values for the synthesized NP1, NP2 and NP3 are 1.488, 0.821, and 0.154, respectively. These values clearly indicate that the increased hydrophobicity, brought about by the introduction of the aromatic amino acid in the synthesized NPs, significantly contributes to the induction of the gelation properties. The difference in the aggregation behavior of the synthesized NPs is attributed to the variation in the CLog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values.
P values.
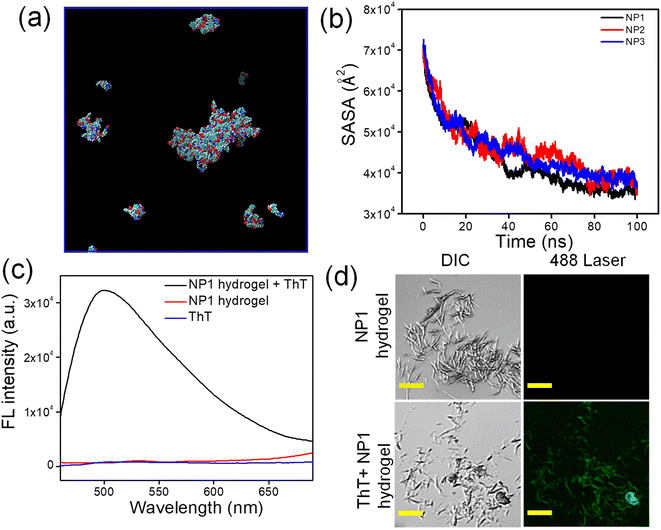
Furthermore, the aggregation behavior of the synthesized nucleopeptides is determined by simulation studies. All the synthesized NPs were subjected to simulation studies using Amber22. The simulation was conducted over a time period of 100 ns. Over time, the synthesized NPs began to aggregate. The rate of aggregation increased with the duration of the simulation. Among the NPs, NP1 showed a higher rate of aggregation compared to NP2 and NP3 (Fig. 1a and Fig. S2†). The solvent accessible surface area (SASA) analysis is used to determine the supramolecular aggregation propensity.51 A decrease in the SASA implies the aggregation behavior of the NPs. It has been observed that the decrease in SASA was more significant for NP1 as compared to NP2 and NP3 (Fig. 1b). Consequently, the increased aggregation in NP1 led to a higher order of self-assembly, which helped in the formation of the hydrogel.52,53 To further support the aggregation observation from the simulation studies, we conducted diffusion ordered NMR Spectroscopy (DOSY) of the synthesized NPs. The size of the aggregates, formed by the NPs, was determined through the DOSY NMR experiment. This experiment helps in determining the diffusion co-efficient of the supramolecular aggregates. The diffusion constant (m2 s−1) values for the C8 proton of the synthesized NP1, NP2 and NP3 are 2.03 × 10−13, 1.91 × 10−9 and 3.15 × 10−10, respectively. Among the synthesized NPs, NP1 had the lowest diffusion constant compared to the other NPs, indicating a higher tendency to form aggregates. The simulation results support the experimental observation (Fig. S3–S5†).
Guanine, which has several H-bonding donor and acceptor sites, can participate in H-bonding interactions among themselves to form the secondary structure of DNA. This helps in the formation of hydrogel.54 Circular dichroism (CD) spectroscopy is an important technique used to determine the presence of secondary structures and the folding of proteins and other biomacromolecules. Therefore, CD spectroscopy was used to investigate whether a secondary structure is present or not. The NP1 hydrogel (30 mM) was diluted to 400 μM using water. The spectra were recorded and showed a positive peak at 277 nm and a negative peak at 233 nm. These peaks indicate the presence of the DNA secondary structure within the hydrogel.55 A peak at 222 nm is due to the π–π* transition for the phenyl moieties in the synthesized NP1 (Fig. S6†).56 Therefore, the CD spectrum suggests that the DNA secondary structure is present within the hydrogel components.
Thioflavin T (ThT) is a benzothiazole-derived fluorophore, which typically used to identify the secondary structure of nucleic acids. Although ThT is not fluorescent in water by itself, it exhibits strong fluorescent emission maxima when it binds with macromolecules such as DNA and RNA. ThT is composed of a benzothiazole ring and benzyl amine, which are connected by a C–C bond. Theoretical studies suggest that the free rotation around this C–C single bond results in fluorescence quenching in the solution. When ThT molecules are added to biomolecules, they attach and restrict free rotation, resulting in strong fluorescence.57 The ThT solution was added to the NP1 hydrogel during its formation. The fluorescence spectrum was recorded for the ThT incorporated hydrogel, the blank hydrogel, and the blank ThT. The excitation wavelength was set at 450 nm. No prominent peak was observed for the NP1 hydrogel and the blank ThT solution. However, a strong fluorescence maximum at 494 nm clearly suggested the presence of secondary structures of nucleic acid, which facilitate the formation of NP1 hydrogel (Fig. 1c). A microscopic evaluation was carried out to corroborate the spectroscopic result. In this process, the hydrogel was incubated with a ThT solution.58 The dye bonded strongly with the supramolecular nano-architecture within the hydrogel, displaying fluorescence that was clearly visible under a confocal laser scanning microscope (CLSM) (Fig. 1d).
A wide-angle powder X-ray diffraction experiment (PXRD) was conducted to examine the self-assembly in the NP1 hydrogel. PXRD spectra were recorded for the xerogel (made from the NP1 hydrogel) and the synthesized NP1. A peak was observed at 2θ = 28.36, which corresponds to a distance of 3.15 Å. This is attributed to the π–π stacking distance (Fig. S7a†).59 The spectra were also recorded for NP1, and its crystalline nature was observed (Fig. S7b†). Therefore, the PXRD experiment also revealed the presence of supramolecular stacking within the NP1 hydrogel, which helps in forming a stable, self-assembled NP1 hydrogel.
