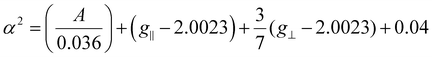Antiproliferative activity of a series of copper(II) complexes derived from a furan-containing N-acylhydrazone: monomers, dimers, charge status, and cell mechanistic studies on triple negative breast cancer cells†
Olivia
Espindola-Moreno
a,
Fagner
da Silva Moura
b,
Lucía
Santa Maria de la Parra
a,
Carolina
Stellet
b,
Jilder D. P.
Serna
c,
Renata
Diniz
d,
Nicolás A.
Rey
 *b and
Ignacio E.
León
*b and
Ignacio E.
León
 *ae
*ae
aCEQUINOR (UNLP, CCT-CONICET La Plata, asociado a CIC), Departamento de Química, Facultad de Ciencias Exactas, Universidad Nacional de La Plata, Blvd. 120 No. 1465, La Plata (1900), Argentina. E-mail: ileon@biol.unlp.edu.ar
bLABSO-Bio (CTC, PUC-Rio), Departamento de Química, Pontifícia Universade Católica do Rio de Janeiro, Rua Marquês de São Vicente, 225 – Gávea, Rio de Janeiro (22453-900), RJ, Brasil. E-mail: nicoarey@puc-rio.br
cCentro Brasileiro de Pesquisas Físicas, Rua Dr. Xavier Sigaud, 150 – Botafogo, Rio de Janeiro (22290-180), RJ, Brasil
dGrupo de Cristalografia Química (ICEx, UFMG), Departamento de Química, Universidade Federal de Minas Gerais, Av. Presidente Antônio Carlos, 6627 – Pampulha, Belo Horizonte (31270-901), MG, Brasil
eCátedra de Fisiopatología, Departamento de Ciencias Biológicas, Facultad de Ciencias Exactas, Universidad Nacional de La Plata, 47 y 115, La Plata 1900, Argentina
First published on 24th January 2025
Abstract
In this work, we evaluated the anticancer activity of compounds 1 (mononuclear) and 2 (dinuclear) copper(II) coordination compounds derived from the ligand 5-methylsalicylaldehyde 2-furoyl hydrazone (H2L) over MDA-MB-231 Triple-negative breast cancer (TNBC) cells, and compared their activities with that of a newly synthesized, protonated, dinuclear analogue of 2 (complex 3). Here, we report the synthesis of compound 3 and it has been characterized in the solid state (X-ray diffraction, FTIR) and in solution (EPR, UV-Vis, ESI) as well as its electrochemical profile. Complexes 1–3 impaired cell viability from 0.5 to 2.5 μM, with IC50 values around 1.25 μM for complexes 2 and 3 and a slightly higher value of 2.0 μM for the monomer 1. It is important to highlight that the three compounds are more active than cisplatin (CDDP), by a factor of 100 in the case of 2 and 3. Our results indicate that the protonation status of the amide group in H2L plays an important role in the stability of the dimer, being compound 2 (amide-deprotonated) fairly stable in solution so we decided to continue the study of mechanism of action using this compound. Complex 2 increases the ROS production and induces cell programmed death on TNBC cells at very low micromolar concentrations (0.5–1.5 μM). Moreover, the compound decreased the amount of breast CSCs on MDA-MB-231 cells reducing the percentage of CD44+/CD24−/low cells at 1 and 1.5 μM.
Introduction
Breast cancer is one of the most common malignancies in women, with a projected 2.3 million cases by 2030.1 One of the most aggressive types is TNBC, which accounts for around 15% of cases and is associated with a poor prognosis and the chemotherapy commonly used to treat it shows severe side effects.2 Thus, different groups around the world focused their efforts to develop different approaches using new therapeutic agents to improve and optimize the treatment.3Transition metal complexes are a new class of antitumor agents broadly used in the treatment of several solid tumors including ovarian, breast, colorectal, bone, liver, and lung.4–6 Within the non-platinum-based compounds,7,8 copper complexes have demonstrated promising anticancer activity tested in several experimental models,9–15 both in vitro and in vivo. Moreover, the anti-angiogenic and anti-metastatic properties of copper(II) complexes have been reported.16 It is important to highlight that different copper complexes are active despite their ligands not showing a substantial cytotoxicity, evidencing the central role of the metal ion itself in the antitumor activity.17,18 Besides, hydrazones and, more specifically, N-acylhydrazones and their metal complexes have attracted a remarkable interest due to their potential pharmacological activities.19–21 In this context, our groups designed, synthetized and tested several mono- and dinuclear copper(II)-N-acylhydrazone complexes with antiproliferative activity on several solid tumor lines, including TNBC cells.22–27
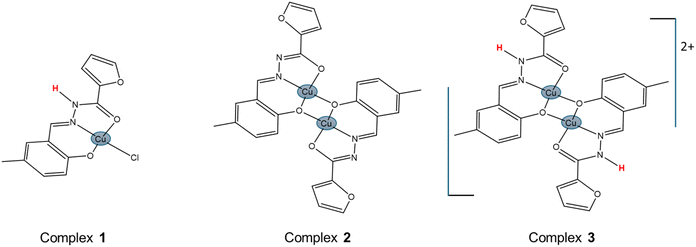
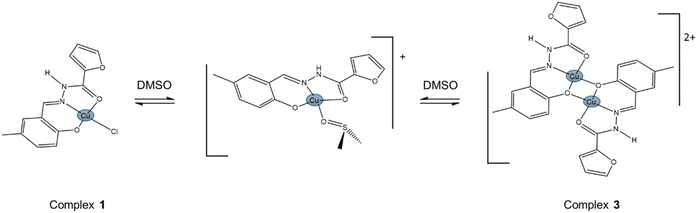
As part of our broader study on the interaction of transition metals with hydrazones, we have recently published a mononuclear (1) and a dinuclear (2) copper(II) coordination compounds derived from the ligand 5-methylsalicylaldehyde 2-furoyl hydrazone (H2L).28 Complex 2 is a deprotonated, electrically neutral dimer of 1. In this work, we assessed the antiproliferative profile of compounds 1 and 2 over MDA-MB-231 TNBC cells, and compared their activities with that of a newly synthesized, protonated, dinuclear analogue of 2 (complex 3). Besides being positively charged in solution, 3 is also, in fact, a more accurate dimer of hydrolyzed (chloride ligand lost) compound 1, in the sense that the hydrazone in both species presents the same protonation status. The expected forms in freshly prepared DMF solutions of complexes 1, 2 and 3 are depicted in Scheme 1. However, solvolysis can quickly modify the structures of 1 and 3, as discussed in the Results section, generating a common derivative. A complete experimental characterization of 3, including single-crystal X-ray diffraction, was performed. FTIR, EPR, electronic absorption spectra and ESI-MS(+) were measured, as well as the electric conductivity and electrochemical profile. As for the previously reported compounds, the interactions of 3 with HSA were studied too.
Working with this structurally-related set of complexes allowed us to get a glimpse of the effect of dimerization and net charge in the anticancer activity. Moreover, we have investigated and report herein, for the first time, the mechanism of action of 2, the most stable of the complexes considered, on MDA-MB-231 cells. Our research was focused on the role of oxidative stress and its effects on apoptosis. Furthermore, we studied the inhibition effect of the compound toward breast cancer stem cells (BCSCs).
Results and discussion
Single-crystal X-ray diffractions study
As in the case of ligand H2L, and complexes 1 and 2, the new coordination compound specifically synthesized for this work, 3, was characterized crystallographically.
