Open Access Article
Open Access ArticleDevelopment of a paper-based fluorescent carbon quantum dots MIPs sensor for selective detection of lumpy skin disease virus
Dalia M. El-Husseinia,
Dalia M. A. Elmasrya,
Eman M. Abo Hatabb and
Samr Kassem *a
*a
aNanomaterials Research and Synthesis Unit, AHRI, ARC, Giza, 12618, Egypt. E-mail: samr.kassem@ahri.gov.eg
bVirology Department, AHRI, ARC, Giza, 12618, Egypt
First published on 29th August 2024
Abstract
Lumpy skin disease (LSD) is a contagious viral disease caused by the Lumpy Skin Disease virus (LSDV), a member of the Capripoxviridae family. Traditional LSDV diagnostic procedures proved to have challenges in terms of cross reactivity as well as limited sensitivity and specificity. Herein, we combined molecularly imprinted polymers (MIPs) and quantum dots (QDs) technology to develop a paper-based turn on fluorescence sensor for rapid, sensitive and selective detection of LSDV. Under optimal conditions, the sensor showed linear enhancement in fluorescence intensity with the increase of LSDV concentration and exhibited a detection limit of 101 log10 TCID50 per ml. It also presented high specificity towards LSDV compared to other viruses viz sheep pox virus (SPV). Furthermore, the proposed sensor was successfully tested with spiked and real LSDV samples, proving its potential to serve as a sensitive selective sensor for LSDV diagnosis. Based on our knowledge, this is the first record of a paper-based diagnostic sensor for LSDV utilizing a CQDs-MIPs turn-on mechanism.
1. Introduction
The OIE lists LSDV as one of the most hazardous animal diseases1 causing considerable economic losses in terms of skin damage, reduced milk output, abortions and deaths in most of the infected ruminants.2 The virion is a brick-shaped particle measuring 170–260 by 300–450 nm in diameter.3 Its capsid contains a linear, non-segmented, double-stranded DNA genome of ∼150 kilobases. The genomes of sheep pox virus (SPV) and goat pox virus (GPV), members of capri pox family, exhibit significant resemblance of >97% nucleotide identity.4 Previously, they were classified solely based on their animal host. However, nowadays, it is possible to differentiate them by analyzing their genomic DNA.Traditional laboratory techniques including virus isolation and serological testing are usually used for LSDV diagnosis.5 These strategies are generally challenging and time-consuming. Molecular techniques including reverse transcription PCR can detect LSDV in clinical samples with sufficient sensitivity.6 However, it is unsuitable for field diagnosis due to the requirement of specialized equipment. Novel quick point-of-care biosensing methods with high sensitivity and specificity are required for early LSDV detection leading to controlling infection spread.7
Nanotechnology can be used to create novel materials and products and is recognized as one of six main enabling technologies that will revolutionize society in the near future.8 CQDs are zero-dimensional carbon nanoparticles that are usually smaller than 10 nm and have a crystal constant of 0.34, which is equivalent to the d-spacing of graphite (002). The carbonyl, carboxyl, epoxy, and hydroxyl functional groups located on the carbon core of CQDs enhances their water solubility and provide a platform for simple functionalization with other substances.9,10 The presence of functional groups on the surface of CQDs enhances their biocompatibility, optical properties, and targeting ability.11
Carbon Quantum dots (CQDs), a superior class of fluorescent nanostructures made from carbon materials, have garnered interest in biosensing applications due to their high biocompatibility, ease of synthesis, outstanding photoluminescence, and tailored surface functionalities. Yet, target selectivity is the main obstacle to CQD-based sensor development.12–14
Recently, CQDs have been widely employed in different fields including biological imaging,15 biochemical sensing,16–19 biomedicine,20 and catalysis.21 Furthermore, notable advancements were achieved in the identification of various molecules through the incorporation of CQDs.17–19 Various types of QDs were developed for the viral detection, for instance, cadmium telluride and carbon QDs were incorporated in immunosensor platform for the detection of human immune deficiency virus while molybdenum trioxide QDs were used for detection of influenza A virus.22–24
The selectivity of CQD-based sensors can be improved by Molecularly Imprinted Polymers, or MIPs. These sensors have proven to be very effective at detecting traces of chemicals in complex matrices.25,26 MIPs have advanced significantly as a result of the desire to enhance disease diagnosis and prevention through the nanoscale detection of harmful viruses and other biological targets. Although there are still many obstacles to overcome, imprinting techniques can quickly and accurately identify viruses, making them useful as novel sensing materials.27
MIPs are able to recognize templates selectively28 by creating templates with functional monomers and cross-linking monomers. After the eluting of the templates, the cavities with high specificity and affinity for the target molecule become apparent.29,30 MIPs components (template, monomers, crosslinker, polymerization initiator, and solvent) and the nature of their interactions with one another have a significant impact on the efficacy, affinity, and selectivity of the recognition sites of the MIPs. Therefore, choosing the right components for the MIPs during design is essential to achieving the required qualities for a particular application.31
MIPs have been employed extensively in the sensor area in recent years due to their inexpensive cost of preparation, simplicity, and durability.32,33 So, MIPs have been synthesized on the surface of CQDs to improve their poor selectivity as graphene QDs,34 CdSe QDs,35 CdTe QDs,36 and CQDs.37
To our knowledge, a paper-based fluorescent CQDs- MIP-sensor for LSDV determination has not yet been reported. In this study, we combined the high selectivity of MIPs with the strong stable fluorescence of CQDs to develop a simple method for LSDV detection.