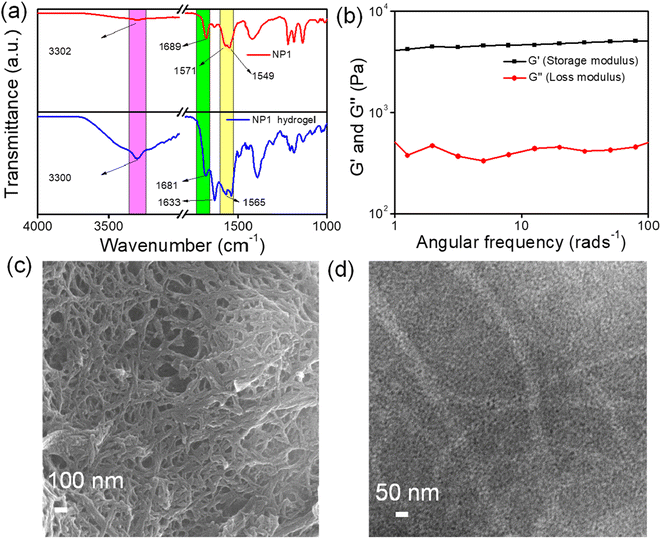
To gain more detailed information about the molecular interactions in the self-assembled stable NP1 hydrogel, we performed FT-IR spectroscopy. The spectra for both the NP1 hydrogel and the synthesized NP1 were recorded. We used FT-IR spectroscopy to investigate the existence of the amide I and amide II bands. Amide I represent the stretching frequencies of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond, while amide II represents the stretching frequencies of the N–H bond.60 The peaks at 1681 cm−1 and 1633 cm−1 correspond to the –C
O bond, while amide II represents the stretching frequencies of the N–H bond.60 The peaks at 1681 cm−1 and 1633 cm−1 correspond to the –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching frequencies. Other peaks at 1570 cm−1 and 3300 cm−1 represent the N–H bending and O–H stretching (Fig. 2a). In the spectrum, the shift of the –C
O stretching frequencies. Other peaks at 1570 cm−1 and 3300 cm−1 represent the N–H bending and O–H stretching (Fig. 2a). In the spectrum, the shift of the –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching frequency in the amide I region and the change of the N–H bending in the amide II region of the hydrogel from its corresponding gelator precursor may be attributed to the participation of the carbonyl group in the hydrogen bonding interactions. The increase in intensity and shift of the O–H stretching indicates that the O–H of the carboxylic acid is involved in the H-bonding interaction. Therefore, the FTIR spectra clearly indicate that the H-bonding interaction in NP1 contributes to the formation of the supramolecular aggregate, which in turn facilitates the formation of NP1 hydrogel.
O stretching frequency in the amide I region and the change of the N–H bending in the amide II region of the hydrogel from its corresponding gelator precursor may be attributed to the participation of the carbonyl group in the hydrogen bonding interactions. The increase in intensity and shift of the O–H stretching indicates that the O–H of the carboxylic acid is involved in the H-bonding interaction. Therefore, the FTIR spectra clearly indicate that the H-bonding interaction in NP1 contributes to the formation of the supramolecular aggregate, which in turn facilitates the formation of NP1 hydrogel.
The mechanical properties of the newly developed NP1 hydrogel were estimated with the help of rheological studies.61 The area of deformation was investigated using an amplitude sweep experiment, keeping a constant value of frequency of 10 rad s−1 throughout the entire experiment. The values of the storage modulus (G′) and loss modulus (G′′) were constant at lower strain levels. However, the storage modulus values gradually decreased from 4133 Pa as the strain increased (Fig. S8†). This experiment reveals that the microstructure deforms at higher strains, resulting in the transformation of the hydrogel into a solution. Subsequently, a frequency sweep experiment was conducted at a constant strain of 0.5%. This experiment also demonstrated that the storage modulus values consistently exceeded the loss modulus values across all frequency ranges (Fig. 2b). Therefore, the rheological experiment reveals the formation of a stable and stiff NP1 hydrogel.
The morphological characterization of the synthesized NP1 hydrogel was carried out using various electron microscopy techniques.62,63 SEM images showed the presence of densely entangled nanofiber structures within the NP1 hydrogel (Fig. 2c). The average width of these nanofibers is 29 nm. Additionally, a transmission electron microscopy experiment was also performed. TEM images also suggest the presence of nanofiber within the hydrogel (Fig. 2d). The nanofiber within the hydrogel has an average width of 25 nm. Therefore, microscopic images also indicate the existence of dense nanofibers within the hydrogel. Consequently, the nanofiber within the NP1 hydrogel contributes to the development of a stable NP1 hydrogel.