Table 1 lists the crystallographic details for 3. The crystal structure investigation of this complex indicates that it crystallizes in the triclinic system, space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) , with two asymmetric units formed by one partially deprotonated (HL−) hydrazonic ligand, one nitrate ion, one cupric center and a water molecule (Fig. 1a) per unit cell. Asymmetric units are arranged into dimeric structures (Fig. 1b), for which Table 2 displays some selected geometrical parameters. Each metal site in the dimer is pentacoordinate in a slightly distorted square pyramidal geometry (τ equal to 0.045). Similar to the structure previously described by us for the analogue 2, the N-acylhydrazone performs as a tridentate equatorial ligand through the carbonyl oxygen O2, azomethine nitrogen N2 and (ionized) phenolate oxygen O3, which acts as a bridge between both copper(II) centres. Cu⋯Cu distance is 2.9846(19) Å, a little bit shorter than the one reported by us for complex 2. This coordination plane is completed by the bridging-phenolate oxygen of a second asymmetric unit (O3i, symmetry code: −x, 1 − y, 1 − z), thus establishing the dimeric Cu2+-(μ-phenoxo)2-Cu2+ scaffold. Average Cu-equatorial donor distance is 1.954(5) Å. An oxygen atom (O5) from a nitrate ion occupies the fifth, apical position, in each of the metal sites, where they point out to different sides of the dimer plane. Cu–O5 distance is 2.314(6) Å due to the Jahn–Teller effect.
, with two asymmetric units formed by one partially deprotonated (HL−) hydrazonic ligand, one nitrate ion, one cupric center and a water molecule (Fig. 1a) per unit cell. Asymmetric units are arranged into dimeric structures (Fig. 1b), for which Table 2 displays some selected geometrical parameters. Each metal site in the dimer is pentacoordinate in a slightly distorted square pyramidal geometry (τ equal to 0.045). Similar to the structure previously described by us for the analogue 2, the N-acylhydrazone performs as a tridentate equatorial ligand through the carbonyl oxygen O2, azomethine nitrogen N2 and (ionized) phenolate oxygen O3, which acts as a bridge between both copper(II) centres. Cu⋯Cu distance is 2.9846(19) Å, a little bit shorter than the one reported by us for complex 2. This coordination plane is completed by the bridging-phenolate oxygen of a second asymmetric unit (O3i, symmetry code: −x, 1 − y, 1 − z), thus establishing the dimeric Cu2+-(μ-phenoxo)2-Cu2+ scaffold. Average Cu-equatorial donor distance is 1.954(5) Å. An oxygen atom (O5) from a nitrate ion occupies the fifth, apical position, in each of the metal sites, where they point out to different sides of the dimer plane. Cu–O5 distance is 2.314(6) Å due to the Jahn–Teller effect.
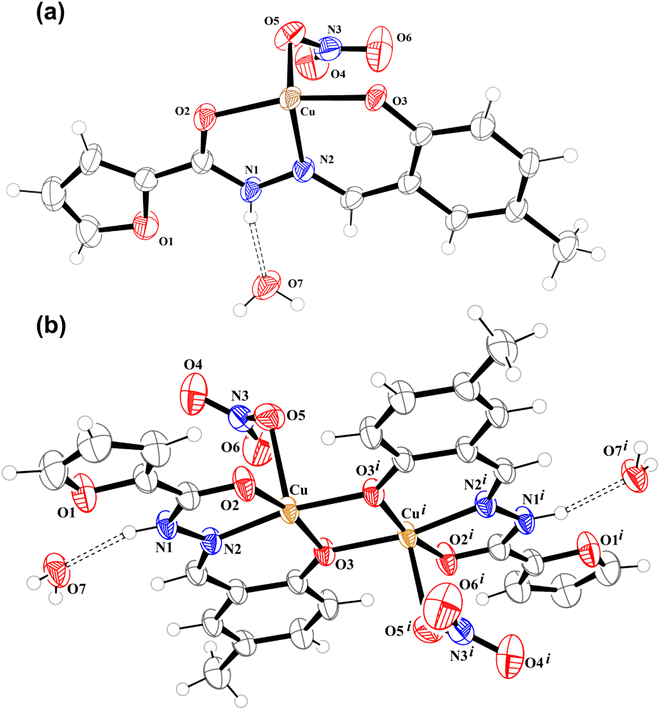 | ||
| Fig. 1 Crystal structure representations of 3: (a) asymmetric unit and (b) dimeric structure. Ellipsoids drawn at 50% probability level. Symmetry code: i (−x, 1 − y, 1 − z). | ||
| Formula of the monomer | C26H26N6O14Cu2 |
| M r/g mol−1 | 773.60 |
| Crystal system, space group | Triclinic, P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
| Temperature/K | 298 |
| a/Å | 7.1609(1) |
| b/Å | 10.7439(2) |
| c/Å | 11.0077(3) |
| α/° | 61.876(3) |
| β/° | 84.989(2) |
| γ/° | 81.589(2) |
| V/Å3 | 1774.5(2) |
| Z | 2 |
| Radiation type | Cu Kα |
| μ/mm−1 | 2.516 |
| Crystal size/mm | 0.03 × 0.07 × 0.19 |
| Measured/independent reflections | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 781/3153 781/3153 |
| Observed reflections/Rint | 2937/0.0630 |
| Parameters | 222 |
| R obs, Rall | 0.0659, 0.0684 |
| wRabs, wRall, S | 0.2277, 0.2287, 1.157 |
| Δρmax, Δρmin/e Å−3 | 1.286, −0.539 |
| Symmetry code: i (−x, 1 − y, 1 − z). | ||||
|---|---|---|---|---|
| Bond distances/Å | ||||
| Cu–O2 | 1.976(5) | |||
| Cu–O3 | 1.941(4) | |||
| Cu–O3i | 1.960(4) | |||
| Cu–N2 | 1.939(5) | |||
| Cu–O5 | 2.314(6) | |||
| Cu⋯Cui | 2.9846(19) | |||
| Bond angles/° | ||||
| O2–Cu–O3 | 167.0(2) | |||
| O2–Cu–O3i | 105.44(19) | |||
| O2–Cu–O5 | 87.0(2) | |||
| O2–Cu–N2 | 81.0(2) | |||
| O3–Cu–O3i | 80.20(19) | |||
| O3–Cu–O5 | 87.0(2) | |||
| O3–Cu–N2 | 91.9(2) | |||
| O5–Cu–O3i | 94.1(2) | |||
| O5–Cu–N2 | 94.3(2) | |||
| N2–Cu–O3i | 169.7(2) | |||
| Cu–O3–Cui | 99.80(19) | |||
| H-bonding parameters | ||||
| D–H⋯A | D–H/Å | H⋯A/Å | D⋯A/Å | D–H⋯A/° |
| O7–H7A⋯O5 | 0.85 | 2.01 | 2.843(9) | 165.0 |
| O7–H7B⋯O4 | 0.85 | 2.01 | 2.784(10) | 150.0 |
| N1–H1⋯O7 | 0.86 | 1.90 | 2.729(9) | 163.0 |
As mentioned above, two crystallization water molecules per dimer are also present in the structure, which are involved in hydrogen bonding with the nitrate ion and HL−, giving rise to a bidimensional (2D) network arrangement along the ab plane. The average of O⋯O and N⋯O distances (Table 2) are 2.814(10) and 2.729(9) Å, respectively.