2. Experimental section
2.1 Equipment
Attenuated total Reflectance-Fourier transform infrared spectroscopy (ATR-FTIR), PerkinElmer, the UK, Atomic force microscope by a Nanosurf C 3000 Atomic force microscope (AFM), Switzerland and all AFM images were operated in contact mode using a Nanosurf SNL-10 silicon tip, Field emission scanning electron Microscope (FE-SEM) (Quanta FEG250, Czech), transmission electron Microscope (TEM) (JEOL JEM-1400, USA), NANOTRAC WAVE ‖ Dynamic light scattering (DLS), USA, X-ray photoelectron spectroscopy (XPS) (Escalab250Xi, Thermo Fisher, USA), Muffle furnace (Nbertherm, Germany) and BioTek Synergy HTX Multimode Reader, Agilent, USA were used in this study.2.2 Synthesis of surface modified APTES-CQDs
CQDs was synthesized and surface functionalized by mixing 0.5 g of anhydrous citric acid (CAS No.: 77-92-9) with 10 ml of 3-Aminopropyl triethoxysilane (APTES) monomer (CAS No.: 919-30-2) according to.12 The mixture was transferred into a 50 mL Teflon-lined autoclave and kept in a muffle at 210 °C for 5 h for carbonization and surface functionalization. The colour of the solution turned colourless confirming the synthesis of the surface modified APTES-CQDs. The solution was then filtered through 0.45 μm filter membrane and stored at 4 °C for further use. DLS, UV-vis absorption microplate reader, XPS, ATR-FTIR and TEM were used for APTES-CQDs characterization. All used glassware was washed by detergent and tap water several times, then rinsed by deionized water and ethanol, then dried in a hot air oven.2.3 Fabrication of the paper-based CQDs- MIP-sensor
MIPs were synthesized using the following polymeric mixture; 13.0 mg of acrylamide (CAS No.: 79-06-1), 10.4 μl of methacrylic acid (CAS No.: 79-41-4), 6.4 μl of methyl methacrylate (CAS No.: 80-62-6) and 6 μl of N-vinyl pyrrolidone (CAS No.:88-12-0) monomer were mixed with 48 mg of N,N-(1,2-dihydroxyethylene) bisacrylamide (CAS No.: 868-63-3) as a crosslinker. The prepared mixture was dissolved in 300 μl of DMSO (CAS No.: 67-68-5) containing 2.4 μl of 2, 5-BIS (tert-butyl peroxy)-2, 5-dimethylhexane (CAS No.: 78-63-7) as an initiator mixed with 10 μl of APTES-CQDs. MIPs synthesis and polymerization were conducted using similar methodology to Kassem et al.37 with modifications including the use of CQDs as a cost-effective fluorescence source instead of N-fluorescein acrylamide monomer, and we also used APTES monomer to functionalize the CQDs surface to ensure proper binding of other monomers forming the MIPs.Afterwards, the mixture was subjected to pre-polymerization at 80 °C for 1 h under stirring followed by 1 h at room temperature. 7 μl of 0.25 mg μl−1 LSDV (provided by Vaccine and Serum Research Institute, Egypt) was pipetted on a sterile glass slide surface and allowed to dry at room temperature in a sterile, clean laminar flow cabinet in order to prepare the LSDV-template. The nitrocellulose membrane (NCM, HF180MC100, Millipore, Germany) was cut into 6 mm circular pieces using a normal sterile paper puncher. NCM pieces were dipped into the pre-polymerization solution and pressed directly onto the dried virus drop to prepare the CQDs-MIPs. Whereas to prepare the negative control CQDs-NIPs, the dipped NCM was pressed onto an empty sterile glass slide. Both NIPs and MIPs were exposed overnight to UV radiation with a wavelength of 254 nm until the NCM surface showed signs of a thin layer of polymerized film. Subsequently, they were submerged in 10% HCL (CAS No.: 7647-01-0, Sigma-Aldrish, UK) for ten seconds to denature and eliminate the viral molecules followed by their incubation for 2 h at 45 °C in deionized water under shaking conditions before being dried on a filter paper.