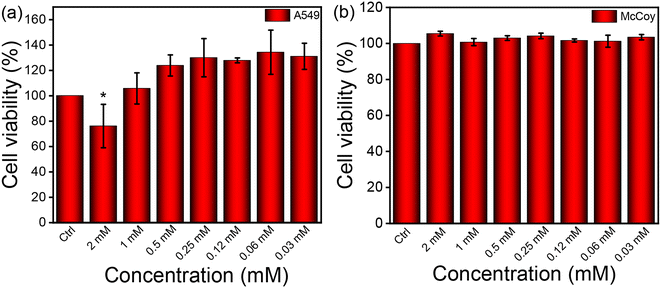
Recent studies have shown that peptide-based nanofibrous hydrogels are used in fields such as tissue engineering, stem cell proliferation and drug delivery. This is because they are biocompatible and can hold a large amount of water molecules. Therefore, biocompatibility is a primary requirement for using this soft functional material in biomedicine.64 The colorimetric MTT experiment was conducted using the synthesized stiff, nanofibrous NP1 hydrogel to evaluate its impact on cells. The cells were incubated with the hydrogel for a duration of 24 h. We selected two different cell lines to determine the number of metabolically active cells after the incubation period. The cancerous cell line A549 and the fibroblast cell line (McCoy) were used to evaluate biocompatibility. The cells were treated with varying concentrations of the synthesized nanofibrous NP1 hydrogel. For the A549 cell, the synthesized NP1 hydrogel demonstrated minimal toxicity when higher concentrations (2 mM and 1 mM) were applied to the cells, compared to the control experiment (Fig. 3a). The control experiment was carried out using only the media (the composition of the media is mentioned in the Experimental section). For the McCoy cell, the hydrogel showed no toxicity. Instead, proliferation was observed with the McCoy cell line (Fig. 3b). Previous studies have also reported minimal cytotoxicity of hydrogels.65 Based on the MTT data, we used a 0.1 mM concentration in our subsequent cell-based experiments.
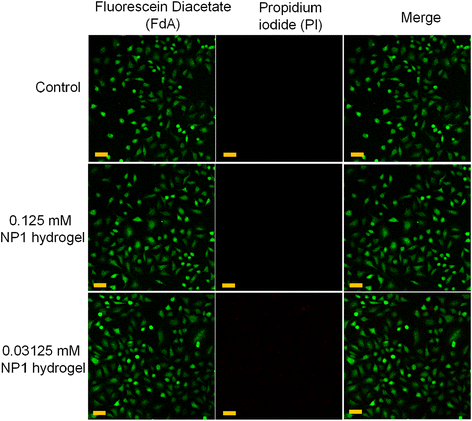
To further confirm the findings from the MTT assay, we performed live–dead cell imaging. This imaging was conducted using the NP1 hydrogel on the McCoy fibroblast cell line and the A549 epithelial cell line. Different concentrations of the hydrogel (0.125 mM and 0.03125 mM) were used for the live–dead cell imaging experiment. The hydrogel was left to incubate in the cells for a period of 24 h. After the incubation period ended, the cells were stained with fluorescein diacetate (FdA). The bipolar side chains in FdA can help to permeate through the cell membrane. FdA is a non-fluorescent dye. The esterases, which are enzymes that catalyze the hydrolysis of ester bonds, transform the nonfluorescent FdA into fluorescent by non-specific cleavage of the acetate groups present in the FdA. The cells, which are metabolically active and have undamaged cell membranes, could be observed under laser light. Dead cells were visualized by staining them with propidium iodide.66 In both cell lines, no red fluorescence was observed (Fig. 4 and Fig. S9†). This clearly indicated the biocompatible nature of the hydrogel. As a result, this biocompatible hydrogel could potentially be used for wound healing. The results of this experiment are also consistent with our MTT data, which show minimal cell death due to cytotoxicity.
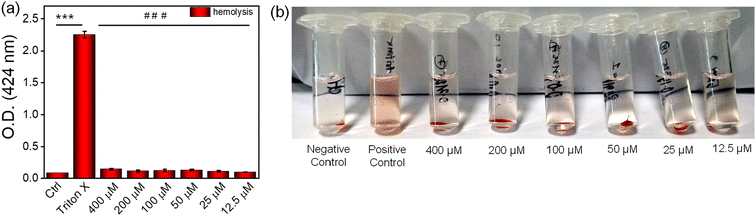
Inflammation, proliferation, and reorganization of tissue layers are all components of the complex and dynamic process of wound healing. This process involves step-by-step, time-consuming restoration of damaged cellular structures.67 Hemolysis occurs when the hydrogel comes into contact with blood cells. This happens when the blood cell membrane ruptures, releasing hemoglobin and other internal components into the surrounding fluid. The presence of a pink to crimson red color in the serum can be used to visually identify it. This color change can be caused by a variety of medical issues, including microbial infections, autoimmune and genetic diseases, and low solute concentration. The term ‘hemolytic potential’ refers to the ability of a substance to cause hemolysis, or the breakdown of red blood cells, to a certain extent when it comes into contact with blood. The hemolysis experiment demonstrated that the nanofibrous supramolecular nucleopeptide hydrogel was nonhemolytic. Fig. 5a shows the hemolysis of blood cells when exposed to different hydrogel samples for an hour at 37 °C. TritonX was used as a positive control, while only a buffer was used as a negative control. An optical image shows the hemolytic activity of the NP1 hydrogel (Fig. 5b). All tested concentrations of the hydrogel were found to be nonhemolytic, with hemolysis levels below the acceptable standard. These findings demonstrate the high compatibility of the supramolecular nucleopeptide hydrogel with blood.
A nucleopeptide gelator consisting of amino acids tends to undergo hydrolysis when exposed to proteolytic enzymes, which is associated with a significant rise in proteolytic enzymes in chronic wounds. Therefore, the stability of the nucleopeptide hydrogel was tested in the presence of the proteolytic enzymes. The biostability was tested in the presence of the proteolytic enzymes α-chymotrypsin and proteinase K.68 The breakdown of the peptide by enzymes was evaluated at different time intervals using RP-HPLC analysis (Fig. S10a and S10b†). The proportion of the nucleopeptide that remained undecomposed was estimated by measuring the area under the curve. The nucleopeptide hydrogel showed a degradation of 30–35% at 30 h (Fig. S11a and S11b†).