Electronic absorption spectroscopy – a complexation study
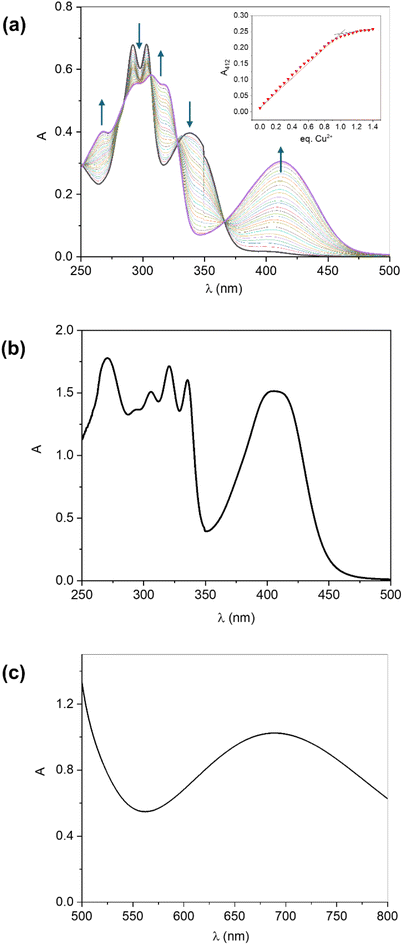
The UV-Vis spectrum of ligand H2L was already reported and fully discussed by us in a previous paper.28 When titrating a 2 × 10−5 mol L−1 solution of H2L in MeOH with successive 0.05 eq. increments of a copper(II) nitrate trihydrate methanolic solution, important spectral changes are observed (Fig. 2a). A series of isosbestic points is clearly present at 283, 305, 327 and 367 nm, indicating the coexistence of only two absorbent species in equilibrium, i.e. free and copper-bound N-acylhydrazone. Plotting the absorbance at 412 nm as a function of the number of Cu2+ equivalents give rise to a classical saturation pattern, which points to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry (Fig. 2a, inset). As the reaction medium is the same used during the synthesis of complex 3, this result provides a mechanism by which the dinuclear species can be obtained: the initial formation of [Cu(HL)(MeOH)]+ followed by a dimerization equilibrium into the form [Cu(HL)]22+. However, the latter step would only occur after days of slow solvent evaporation, increasing [Cu(HL)(MeOH)]+ concentration to the point where dimerization is favored and dimer precipitation occurs. When the synthesized solid is dissolved in DMF, almost the same absorption profile is observed, although the N-acylhydrazone-related bands, present between 280 and 325 nm in methanolic solution, are bathochromically shifted (Fig. 2b and Table 3) due to the different nature of the solvent (i.e., protic vs. aprotic).
1 stoichiometry (Fig. 2a, inset). As the reaction medium is the same used during the synthesis of complex 3, this result provides a mechanism by which the dinuclear species can be obtained: the initial formation of [Cu(HL)(MeOH)]+ followed by a dimerization equilibrium into the form [Cu(HL)]22+. However, the latter step would only occur after days of slow solvent evaporation, increasing [Cu(HL)(MeOH)]+ concentration to the point where dimerization is favored and dimer precipitation occurs. When the synthesized solid is dissolved in DMF, almost the same absorption profile is observed, although the N-acylhydrazone-related bands, present between 280 and 325 nm in methanolic solution, are bathochromically shifted (Fig. 2b and Table 3) due to the different nature of the solvent (i.e., protic vs. aprotic).
| λ max (nm) | ε (L mol−1 cm−1) | Assignment |
|---|---|---|
| 270 | 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 ± 2000 500 ± 2000 |
Intra-ligand |
| 305 | 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 ± 405 400 ± 405 |
Intra-ligand |
| 320 | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ± 780 000 ± 780 |
Intra-ligand |
| 336 | 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 ± 1020 900 ± 1020 |
Intra-ligand |
| 406 | 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150 ± 1140 150 ± 1140 |
LMCT |
| 688 | 202 ± 1 | d–d transition |
In DMF solution, 3 presents a d–d absorption centred at around 688 nm, which is much closer to the one of the mononuclear complex 1 (678 nm) than to the dinuclear compound 2 (630 nm), suggesting that 3 undergoes dissociation to its monomers in solution (Fig. 2c).
EPR, electrochemical and ESI-MS profiles: supporting dimer dissociation
Complex 3 is virtually EPR-silent in the solid state, due to an antiferromagnetic coupling between the copper(II) centres at close proximity [2.9846(19) Å]. Conversely, frozen DMF and DMSO solutions of this complex exhibit axial spectra with well-resolved parallel and partially resolved perpendicular copper-related hyperfine splitting patterns (Fig. 3), indicating, once again, that the dimer is broken to [Cu(HL)DMF]+ and [Cu(HL)DMSO]+ upon dissolution in DMF and DMSO, respectively. EPR data are summarized in Table 4.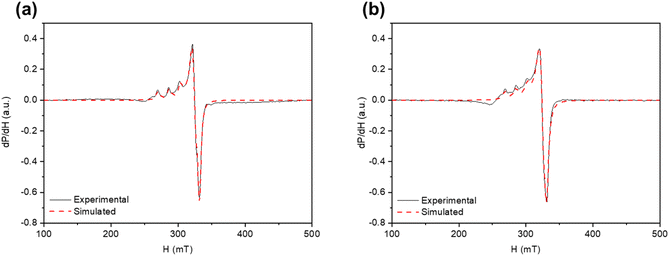 | ||
| Fig. 3 X-band EPR spectra of complex 3 in frozen (77 K) solution of (a) DMF and (b) DMSO. Simulated profiles are given as dashed red lines. | ||
| Solvent | g ∥ | g ⊥ | A ∥ (10−4 cm−1) | A ⊥ (10−4 cm−1) |
|---|---|---|---|---|
| DMF | 2.26 | 2.05 | 165 | 11.7 |
| DMSO | 2.27 | 2.05 | 163 | 2.8 |
In both cases, the fact that g∥ > g⊥ > 2.0023 constitutes an indicative of unpaired electron localization in the dx2−y2 orbital of copper, which points to a tetragonal symmetry around the metal centre, although heavily distorted, since Sakaguchi29 ratios, i.e., g∥/A∥, equal to 137 cm (DMF) and 139 cm (DMSO) were calculated. We also estimated the covalency parameter α2, given by the equation proposed by Kivelson and Neiman:30
If the value of α2 is 0.5, it suggests a complete covalent bonding, while a value of 1.0 means an entire ionic bonding. We obtained α2 = 0.78 for both solvents, which indicate that the donor-metal bonds in the complexes have an intermediate covalent character.
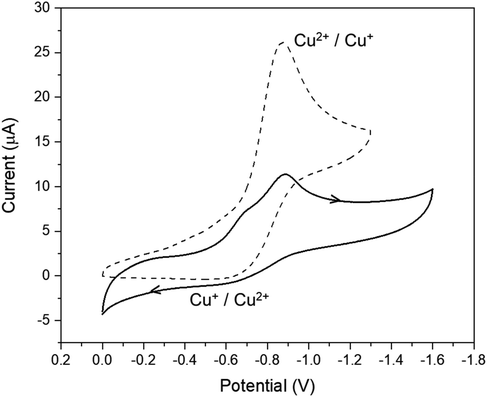
The same conclusion, regarding dimer dissociation, can be taken from electrochemical data measured in DMF. Fig. 4 presents the cyclic voltammogram of 3 at 100 mV s−1. It is possible to observe a major cathodic peak at −0.88 V vs. Ag/AgCl, related to a one-electron process, altogether with a poorly defined shoulder at less negative potentials (around −0.70 V vs. Ag/AgCl). On the other hand, only an anodic peak (at −0.60 V vs. Ag/AgCl) was identified, which involves a very low current. In order to better interpreting the results obtained, we compared this electrochemical profile with the one previously published by us for the mononuclear complex 1 under the same conditions (dashed voltammogram in the figure). It became clear that the redox processes at −0.88 V and −0.60 V vs. Ag/AgCl are related to the presence of the solvolyzed form of 1, [Cu(HL)(DMF)]+, indicating that most of the dimeric nature of compound 3 is lost through dissociation in solution. The unresolved reduction shoulder observed at around −0.70 V vs. Ag/AgCl is probably associated with a remnant of the cationic dimer in DMF, although this could not be completely established.