The chemical composition and binding of the functional groups between LSDV and CQDs-MIPs were evaluated using ATR-FTIR at the scanning range of 4000–400 cm−1. The surface morphology of the CQDs-NIPs and CQDs-MIPs NCM were examined using AFM and FE-SEM.
2.4 LSDV-CQDs-MIP sensor validation
The LSDV-CQDs-MIP sensor's sensitivity was assessed by measuring the fluorescence intensity that was observed at various LSDV concentrations (101–106 log10 TCID50 per ml). In summary, a 96-well black fluorescent microplate containing CQDs-NIPs and CQDs-MIPs NCM was loaded with 20 μl of each LSDV dilution. To ensure that the viral particles were evenly distributed across the NCM, the reaction was incubated for 15 min under orbital shaking. The fluorescence intensity was then measured at 485/20 nm for excitation and 528/20 nm for emission.Similar procedures were used to determine the assay's specificity, with the exception of the virus types, SPV, FMDV, and BVDV (they were provided by virology department, Animal Health Research Institute, Egypt) at 105 log10 TCID50 per ml. In order to assess the effectiveness of the sensor in identifying LSDV within the complex matrix of field real samples, 15 samples including viral-free blood buffy coat, serum, and skin nodules were spiked with varying amounts of LSDV (101–106 log10 TCID50 per ml). For confirmation and sensitivity comparison, Viral DNA was extracted following QIAamp DNA extraction kit instructions (Cat no. 51306, Qiagen, USA) then subjected to Real-Time PCR (qPCR) testing using LSDV dtec-qPCR kit (F100, GPS™, Spain) according to instruction manual. All experiments were run in triplicates.
3. Results and discussion
3.1 Characterization of surface modified APTES-CQDs
CQDs have emerged as a promising class of quantum dots. CQDs have garnered significant attention in recent years due to their numerous advantages. These include excellent photo-luminescence properties, straightforward synthesis methods, cost-effective precursors, solubility, low toxicity, chemical stability, and ease of functionalization. As a result, they have been utilized in point-of-care sensors.38,39The synthetic pathways of CQDs can be classified into “top-down” and “bottom-up” techniques based on the carbon source utilized during the synthesis.40 The “top-down” strategy, often uses physical or chemical processes, results in small-sized CQDs when a large-scale carbon source is used such as graphite rod, carbon fibers, and carbon nanotubes. On the other hand, “bottom-up” approach always begins with carbon sources of small molecules, such as folic acid, glucose, and citric acid.41 Bottom-up methodologies for synthesizing CQDs include hydrothermal,42 solvothermal,43 microwave-assisted,44 ultrasonic-assisted,45 and combustion46 procedures. The hydrothermal method usually results in CQDs with high quantum yield and strong fluorescence intensity when compared to those derived from the “top-down” approach. Hence, in recent years, this approach has mostly been used in the production of CQDs.41,47
In our study, Surface modified APTES-CQDs were obtained via a simple one-pot hydrothermal process. The acylation reaction and surface silanization occurred directly in the presence of APTES. In this process, CQDs were synthesized from anhydrous citric acid which was carbonized under temperature of 210 °C for 5 h. The amine groups in APTES acylated with the carboxyl groups on the surface of the pyrolyzed organics to provide surface APTES-functionalized CQDs with fluorescent characteristics. Subsequently, CQDs-MIPs were successfully obtained by the polymerization assay using a co-polymer mixture consisted of acrylamide, APTES-CQDs, methacrylic acid, methyl methacrylate and N-vinyl pyrrolidone and N,N-(1,2-dihydroxyethylene) bisacrylamide with adjusted ratios to enhance sensitivity and specificity of the developing sensor.38