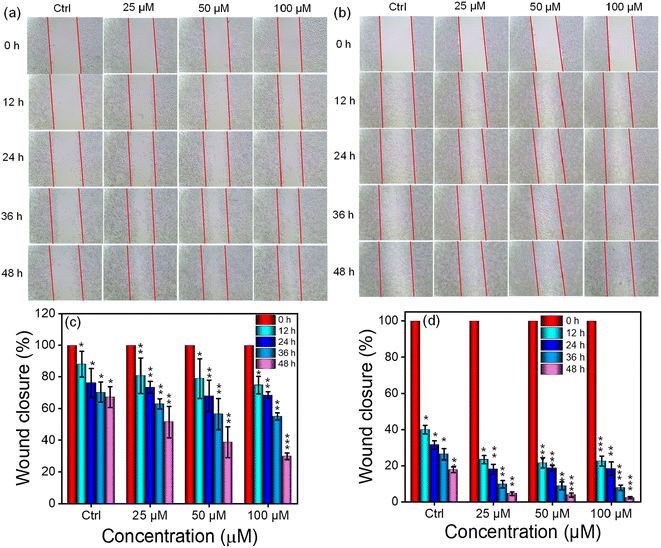
In addition, we examined the impact of the NP1 hydrogel on cell migration by performing a scratch wound test, as cell proliferation and migration are crucial factors in the wound healing process. After making a small scratch on the cell layer, we then incubated them with the NP1 hydrogel. The concentration dependent effect of the hydrogel was studied. Images of the scratch were captured at different times (0, 12, 24, 36 and 48 h), revealing that the cells treated with a higher concentration of hydrogel showed improved migration, effectively closing the gap within 48 h (Fig. 6a and b). When a higher concentration of the hydrogel was applied, about 70–75% of the wounds closed in the A549 cell line, while 95–98% of the scratch wound healed in the McCoy cell line (Fig. 6c and d). The percentage wound closure was used to represent the amount of wound closure in response to our hydrogel. In this method, the width of the wound at 0 h was considered to be 100%, and as time increased and the wound area decreased, the percentage of the wound area also decreased from 100%. The wound area at respective time points was calculated with respect to 0 h of the respective groups. The higher migration in the keratinocytes (McCoy) shows NP1's superior wound healing capability in a natural setup. A previous study by Sparks et al. also reported the enhanced wound healing potential of peptide hydrogels.69
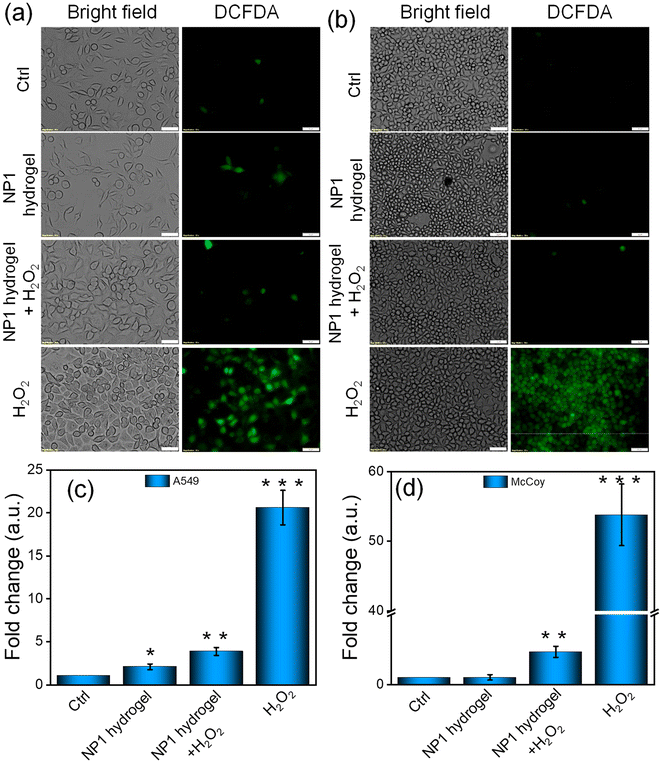
The main barrier to tissue healing of a scratch wound is the elevated levels of reactive oxygen species (ROS).70 In this study, H2O2 was used to stimulate the production of ROS and induce oxidative stress in the McCoy fibroblasts and A549 epithelial cell lines. The objective was to evaluate the effect of the NP1 hydrogel on the ROS levels in these cell lines when they are under stress from H2O2. The hydrogel was used to protect against cellular damage by inhibiting the formation of ROS. The nonfluorescent compound DCFH-DA transforms into the fluorescent compound DCF when it comes into contact with ROS. This transformation was employed as an indirect method to measure the production of ROS within the cells. For qualitative analysis, the McCoy fibroblasts and A549 epithelial cell lines were examined using a fluorescent microscope to investigate the intracellular location of ROS. The image demonstrates that cells treated with H2O2 exhibited a significant amount of green fluorescence, indicating a rise in the intracellular ROS levels (Fig. 7a and b). In contrast, the control and the NP1 hydrogel alone showed much less green fluorescence in both cell lines. These data suggest that the gel has a significant effectiveness as a free radical scavenger. The measurement of fluorescence intensity also demonstrated a significant increase in the intracellular ROS levels after H2O2 treatment. However, the use of the NP1 hydrogel effectively mitigated the formation of ROS generated by H2O2. The fluorescence intensity of cells decreased by about 50% after being exposed to hydrogel, compared to cells that were treated with H2O2 (Fig. 7c and d). These results further validated the exceptional ability of the hydrogel to scavenge reactive oxygen species (ROS). After comparing the ROS scavenging capacity of the NP1 hydrogel with that in the literature, we conclude that our findings are consistent with previously published results. Several groups in previous studies have also demonstrated the ROS scavenging properties of various hydrogels, including peptide hydrogels.71 It is noteworthy to mention that the guanine moiety in NP1 plays a significant role to scavenge free radicals. Among the nucleobases, guanine has the lowest reduction potential (1.29 V). Therefore, it is the best electron donor and is preferentially oxidized. Hence, hydroxyl and super oxide radicals react with guanine that results in a decrease in ROS.72