ESI-MS(+) studies performed after dissolution of 3 in DMSO (as in the cytotoxicity studies discussed below) and subsequent 200x dilution in MeOH (Fig. 5), show that only one major species can be distinguished in the range between 300 and 800 Da, at (m/z) 384.02, corresponding to the mononuclear, cationic compound [Cu(HL)(DMSO)]+. This was clearly confirmed based on the isotopic distribution analysis presented in the inset of Fig. 5. The detection of this DMSO-containing complex indicates a strong affinity of the copper centre by dimethyl sulfoxide, even in a methanol rich medium.
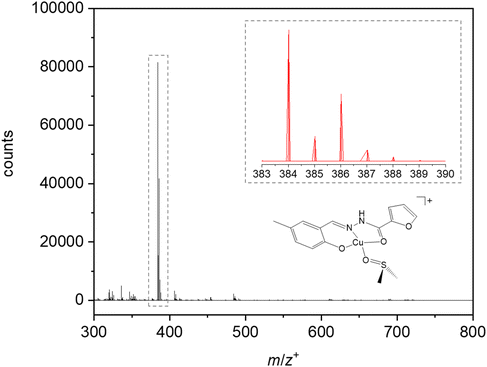 | ||
Fig. 5 ESI-MS spectrum (positive mode) of 3 in a DMSO/methanol (0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 99.5) mixture. Inset: isotope distribution pattern for [Cu(HL)(DMSO)]+ (C15H17N2O4SCu+). 99.5) mixture. Inset: isotope distribution pattern for [Cu(HL)(DMSO)]+ (C15H17N2O4SCu+). | ||
HSA-binding assessment
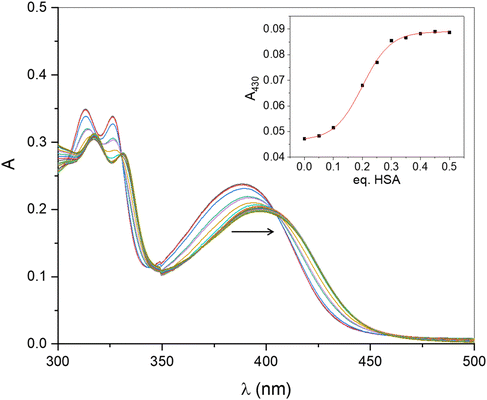
Fig. 6 shows the spectral changes in the 300–500 nm region produced by the addition of successive amounts of HSA to a 2 × 10−5 mol L−1 solution of complex 3. We observe a clear bathochromic effect in all the bands of the spectrum as the titration progresses. Isosbestic points could be clearly identified at 330, 345 and 404 nm, indicating the presence of only two absorbent species in equilibrium, i.e., free and bound forms of 3. Plotting the absorbance at 430 nm as a function of HSA equivalents (Fig. 6, inset) affords a sigmoidal curve that stabilizes at ca. [HSA]/[3] = 0.50, indicating that HSA can host up to two dimers or, alternatively, up to four units of the mononuclear complex derived from the dissociation of 3. The last hypothesis is in accordance to the behaviour previously observed for the interaction of complex 1 with HSA.28 In fact, the spectra in Fig. 6 are identical to those obtained when studying the system HSA-1.Cytotoxicity and cell proliferation studies
Cytotoxicity studies were determined by the MTT assay for complexes 1–3 on MDA-MB-231 (triple negative breast adenocarcinoma). The well-known clinical agent cisplatin (CDDP) was used as a reference for antiproliferative activity.Table 5 shows that all the three copper compounds decreased cell viability in the submicromolar range of concentrations (0.5–2.5 μM), with similar IC50 values for both dimers (complexes 2 and 3, around 1.25 μM) and a slightly higher value of 2.0 μM for the monomer 1. Given the good activity found, it seems that the charged nature of 3 does not prevent its entrance in the cell, so one can conclude that some transporter may be involved in the process. Furthermore, the fact that the antiproliferative potential of this compound is, considering standard-deviation involved, exactly double that observed for the mononuclear derivative 1 constitutes another clue supporting dissociation of 3, previously demonstrated through EPR, electrochemical and ESI-MS, since both compounds mostly share a common derivative after their initial dissolution in DMSO (Scheme 2). Noteworthy, all the compounds are more active than CDDP, since it showed an IC50 value of 131 μM on MDA-MB-231 cells. In addition, the IC50 values obtained for 1–3 towards MDA-MB-231 cells are comparable with those of the clinical drug reference doxorubicin (DOX).31
| Compound | IC50 [μM] |
|---|---|
| H2L | >50 |
| Cu(NO3)2·3H2O | >50 |
| 1 | 2.0 ± 0.1 |
| 2 | 1.3 ± 0.1 |
| 3 | 1.2 ± 0.1 |
It must be pointed out that stability studies under conditions similar to those employed in the cell experiments (0.5% DMSO solution in PBS, pH 7.4) were performed for complexes 2 and 3, nominally the most active ones. Upon dissolution, both compounds demonstrated to be fairly stable over a time period of 24 h (UV-Vis spectra taken over time showed only slight variations for 2 and no noticeable changes for 3, as presented in Fig. SM1†).
On the other hand, the IC50 values of H2L and Cu2+ (free metal cation) are greater than 50 μM on the MDA-MB-231 strain, hence revealing the key role of complexation to modulate the anticancer activity of hydrazone-containing copper(II) complexes. This kind of effect has been reported for other copper complexes and other metal-based compounds.32–34
Previously, we have reported a series of copper(II) complexes with N-acylhydrazones as ligands, including [Cu(N-N-Tph)(H2O)]NO3 (or CuHL1) (IC50 = 1.6 ± 0.2 μM),35 [Cu(N-N-BzOMe)(H2O)2]NO3 (CuHL2) (IC50 = 1.6 ± 0.1 μM),23 [Cu(N-N-BzOH)(H2O)2]NO3 (CuHL3) (IC50 = 5.8 ± 0.4 μM),18 [Cu(N-N-BzOH)(o-phen)] (CuHL3/phen) (IC50 = 5.3 ± 0.2 μM)36 and [Cu(N-N BzOH)(bipy)]NO3 (CuHL3/bipy) (IC50 = 11.4 ± 0.6 μM).36 Complexes CuHL1 and CuHL2 displayed a similar anticancer activity in comparison with 2 and 3. Nevertheless, CuHL3 exhibited less activity than CuHL1 and CuHL2, showing the importance of methoxi above hydroxy in the benzohydrazide. Besides, the presence of coligands as o-phen and bipy did not improve the anticancer actions of CuHL3. Several copper(II) complexes with vanillin derivatives and naproxen were synthetized by Lu and coworkers. Among them, the most effective compound showed a moderate cytotoxicity against bulk breast cancer cells and breast cancer stem cells, with a medium micromolar cytotoxicity (37.6 ± 3.3 and 36.0 ± 4.6 μM respectively).37
Low and coworkers described the antitumor action of Cu2+ complexes containing the Schiff base N′-[1-(2-oxo-2H-chromen-3-yl)-ethylidene]-hydrazinecarbodithioic acid benzyl ester on MDA-MB-231 and MCF-7 cells, reporting IC50 values around 20 μM.38
Due to the higher activity and solution stability of complex 2, we decided to continue the study of mechanism of action using this compound on MDA-MB-231 cells.
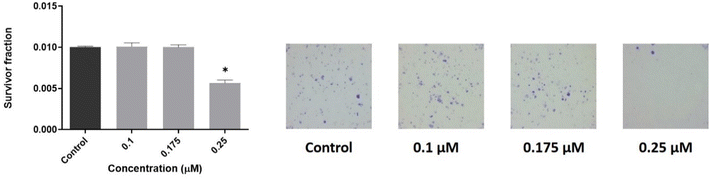
To evaluate the effect of 2 on the cellular reproductive potential, a clonogenic assay was performed. Fig. 7 shows a strong reduction of cell proliferation on the MDA-MB-231 strain, affecting colony formation at 0.25 μM (p < 0.01). Concentrations of 0.1 and 0.175 μM, instead, were unable to change basal conditions. Results agree with the cell viability assay.
ROS production
Several copper drugs can initiate pseudo-Fenton reactions intracellularly, resulting in the production of ROS.39,40 For a good consideration of the possible mechanism involved in the cytotoxicity of complex 2 on TNBC cells, we explored the effect of this dinuclear compound (1–15 μM) on oxidative stress using a specific probe (DHR123).41The results (Table 6) evidenced that 2 increased ROS production of 242, 447 and 530% over basal levels on MDA-MB-231 cells for concentrations of 5, 10 and 15 μM after 3 h of incubation, thus suggesting that ROS production could be considered as one of the relevant mechanisms of action of compound 2. Several reports show that many copper(II) complexes have ROS generation as an important mechanism of action related to their anticancer activity42–44 and associated with cell death.
| Complex 2 concentration [μM] | % ROS level |
|---|---|
| 0 | 100.5 ± 3.2 |
| 1 | 97.6 ± 4.1 |
| 2.5 | 206.2 ± 18.6 |
| 5 | 242.2 ± 34.4 * |
| 10 | 446.9 ± 55.8 ** |
| 15 | 530.4 ± 44.1 ** |
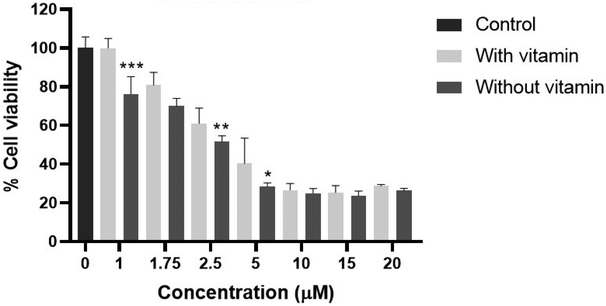
To confirm the importance of ROS as a mechanism of cell death by complex 2, we carried out a cell viability assay using radical scavengers, such as a mixture of vitamins C and E, under the same experimental conditions. Fig. 8 shows that cell viability was completely recovered at [2] = 1 μM, whilst the deleterious effects of this complex were only partially overcome at the higher concentrations of 2.5 and 5 μM.
Apoptosis
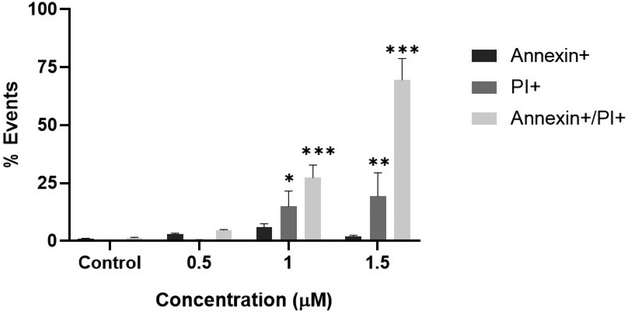
With the aim to evaluate if 2 induces programmed cell death, we performed cell cytometry assays using annexin V (early apoptotic biomarker) and propidium iodide (PI). Fig. 9 and SM2 show the flow cytometry results of the apoptotic process in the presence of different concentrations of compound 2 (0.5, 1, 1.5 μM) on MDA-MB-231 cells. Complex 2 augmented the late apoptotic (Annexin V+/PI+) and necrotic (Annexin V−/PI+) fractions from 1 μM on. The basal condition showed 1.2% of V+/PI+ cells whilst after incubation with 1 and 1.5 μM of 2 this value increases until 31.7% and 69.6%, respectively. Also, at [2] = 1.5 μM, the necrotic population (V−/PI+) increased until 19.3%.Our results are in agreement with the cell viability studies, suggesting that 2 induced cell apoptosis in a concentration-dependent manner. It is important to highlight that apoptosis cell death have also been reported as the leading mechanism of action for many copper complexes with antitumor activity toward different cancer cell lines.4
Cancer stem cell studies
Solid tumors are composed of diverse cancer cell types with varying hierarchical structures.45 In this respect, cancer stem cells (CSCs) constitute an immortal subset of the total malignant cell population with the ability to accumulate mutations that drive tumor growth.46 CSCs play an important role in tumor aggressiveness due to their self-renewal and stemness potential. As a result, in recent years, there has been a lot of interest in developing anti-CSC medicines, and CSCs have emerged as an important and specialized cell target.47In order to evaluate the anti-CSCs activity of compound 2, we performed flow cytometry experiments using CD44 and CD24 biomarkers. Table 7 and Fig. SM3† show that 2 decreased the amount of breast CSCs on MDA-MB-231 cells. The basal condition resulted on 99.1% CD44+/CD24− whilst this value is reduced after incubation with 2, displaying 95.1% (1.0 μM) and 91.6% (1.5 μM). However, another copper(II)-hydrazone compound, CuHL2, exhibited better anti-CSCs activity than complex 2 on MDA-MB-231 cells, with a reduction of CD44+/CD24− population from 98.6% (basal condition) to 68.3% (at 1.5 μM).23 Furthermore, Boodram et al. and Laws described the anti-CSCs activity of copper(II)-non-steroidal complexes and copper(II)-phenanthroline on breast cancer cells.48,49
| Compound [μM] | % CD44HIGH/CD24LOW |
|---|---|
| 0 | 99.1 |
| 0.5 | 99.1 |
| 1.0 | 95.1 * |
| 1.5 | 91.6 * |
Conclusions
The synthesis of a new dimeric complex containing the phenol-deprotonated/amide-protonated form of H2L (5-methylsalicylaldehyde 2-furoyl hydrazone), i.e. compound 3 – [Cu(HL)(NO3)]2·2H2O, allowed us to establish a link between the previously published complexes 1 – [Cu(HL)Cl]·H2O (mononuclear, phenol-deprotonated/amide-protonated) and 2 – [Cu(L)(i-PrOH)]2 (dimeric, fully deprotonated form of the ligand). Our results indicate that the protonation status of the amide group in H2L plays an important role in the stability of the dimer, being compound 2 (amide-deprotonated) fairly stable in solution. Conversely, complex 3, in which the amide group remains protonated, mainly dissociates into monomers upon dissolution, as proved by UV-Vis, EPR and cyclic voltammetry. When a coordinating solvent is employed, compounds 1 and 3 generate virtually the same species: [Cu(HL)Solv]+.The anticancer activities of complexes 1 and 2 on MDA-MB-231 TNBC cells were assessed, and compared with that of the new analogue 3. All the three compounds reduced cell viability in the submicromolar range of concentrations (0.5–2.5 μM), with IC50 values around 1.25 μM for complexes 2 and 3 and a slightly higher value of 2.0 μM for the monomer 1 (which almost double the one observed for the dimers). It seems that the charged nature of the solvolysis/dissociation products of 1 and 3 does not prevent their entrance in the cell, so one can conclude that some transporter may be involved in the process. Remarkably, all the compounds are more active than the clinically used drug CDDP, by a factor of 100 in the case of 2 and 3. Due to the high activity and solution stability of complex 2, we decided to continue the study of mechanism of action using this compound.