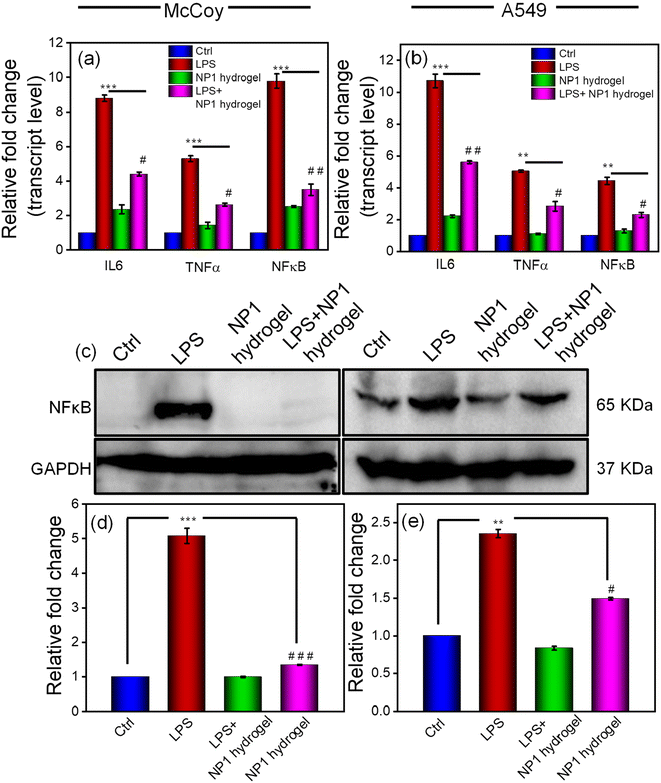
In addition to the ROS scavenging, wound filling and antibacterial are crucial properties of the gel, and it is important to investigate its anti-inflammatory properties. This is because the formation of a wound triggers an inflammatory response. To investigate the anti-inflammatory properties of the NP1 hydrogel, we determined the transcript (IL6, TNFα and NFκB) and protein (NFκB) levels of inflammatory markers using the qRT-PCR and western blot methods, respectively (Fig. 8). Our results indicate that treating cells exposed to LPS with the NP1 hydrogel significantly (p < 0.05) reduces the transcript levels of IL6, TNFα and NFκB in the LPS + gel group compared to the LPS treated group in both fibroblast and lung epithelial cells (Fig. 8a and b). Furthermore, the gel by itself does not exhibit any inflammatory properties, as evidenced by the levels of these markers in the gel treatment group (Fig. 8a and b). We have also evaluated the protein level of NFκB, the primary regulator of inflammation. In the McCoy cell line, compared to the LPS treatment group, the LPS + gel group showed a diminished expression of NFκB (p < 0.0001). However, in the A549 cell line, the decrease in NFκB expression was less compared to that in the McCoy cell line (p < 0.05) (Fig. 8c–e). The results from the anti-inflammatory study suggest that the NP1 hydrogel possesses anti-inflammatory properties in addition to the other properties mentioned. Previous studies by Chen et al. have also demonstrated the reduction of IL6 and TNFα in mice treated with hydrogel.73 Additionally, another study involving a peptide-ibuprofen amphiphile based hydrogel was able to reduce inflammation induced by LPS, as indicated by the reduction of IL6, TNFα and NFκB.74
Microbial infections and the growth of biofilms on biomaterial surfaces pose significant worldwide challenges and health concerns. Antibacterial hydrogels have inherent properties that effectively hinder bacterial contaminations, making them a viable option in combating drug-resistant pathogens. Bacterial infections in the wound area can lead to chronic conditions and the healing process becomes slow.75 The effective wound healable biomaterials should effectively eradicate biological contaminations without causing undesirable allergic responses. The inherent propensity of nucleopeptide-based materials to self-assemble confers significant benefits in treating bacterial infections. The intrinsic antibacterial activities of the NP1 hydrogel were estimated by measuring the optical density at 625 nm against harmful Gram-positive (B. subtilis) and Gram-negative (E. coli) bacteria. The results of antibacterial tests indicate that the NP1 hydrogel effectively suppresses the proliferation of Gram-positive and Gram-negative bacteria. The higher concentration of the NP1 hydrogel was found to be more effective against the bacteria (Fig. S12a and S12b†). FE-SEM images were captured after a 24 h incubation period of the NP1 hydrogel with the Gram-negative bacteria E. coli. The images clearly show the breakdown of the bacterial cell membrane when the hydrogel was applied (Fig. S13†). Therefore, the NP1 hydrogel can be potentially used for wound dressing purposes.