Our findings highlighted that the main mechanism of action of 2 involves the ROS production and apoptosis induction. Also, this compound decreased the amount of breast CSCs, reducing the percentage of CD44+/CD24− from 1.0 μM on. Taking into account the antiproliferative activity of the dimer 2 on MDA-MB-231 cells, we strongly believe that this complex is an engaging candidate for potential antitumor therapies and it would be attractive to further test it on TNBC in vivo experimental models.
Experimental part
Materials
Solvents and reactants were purchased from commercial sources. Human serum albumin (HSA) was bought from Sigma-Aldrich. All chemicals were used without any type of pretreatment or further purification.Dulbecco's modified Eagle's médium (DMEM) and TrypLE ™ were purchased from Gibco (Gaithersburg, MD, USA), and fetal bovine serum (FBS) was purchased from Internegocios (Argentina). Tissue culture materials were purchased from Corning (Princeton, NJ, USA). Annexin V, fluorescein isothiocyanate (FITC), propidium iodide (PI), DHR123 and Dihydroetidium were from Invitrogen (Buenos Aires, Argentina). CD44 and CD24 antibodies were purchased by BD (USA).
Methods
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)]; 1628 [ν(C
O)]; 1628 [ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)hydrazone]; 1495 [δ(C–N–H)hydrazone]; 1326 [ν(C–O)phenol]; 1164 [ν(N–N)].
N)hydrazone]; 1495 [δ(C–N–H)hydrazone]; 1326 [ν(C–O)phenol]; 1164 [ν(N–N)].
[Cu(C13H11O3N2)(NO3)]2·2H2O (3). 242 mg of Cu(NO3)2·3H2O (1.0 mmol) were dissolved in 10 mL MeOH and dropwise added to 15 mL of a H2L methanolic solution (262 mg, 1.0 mmol). The reactants were stirred at room temperature for 1 h, affording a dark green solution. After a few days on the bench, green crystals suitable for XRD analysis were isolated. Yield = 310 mg (0.40 mmol of dimer, 80%). Elemental analysis (%) − calculated for [Cu(C13H11O3N2)(NO3)]2·2H2O (773.60 g mol−1): C − 40.4, H − 3.4, N − 10.9; found: C − 39.9, H − 3.0, N − 10.6. ICP-OES (%) − calculated: Cu − 16.4; found: Cu − 16.2. TGA: a weight loss of approximately 4.6% (calcd 4.6%) between 110 and 145 °C, consistent with the removal of two hydration water molecules per dimer. Mid IR (KBr, cm−1): 3198 [ν(N–H)hydrazone]; 1630 [ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)]; 1610 [ν(C
O)]; 1610 [ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)hydrazone]; 1610 [ν(C
N)hydrazone]; 1610 [ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)hydrazone]; 1384 [ν(NO2)as nitrate]; 1345 [ν(C–O)phenol]; 1305 [ν(NO2)sym nitrate]; 1118 [ν(N–N)].
N)hydrazone]; 1384 [ν(NO2)as nitrate]; 1345 [ν(C–O)phenol]; 1305 [ν(NO2)sym nitrate]; 1118 [ν(N–N)].
Molar conductivity in DMF: 160.4 ohm−1 cm2 mol−1 (equivalent to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 electrolyte system),50 indicating that both nitrate ions are dissociated upon dissolution. In DMSO, the value of 112 ohm−1 cm2 mol−1 was obtained, also in agreement with a 1
2 electrolyte system),50 indicating that both nitrate ions are dissociated upon dissolution. In DMSO, the value of 112 ohm−1 cm2 mol−1 was obtained, also in agreement with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2-type electrolyte (expected range: 110–195 ohm−1 cm2 mol−1, according to the reference by Ali et al.)51
2-type electrolyte (expected range: 110–195 ohm−1 cm2 mol−1, according to the reference by Ali et al.)51
Physicochemical analyses
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 counts, gated on an FSC vs. SSC dot plot, were recorded.
000 counts, gated on an FSC vs. SSC dot plot, were recorded.
Data availability
The data supporting this article have been included as part of the ESI (Fig. SM1 and Fig. SM2†).Crystallographic data for complex 3 has been deposited with the Cambridge Crystallographic Data Centre as Suppl. Publications CCDC 2370060.†
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by CONICET (PIP 2051), UNLP (EX005) and ANPCyT (PICT 2019-2322), Argentina. IEL is a member of the Research Career of CONICET. OEM and LSMdlP have a fellowship from CONICET. Prof. NAR wishes to thank FAPERJ (Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro, Brazil) as well as CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil) for the research fellowships awarded.References
- I. O. Alanazi and Z. Khan, Understanding EGFR Signaling in Breast Cancer and Breast Cancer Stem Cells: Overexpression and Therapeutic Implications, Asian Pac. J. Cancer Prev., 2016, 17(2), 445–453, DOI:10.7314/APJCP.2016.17.2.445.
- S. Wen, L. Manuel, M. Doolan, J. Westhuyzen, T. P. Shakespeare and N. J. Aherne, Effect of Clinical and Treatment Factors on Survival Outcomes of Triple Negative Breast Cancer Patients, Breast Cancer: Targets Ther., 2020, 12, 27–35, DOI:10.2147/BCTT.S236483.
- R. Nanda, Novel therapies for triple-negative breast cancer, Clin. Adv. Hematol. Oncol., 2020, 18(12), 799–801 Search PubMed.
- L. M. Balsa, E. J. Baran and I. E. León, Copper Complexes as Antitumor Agents: In vitro and In vivo Evidence, Curr. Med. Chem., 2023, 30(5), 510–557, DOI:10.2174/0929867328666211117094550.
- V. Ferretti and I. León, An Overview of Vanadium and Cell Signaling in Potential Cancer Treatments, Inorganics, 2022, 10(4), 47, DOI:10.3390/inorganics10040047.
- E. J. Anthony, E. M. Bolitho and H. E. Bridgewater, et al., Metallodrugs are unique: opportunities and challenges of discovery and development, Chem. Sci., 2020, 11(48), 12888–12917, 10.1039/D0SC04082G.
- I. Ott and R. Gust, Non Platinum Metal Complexes as Anti–cancer Drugs, Arch. Pharm., 2007, 340(3), 117–126, DOI:10.1002/ardp.200600151.
- I. Leon, J. Cadavid-Vargas, A. Di Virgilio and S. Etcheverry, Vanadium, Ruthenium and Copper Compounds: A New Class of Nonplatinum Metallodrugs with Anticancer Activity, Curr. Med. Chem., 2017, 24(2), 112–148, DOI:10.2174/0929867323666160824162546.
- D. Denoyer, S. A. Clatworthy and M. A. Cater, Metallo-Drugs: Development and Action of Anticancer Agents, ed. A. Sigel, H. Sigel, E. Freisinger and R.KO. Sigel, De Gruyter, 2018, vol. 18. DOI:10.1515/9783110470734.
- V. Gandin, C. Ceresa and G. Esposito, et al., Therapeutic potential of the phosphino Cu(I) complex (HydroCuP) in the treatment of solid tumors, Sci. Rep., 2017, 7(1), 13936, DOI:10.1038/s41598-017-13698-1.
- C. Santini, M. Pellei, V. Gandin, M. Porchia, F. Tisato and C. Marzano, Advances in Copper Complexes as Anticancer Agents, Chem. Rev., 2014, 114(1), 815–862, DOI:10.1021/cr400135x.
- R. Alemón-Medina, M. Breña-Valle, J. L. Muñoz-Sánchez, M. I. Gracia-Mora and L. Ruiz-Azuara, Induction of oxidative damage by copper-based antineoplastic drugs (Casiopeínas®), Cancer Chemother. Pharmacol., 2007, 60(2), 219–228, DOI:10.1007/s00280-006-0364-9.
- J. F. Cadavid-Vargas, I. E. Leon, S. B. Etcheverry, E. Santi, M. H. Torre and A. L. Di Virgilio, Copper(II) Complexes with Saccharinate and Glutamine as Antitumor Agents: Cytoand Genotoxicity in Human Osteosarcoma Cells, Anticancer Agents Med. Chem., 2017, 17(3), 424–433, DOI:10.2174/1871520616666160513130204.
- L. M. Balsa, M. C. Ruiz, L. Santa Maria de la Parra, E. J. Baran and I. E. León, Anticancer and antimetastatic activity of copper(II)-tropolone complex against human breast cancer cells, breast multicellular spheroids and mammospheres, J. Inorg. Biochem., 2020, 204, 110975, DOI:10.1016/j.jinorgbio.2019.110975.
- L. M. Balsa, V. Ferretti and M. Sottile, et al., New copper(II) and oxidovanadium(IV) complexes with a vitamin B6 Schiff base: mechanism of action and synergy studies on 2D and 3D human osteosarcoma cell models, Dalton Trans., 2024, 53(7), 3039–3051, 10.1039/D3DT02964F.
- X. Shi, Z. Chen, Y. Wang, Z. Guo and X. Wang, Hypotoxic copper complexes with potent anti-metastatic and anti-angiogenic activities against cancer cells, Dalton Trans., 2018, 47(14), 5049–5054, 10.1039/C8DT00794B.
- M. R. Rodríguez, L. M. Balsa and J. Del Plá, et al., Synthesis, characterization, DFT calculations and anticancer activity of a new oxidovanadium(IV) complex with a ligand derived from o-vanillin and thiophene, New J. Chem., 2019, 43(29), 11784–11794, 10.1039/C9NJ02092F.
- Y. Burgos-Lopez, J. Del Plá and L. M. Balsa, et al., Synthesis, crystal structure and cytotoxicity assays of a copper(II) nitrate complex with a tridentate ONO acylhydrazone ligand. Spectroscopic and theoretical studies of the complex and its ligand, Inorg. Chim. Acta, 2019, 487, 31–40, DOI:10.1016/j.ica.2018.11.039.
- S. D. Joshi, D. Kumar and S. R. Dixit, et al., Synthesis, characterization and antitubercular activities of novel pyrrolyl hydrazones and their Cu-complexes, Eur. J. Med. Chem., 2016, 121, 21–39, DOI:10.1016/j.ejmech.2016.05.025.
- A. K. Tanoli, Z. Khan, T. Kamal, M. Ali, M. Latif and Z. T. Maqsood, Metal-based biologically active compounds: Synthesis, spectral and antioxidant studies of transition metal complexes with hydrazone derivatives, Pak. J. Pharm. Sci., 2019, 32(1), 103–108 CAS.
- S. Thota and D. A. Rodrigues, Pinheiro P de SM, Lima LM, Fraga CAM, Barreiro EJ, N-Acylhydrazones as drugs, Bioorg. Med. Chem. Lett., 2018, 28(17), 2797–2806, DOI:10.1016/j.bmcl.2018.07.015.
- L. Santa Maria de la Parra, A. I. B. Romo and J. Rodríguez-López, et al., Promising Dual Anticancer and Antimetastatic Action by a Cu(II) Complex Derived from Acylhydrazone on Human Osteosarcoma Models, Inorg. Chem., 2024, 63(11), 4925–4938, DOI:10.1021/acs.inorgchem.3c04085.
- L. M. Balsa, V. Ferraresi-Curotto and M. J. Lavecchia, et al., Anticancer activity of a new copper(II) complex with a hydrazone ligand. Structural and spectroscopic characterization, computational simulations and cell mechanistic studies on 2D and 3D breast cancer cell models, Dalton Trans., 2021, 50(28), 9812–9826, 10.1039/D1DT00869B.
- M. R. Rodríguez, L. M. Balsa and O. E. Piro, et al., Synthesis, Crystal Structure, Spectroscopic Characterization, DFT Calculations and Cytotoxicity Assays of a New Cu(II) Complex with an Acylhydrazone Ligand Derived from Thiophene, Inorganics, 2021, 9(2), 9, DOI:10.3390/inorganics9020009.
- JP Rada, J Forté and G Gontard, et al., Novel luminescent benzopyranothiophene- and BODIPY-derived aroylhydrazonic ligands and their dicopper(II) complexes: syntheses, antiproliferative activity and cellular uptake studies, Anticancer Agents Med. Chem., 2021, 26(6), 675–688, DOI:10.1007/s00775-021-01885-5.
- J. P. Rada, J. Forté, G. Gontard, V. Corcé, M. Salmain and N. A. Rey, Isoxazole–Derived Aroylhydrazones and Their Dinuclear Copper(II) Complexes Show Antiproliferative Activity on Breast Cancer Cells with a Potentially Alternative Mechanism Of Action, ChemBioChem, 2020, 21(17), 2474–2486, DOI:10.1002/cbic.202000122.
- J. P. Rada, B. S. M. Bastos and L. Anselmino, et al., Binucleating Hydrazonic Ligands and Their μ-Hydroxodicopper(II) Complexes as Promising Structural Motifs for Enhanced Antitumor Activity, Inorg. Chem., 2019, 58(13), 8800–8819, DOI:10.1021/acs.inorgchem.9b01195.
- F. d. S. Moura, Y. S. Sobrinho and C. Stellet, et al., Copper(II) complexes of a furan-containing aroylhydrazonic ligand: syntheses, structural studies, solution chemistry and interaction with HSA, Dalton Trans., 2023, 52(47), 17731–17746, 10.1039/D3DT02597G.
- U. Sakaguchi and A. W. Addison, Spectroscopic and redox studies of some copper(II) complexes with biomimetic donor atoms: implications for protein copper centres, J. Chem. Soc., Dalton Trans., 1979,(4), 600, 10.1039/dt9790000600.
- D. Kivelson and R. Neiman, ESR Studies on the Bonding in Copper Complexes, J. Chem. Phys., 1961, 35(1), 149–155, DOI:10.1063/1.1731880.
- N. Ayoub, K. Al-Shami, M. A. Y. Alqudah and N. Mhaidat, Crizotinib, a MET inhibitor, inhibits growth, migration, and invasion of breast cancer cells in vitro and synergizes with chemotherapeutic agents, OncoTargets Ther., 2017, 10, 4869–4883, DOI:10.2147/OTT.S148604.
- MR Rodríguez, J Del Plá and LM Balsa, et al., Cu(II) and Zn(II) complexes with a poly-functional ligand derived from o-vanillin and thiophene. Crystal structure, physicochemical properties, theoretical studies and cytotoxicity assays against human breast cancer cells, New J. Chem., 2019, 43(18), 7120–7129, 10.1039/C8NJ06274A.