Conclusion
The nucleopeptide derivatives (NPs) were synthesized and the ability of these synthesized NPs to form hydrogels was tested. The aggregation behavior of these NPs was analyzed using a molecular dynamics simulation study. The DOSY NMR experiment confirmed the higher level of aggregation of NP1. Furthermore, the NP1 hydrogel was characterized using CD, PXRD, and ThT-dye binding experiments. SEM and TEM experiments revealed the existence of nanofibers within the NP1 hydrogel. The NP1 hydrogel exhibited minimal cytotoxicity, as demonstrated by MTT and cell death studies. The NP1 hydrogel was also found to be biostable against the proteolytic enzymes proteinase K and α-chymotrypsin. Furthermore, the NP1 hydrogel showed efficacy against B. subtilis and E. coli bacteria. It also demonstrated properties of ROS scavenging and antibacterial and anti-inflammatory activities in epithelial cells and keratinocytes. Notably, the hydrogel showed enhanced activity in keratinocytes compared to epithelial cells. In conclusion, the NP1 hydrogel could be an efficient candidate for wound healing applications. In addition to our findings, the molecular mechanism underlying the wound healing induced by the hydrogel could be studied in more detail.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
A. K. D. sincerely acknowledges CSIR, New Delhi, Government of India (Project No. 01/3120/23/EMR-II) for financial support. S. B. sincerely acknowledges DST-Inspire for his doctoral fellowship. B. B. acknowledges UGC for the fellowship. T. R. acknowledges PMRF for the fellowship. The authors are also thankful to SAIF, IIT Bombay for the TEM facility, the DST-FIST 500 MHz NMR facility, the Department of Chemistry and SIC IIT Indore for providing the rest of the facilities. Dr Shrikant Kunjir is acknowledged for DOSY NMR data analysis. The authors wish to express their gratefulness for the assistance of molecular dynamics simulation offered by Prof. Satya S. Bulusu. We acknowledge BRAF, CDAC Pune for the HPC facility.References
- G. Kaur, G. Narayanan, D. Garg, A. Sachdev and I. Matai, ACS Appl. Bio Mater., 2022, 5, 2069–2106 CrossRef CAS PubMed.
- T. Guan, J. Li, C. Chen and Y. Liu, Adv. Sci., 2022, 9, e2104165 CrossRef PubMed.
- Y. P. Liang, J. H. He and B. L. Guo, ACS Nano, 2021, 15, 12687–12722 CrossRef CAS PubMed.
- C. Togo, A. P. Zidorio, V. Goncalves, P. Botelho, K. de Carvalho and E. Dutra, Nutrients, 2022, 14, 111 CrossRef CAS PubMed.
- X. D. Tang, X. M. Wang, Y. H. Sun, L. Zhao, D. W. Li, J. H. Zhang, H. C. Sun and B. Yang, Adv. Funct. Mater., 2021, 31, 2105718 CrossRef CAS.
- S. Sharifi, M. J. Hajipour, L. Gould and M. Mahmoudi, Mol. Pharmaceutics, 2021, 18, 550–575 CrossRef CAS PubMed.
- H. Montazerian, E. Davoodi, A. Baidya, S. Baghdasarian, E. Sarikhani, C. E. Meyer, R. Haghniaz, M. Badv, N. Annabi, A. Khademhosseini and P. S. Weiss, Chem. Rev., 2022, 122, 12864–12903 CrossRef CAS PubMed.
- W. Huang, Y. Wang, Z. Huang, X. Wang, L. Chen, Y. Zhang and L. Zhang, ACS Appl. Mater. Interfaces, 2018, 10, 41076–41088 CrossRef CAS PubMed.
- R. N. Dong and B. L. Guo, Nano Today, 2021, 41, 101290 CrossRef CAS.
- M. Tavakolian, J. G. Munguia-Lopez, A. Valiei, M. S. Islam, J. M. Kinsella, N. Tufenkji and T. G. M. van de Ven, ACS Appl. Mater. Interfaces, 2020, 12, 39991–40001 CrossRef CAS PubMed.
- G. C. Gurtner, S. Werner, Y. Barrandon and M. T. Longaker, Nature, 2008, 453, 314–321 CrossRef CAS PubMed.
- L. B. Maia and J. J. G. Moura, Chem. Rev., 2014, 114, 5273–5357 CrossRef CAS PubMed.
- S. Naik, S. B. Larsen, N. C. Gomez, K. Alaverdyan, A. Sendoel, S. Yuan, L. Polak, A. Kulukian, S. Chai and E. Fuchs, Nature, 2017, 550, 475–480 CrossRef CAS PubMed.
- N. Ninan, A. Forget, V. P. Shastri, N. H. Voelcker and A. Blencowe, ACS Appl. Mater. Interfaces, 2016, 8, 28511–28521 CrossRef CAS PubMed.
- J. C. J. Holzer-Geissler, S. Schwingenschuh, M. Zacharias, J. Einsiedler, S. Kainz, P. Reisenegger, C. Holecek, E. Hofmann, B. Wolff-Winiski, H. Fahrngruber, T. Birngruber, L. P. Kamolz and P. Kotzbeck, Biomedicines, 2022, 10, 856 CrossRef CAS PubMed.
- I. Pilipenko, A. Murzova, A. Savin, A. A. Mohamed, E. Vladimirova, E. Koshel, O. Shamova and E. Kumacheva, ACS Appl. Bio Mater., 2023, 6, 1896–1905 CrossRef CAS PubMed.
- X. Han, Y. Su, G. Che, Q. Wei, H. Zheng, J. Zhou and Y. Li, ACS Appl. Mater. Interfaces, 2022, 14, 50199–50214 CrossRef CAS PubMed.
- M. C. Giano, Z. Ibrahim, S. H. Medina, K. A. Sarhane, J. M. Christensen, Y. Yamada, G. Brandacher and J. P. Schneider, Nat. Commun., 2014, 5, 4095 CrossRef CAS PubMed.
- L. Zhang, Z. Cao, T. Bai, L. Carr, J.-R. Ella-Menye, C. Irvin, B. D. Ratner and S. Jiang, Nat. Biotechnol., 2013, 31, 553–556 CrossRef CAS PubMed.
- B. Chu, J. M. He, L. L. Liu, C. X. Wu, L. L. You, X. L. Li, S. Wang, C. S. Chen and M. Tu, ACS Biomater. Sci. Eng., 2021, 7, 1100–1110 CrossRef CAS PubMed.