- A. I. Matesanz, E. Jimenez-Faraco and M. C. Ruiz, et al., Mononuclear Pd(II) and Pt(II) complexes with an α-N-heterocyclic thiosemicarbazone: cytotoxicity, solution behaviour and interaction versus proven models from biological media, Inorg. Chem. Front., 2018, 5(1), 73–83, 10.1039/C7QI00446J.
- A. C. González-Baró, V. Ferraresi-Curotto and R. Pis-Diez, et al., A novel oxidovanadium(V) compound with an isonicotinohydrazide ligand. A combined experimental and theoretical study and cytotoxity against K562 cells, Polyhedron, 2017, 135, 303–310, DOI:10.1016/j.poly.2017.07.013.
- L. M. Balsa, M. R. Rodriguez, B. S. Parajón-Costa, A. C. González-Baró, M. J. Lavecchia and I. E. León, Anticancer Activity and Mechanism of Action Evaluation of an Acylhydrazone Cu(II) Complex toward Breast Cancer Cells, Spheroids, and Mammospheres, ChemMedChem, 2022, 17(4) DOI:10.1002/cmdc.202100520.
- Y. Burgos-López, L. M. Balsa and O. E. Piro, et al., Tridentate acylhydrazone copper(II) complexes with heterocyclic bases as coligands. Synthesis, spectroscopic studies, crystal structure and cytotoxicity assays, Polyhedron, 2022, 213, 115621, DOI:10.1016/j.poly.2021.115621.
- C. Lu, A. Eskandari, P. B. Cressey and K. Suntharalingam, Cancer Stem Cell and Bulk Cancer Cell Active Copper(II) Complexes with Vanillin Schiff Base Derivatives and Naproxen, Chem. – Eur. J., 2017, 23(47), 11366–11374, DOI:10.1002/chem.201701939.
- M. L. Low, G. Paulus and P. Dorlet, et al., Synthesis, characterization and biological activity of Cu(II), Zn(II) and Re(I) complexes derived from S-benzyldithiocarbazate and 3-acetylcoumarin, BioMetals, 2015, 28(3), 553–566, DOI:10.1007/s10534-015-9831-2.
- U. Jungwirth, C. R. Kowol, B. K. Keppler, C. G. Hartinger, W. Berger and P. Heffeter, Anticancer Activity of Metal Complexes: Involvement of Redox Processes, Antioxid. Redox Signaling, 2011, 15(4), 1085–1127, DOI:10.1089/ars.2010.3663.
- I. Garcia–Bosch and M. A. Siegler, Copper–Catalyzed Oxidation of Alkanes with H2O2 under a Fenton–like Regime, Angew. Chem., Int. Ed., 2016, 55(41), 12873–12876, DOI:10.1002/anie.201607216.
- M. Yazdani, Concerns in the application of fluorescent probes DCDHF-DA, DHR 123 and DHE to measure reactive oxygen species in vitro, Toxicol. In Vitro, 2015, 30(1 Pt B), 578–582, DOI:10.1016/j.tiv.2015.08.010.
- A. I. B. Romo, V. S. Dibo and D. S. Abreu, et al., Ascorbyl and hydroxyl radical generation mediated by a copper complex adsorbed on gold, Dalton Trans., 2019, 48(37), 14128–14137, 10.1039/C9DT01726G.
- M. C. Ruiz, K. Perelmulter and P. Levín, et al., Antiproliferative activity of two copper(II) complexes on colorectal cancer cell models: Impact on ROS production, apoptosis induction and NF-κB inhibition, Eur. J. Pharm. Sci., 2022, 169, 106092, DOI:10.1016/j.ejps.2021.106092.
- P. Levín, M. C. Ruiz and A. I. B. Romo, et al., Water-mediated reduction of [Cu(dmp)2(CH3CN)]2+: implications of the structure of a classical complex on its activity as an anticancer drug, Inorg. Chem. Front., 2021, 8(13), 3238–3252, 10.1039/D1QI00233C.
- M. C. Cabrera, R. E. Hollingsworth and E. M. Hurt, Cancer stem cell plasticity and tumor hierarchy, World J. Stem Cells, 2015, 7(1), 27–36, DOI:10.4252/wjsc.v7.i1.27.
- M. Z. Ratajczak, Cancer stem cells–normal stem cells “Jedi” that went over to the “dark side”, Folia Histochem. Cytobiol., 2005, 43(4), 175–181 CAS.
- M. De Angelis, F. Francescangeli and A. Zeuner, Breast Cancer Stem Cells as Drivers of Tumor Chemoresistance, Dormancy and Relapse: New Challenges and Therapeutic Opportunities, Cancers, 2019, 11(10), 1569, DOI:10.3390/cancers11101569.
- K. Laws, G. Bineva-Todd, A. Eskandari, C. Lu, N. O'Reilly and K. Suntharalingam, A Copper(II) Phenanthroline Metallopeptide That Targets and Disrupts Mitochondrial Function in Breast Cancer Stem Cells, Angew. Chem., Int. Ed., 2018, 57(1), 287–291, DOI:10.1002/anie.201710910.
- J. N. Boodram, I. J. Mcgregor, P. M. Bruno, P. B. Cressey, M. T. Hemann and K. Suntharalingam, Breast Cancer Stem Cell Potent Copper(II)–Non–Steroidal Anti–Inflammatory Drug Complexes, Angew. Chem., Int. Ed., 2016, 55(8), 2845–2850, DOI:10.1002/anie.201510443.
- W. J. Geary, The use of conductivity measurements in organic solvents for the characterisation of coordination compounds, Coord. Chem. Rev., 1971, 7(1), 81–122, DOI:10.1016/S0010-8545(00)80009-0.
- I. Ali, W. A. Wani and K. Saleem, Empirical Formulae to Molecular Structures of Metal Complexes by Molar Conductance, Synth. React. Inorg., Met.-Org., Nano-Met. Chem., 2013, 43(9), 1162–1170, DOI:10.1080/15533174.2012.756898.
- Rigaku Oxford Diffraction, CrysAlisPRO, version 11713846, Rigaku Corporation, Tokyo, Japan. Published online 2015 Search PubMed.
- G. M. Sheldrick, Crystal structure refinement with SHELXL, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71(1), 3–8, DOI:10.1107/S2053229614024218.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, OLEX2: a complete structure solution, refinement and analysis program, J. Appl. Crystallogr., 2009, 42(2), 339–341, DOI:10.1107/S0021889808042726.
- L. J. Farrugia, WinGX and ORTEP for Windows: an update, J. Appl. Crystallogr., 2012, 45(4), 849–854, DOI:10.1107/S0021889812029111.
- C. F. Macrae, I. J. Bruno and J. A. Chisholm, et al., Mercury CSD 2.0 – new features for the visualization and investigation of crystal structures, J. Appl. Crystallogr., 2008, 41(2), 466–470, DOI:10.1107/S0021889807067908.
- T. Mosmann, Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays, J. Immunol. Methods, 1983, 65(1–2), 55–63, DOI:10.1016/0022-1759(83)90303-4.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. SM1: UV-Vis stability evaluation for complexes 2 and 3 at 2 × 10−5 mol L−1, under conditions similar to those in cell studies (0.5% DMSO/PBS, pH 7.4), monitored over 24 h. Fig. SM2: Apoptosis dot plots on MDA-MB-231 cells treated with complex 2 (0.5, 1 and 1.5 μM) obtained by flow cytometry. Fig. SM3: CSC dot plots on MDA-MB-231 cells treated with complex 2 (0.5, 1 and 1.5 μM) obtained by flow cytometry. CCDC 2370060 (3). For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4dt03445g |
| This journal is © The Royal Society of Chemistry 2025 |