- M. Halder, M. Narula and Y. Singh, Bioconjugate Chem., 2023, 34, 645–663 CAS.
- J. Liu, C. Du, W. Huang and Y. Lei, Biomater. Sci., 2024, 12, 8–56 RSC.
- B. Cheng, Y. Yan, J. Qi, L. Deng, Z. W. Shao, K. Q. Zhang, B. Li, Z. Sun and X. Li, ACS Appl. Mater. Interfaces, 2018, 10, 12474–12484 CrossRef CAS PubMed.
- L. Yang, C. Zhang, C. Ren, J. Liu, Y. Zhang, J. Wang, F. Huang, L. Zhang and J. Liu, ACS Appl. Mater. Interfaces, 2018, 11, 331–339 CrossRef PubMed.
- T. Ghosh and A. K. Das, Coord. Chem. Rev., 2023, 488, 215170 CrossRef CAS.
- S. Mondal, S. Das and A. K. Nandi, Soft Matter, 2020, 16, 1404–1454 RSC.
- D. Pochan and O. Scherman, Chem. Rev., 2021, 121, 13699–13700 CrossRef CAS PubMed.
- A. Brito, S. Kassem, R. L. Reis, R. V. Ulijn, R. A. Pires and I. Pashkuleva, Chem, 2021, 7, 2943–2964 CAS.
- M. J. Webber, E. A. Appel, E. W. Meijer and R. Langer, Nat. Mater., 2016, 15, 13–26 CrossRef CAS PubMed.
- J. Zhou, J. Li, X. Du and B. Xu, Biomaterials, 2017, 129, 1–27 CrossRef CAS PubMed.
- N. C. Carrejo, A. N. Moore, T. L. Silva, D. G. Leach, I.-C. Li, D. R. Walker and J. D. Hartgerink, ACS Biomater. Sci. Eng., 2018, 4, 1386–1396 CrossRef CAS PubMed.
- G. Uzunalli, R. Mammadov, F. Yesildal, D. Alhan, S. Ozturk, T. Ozgurtas, M. O. Guler and A. B. Tekinay, ACS Biomater. Sci. Eng., 2016, 3, 1296–1303 CrossRef PubMed.
- T. Cui, X. Li, S. He, D. Xu, L. Yin, X. Huang, S. Deng, W. Yue and W. Zhong, ACS Biomater. Sci. Eng., 2020, 6, 5001–5011 CrossRef CAS PubMed.
- H. D. Sparks, T. Sigaeva, S. Tarraf, S. Mandla, H. Pope, O. Hee, E. S. Di Martino, J. Biernaskie, M. Radisic and W. M. Scott, ACS Biomater. Sci. Eng., 2021, 7, 265–278 CrossRef CAS PubMed.
- K. S. Hellmund, B. von Lospichl, C. Böttcher, K. Ludwig, U. Keiderling, L. Noirez, A. Weiß, D. J. Mikolajczak, M. Gradzielski and B. Koksch, Pept. Sci., 2021, 113, e24201 CrossRef CAS.
- T. Giraud, P. Hoschtettler, G. Pickaert, M. C. Averlant-Petit and L. Stefan, Nanoscale, 2022, 14, 4908–4921 RSC.
- T. Giraud, S. Bouguet-Bonnet, M. J. Stébé, L. Richaudeau, G. Pickaert, M. C. Averlant-Petit and L. Stefan, Nanoscale, 2021, 13, 10566–10578 RSC.
- N. Bakhtiary, B. Ghalandari, F. Ghorbani, S. N. Varma and C. Liu, Polymers, 2023, 15, 1068 CrossRef CAS PubMed.
- Y. Wang, H. Q. Lv, X. Chao, W. X. Xu, Y. Liu, G. X. Ling and P. Zhang, Mil. Med. Res., 2022, 9, 16 Search PubMed.
- B. O. Okesola, Y. Wu, B. Derkus, S. Gani, D. Wu, D. Knani, D. K. Smith, D. J. Adams and A. Mata, Chem. Mater., 2019, 31, 7883–7897 CrossRef CAS PubMed.
- X. Du, J. Zhou, J. Shi and B. Xu, Chem. Rev., 2015, 115, 13165–13307 CrossRef CAS PubMed.
- A. Biswas, S. Malferrari, D. M. Kalaskar and A. K. Das, Chem. Commun., 2018, 54, 1778–1781 RSC.
- F. D. R. Lapa, M. D. D. Silva, D. D. A. Cabrini and A. R. S. Santos, Purinergic Signalling, 2012, 8, 693–704 CrossRef CAS PubMed.
- M. Kuno, N. Seki, S. Tsujimoto, I. Nakanishi, T. Kinoshita, K. Nakamura, T. Terasaka, N. Nishio, A. Sato and T. Fujii, Eur. J. Pharmacol., 2006, 534, 241–249 CrossRef CAS PubMed.
- T. Ghosh, S. Wang, D. Kashyap, R. G. Jadhav, T. Rit, H. C. Jha, B. G. Cousins and A. K. Das, Chem. Commun., 2022, 58, 7534–7537 RSC.
- B. Baral, D. Kashyap, N. Varshney, T. P. Verma, A. K. Jain, D. Chatterji, V. Kumar, A. Mishra, A. Kumar and H. C. Jha, Genes Dis., 2023, 11, 34–37 CrossRef PubMed.
- B. Baral, V. Saini, A. Tandon, S. Singh, S. Rele, A. K. Dixit, H. S. Parmar, A. K. Meena and H. C. Jha, Apoptosis, 2023, 28, 1596–1617 CrossRef CAS PubMed.
- S. Bhowmik, T. Ghosh, Y. S. Sanghvi and A. K. Das, ACS Appl. Bio Mater., 2023, 6, 5301–5309 CrossRef CAS PubMed.
- K. P. Gan, M. Yoshio, Y. Sugihara and T. Kato, Chem. Sci., 2018, 9, 576–585 RSC.
- D. Datta, Nucleotides Nucleic Acids, 2020, 39, 530–541 CrossRef CAS PubMed.
- P. W. Frederix, G. G. Scott, Y. M. Abul-Haija, D. Kalafatovic, C. G. Pappas, N. Javid, N. T. Hunt, R. V. Ulijn and T. Tuttle, Nat. Chem., 2015, 7, 30 CrossRef CAS PubMed.
- S. Misra, S. Mukherjee, A. Ghosh, P. Singh, S. Mondal, D. Ray, G. Bhattacharya, D. Ganguly, A. Ghosh, V. K. Aswal, A. K. Mahapatra, B. Satpati and J. Nanda, Chem. – Eur. J., 2021, 27, 16744–16753 CrossRef CAS PubMed.
- A. D. Noblett, K. Baek and L. J. Suggs, ACS Biomater. Sci. Eng., 2021, 7, 2605–2614 CrossRef CAS PubMed.
- R. del Villar-Guerra, J. O. Trent and J. B. Chaires, Angew. Chem., Int. Ed., 2018, 57, 7171–7175 CrossRef CAS PubMed.
- R. del Villar-Guerra, R. D. Gray and J. B. Chaires, Nucleic Acid Chem., 2017, 68, 17.8.1–17.8.16 CAS.
- N. Amdursky and M. M. Stevens, ChemPhysChem, 2015, 16, 2768–2774 CrossRef CAS PubMed.
- Y. Dai, Y. Zhang, W. Liao, W. Wang and L. Wu, Spectrochim. Acta, Part A, 2020, 238, 118406 CrossRef CAS PubMed.
- P. Sharma, V. K. Pal, H. Kaur and S. Roy, Biomacromolecules, 2022, 23, 2496–2511 CrossRef CAS PubMed.
- X. Xie, Y. Zhang, Y. Liang, M. Wang, Y. Cui, J. Li and C. Liu, Angew. Chem., Int. Ed., 2022, 61, e202114471 CrossRef CAS PubMed.
- A. Schwaighofer, M. Montemurro, S. Freitag, C. Kristament, M. J. Culzoni and B. Lendl, Anal. Chem., 2018, 90, 7072–7079 CrossRef CAS PubMed.
- V. S. Raghuwanshi and G. Garnier, Adv. Colloid Interface Sci., 2019, 274, 102044 CrossRef CAS PubMed.
- R. Kubota, W. Tanaka and I. Hamachi, Chem. Rev., 2021, 121, 14281–14347 CrossRef CAS PubMed.
- S. Onogi, H. Shigemitsu, T. Yoshii, T. Tanida, ]M. Ikeda, R. Kubota and I. Hamachi, Nat. Chem., 2016, 8, 743–752 CrossRef CAS PubMed.
- J. A. Hunt, R. Chen, T. van Veen and N. Bryan, J. Mater. Chem. B, 2014, 2, 5319–5338 RSC.
- J. Farzanfar, F. Farjadian, A. Roointan, S. Mohammadi-Samani and L. Tayebi, Macromol. Res., 2021, 29, 54–61 CrossRef CAS.
- K. Metzger, D. Dannenberger, A. Tuchscherer, S. Ponsuksili and C. Kalbe, BMC Mol. Cell Biol., 2021, 22, 36 CrossRef CAS PubMed.
- P. K. Gavel, D. Dev, H. S. Parmar, S. Bhasin and A. K. Das, ACS Appl. Mater. Interfaces, 2018, 10, 10729–10740 CrossRef CAS PubMed.
- A. Baral, S. Roy, S. Ghosh, D. Hermida-Merino, I. W. Hamley and A. Banerjee, Langmuir, 2016, 32, 1836–1845 CrossRef CAS PubMed.
- H. D. Sparks, S. Mandla, K. Vizely, N. Rosin, M. Radisic and J. Biernaskie, Sci. Rep., 2022, 12, 14233 CrossRef CAS PubMed.
- Y. Xiong, X. Chu, T. Yu, S. Knoedler, A. Schroeter, L. Lu, K. Zha, Z. Lin, D. Jiang, Y. Rinkevich, A. C. Panayi, B. Mi, G. Liu and Y. Zhao, Adv. Healthcare Mater., 2023, 12, 2300779 CrossRef CAS PubMed.
- Y. E. Kim and J. Kim, ACS Appl. Mater. Interfaces, 2022, 14, 23002–23021 CrossRef CAS PubMed.
- M. Dizdaroglu and P. Jaruga, Free Radical Res., 2012, 46, 382–419 CrossRef CAS PubMed.
- X. Chen, Y. Zhang, W. Yu, W. Zhang, H. Tang and W. E. Yuan, J. Nanobiotechnol., 2023, 21, 129 CrossRef CAS PubMed.
- J. Deng, D. Lin, X. Ding, Y. Wang, Y. Hu, H. Shi, L. Chen, B. Chu, L. Lei, C. Wen, J. Wang, Z. Qian and X. Li, Adv. Funct. Mater., 2022, 32, 2109173 CrossRef CAS.
- P. K. Gavel, N. Kumar, H. S. Parmar and A. K. Das, ACS Appl. Bio Mater., 2020, 3, 3326–3336 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr01149j |
| This journal is © The Royal Society of Chemistry 2024 |