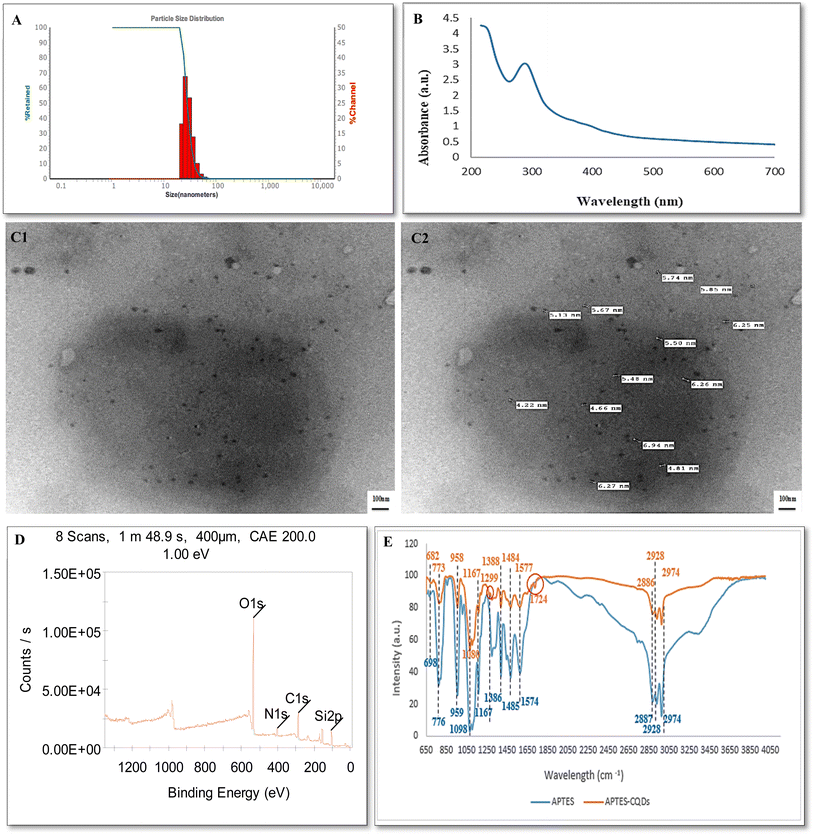
DLS measurement indicated average size of 25.3 ± 2.5 nm with PDI of 0.3 and surface charge of +28 mv (Fig. 1A). The positive charge relates to the presence of amine groups on APTES functionalized CQDs surface. The morphology of APTES-CQDs was characterized by TEM showing the presence of homogenous well dispersed spherical shaped particles with average size of 5.1 ± 0.8 nm (Fig. 1C1 and C2).
FT-IR spectra of APTES and APTES-CQDs are shown in (Fig. 1E) and Table 1. APTES results showed characteristic peaks at 2974, 2928, and 2886 cm−1 which corresponds to the stretching vibrations of O–H group. While the peaks at 1577, and 1484 cm−1 were attributed to the bending vibrations of the N–H group. Peaks at 1388 cm−1 was attributed to the bending vibrations of C–H group. In addition, the peaks at 1167 and 1080 cm− 1 were attributed to the stretching vibration of C–O group. The peaks at 958 and 773 cm−1 indicated the bending vibration of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C-CL groups respectively. Also, the peak at 682 cm−1 was attributed to the stretching vibration of the C
C and C-CL groups respectively. Also, the peak at 682 cm−1 was attributed to the stretching vibration of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C group. Compared with APTES, APTES-CQDs FT-IR spectrum displayed two different absorption peaks at 1724 and 1299 cm−1 in addition to the shifting of most functional groups of O–H, N–H, C–H, C–O and C
C group. Compared with APTES, APTES-CQDs FT-IR spectrum displayed two different absorption peaks at 1724 and 1299 cm−1 in addition to the shifting of most functional groups of O–H, N–H, C–H, C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C groups. The two different peaks at 1724 and 1299 cm−1 which are attributed to the stretching vibration of C
C groups. The two different peaks at 1724 and 1299 cm−1 which are attributed to the stretching vibration of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and the bending vibration of the C–H group, together with the shifting of other functional groups, revealed the formation of an amide bond, which demonstrated the successful acylation reaction between CQDs and APTES, suggesting the successful synthesis of APTES-CQDs.
O and the bending vibration of the C–H group, together with the shifting of other functional groups, revealed the formation of an amide bond, which demonstrated the successful acylation reaction between CQDs and APTES, suggesting the successful synthesis of APTES-CQDs.
| Material | Absorption (cm−1) | Chemical bond | Vibration mode | Functional group |
|---|---|---|---|---|
| APTES | 2974 | O–H | Stretching | Carboxylic acid |
| 2928 | O–H | Stretching | Carboxylic acid | |
| 2886 | O–H | Stretching | Carboxylic acid | |
| 1577 | N–H | Bending | Primary amine | |
| 1484 | N–H | Bending | Secondary amine | |
| 1388 | C–H | Bending | Aldehyde | |
| 1167 | C–O | Stretching | Alcohol | |
| 1080 | C–O | Stretching | Alcohol | |
| 958 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C C |
Bending | Alkene | |
| 773 | C–CL | Bending | Halo compound | |
| 682 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C C |
Stretching | Alkene | |
| APTES functionalized CQDs | 2974 | O–H | Stretching | Carboxylic acid |
| 2928 | O–H | Stretching | Carboxylic acid | |
| 2887 | O–H | Stretching | Carboxylic acid | |
| 1724 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
Stretching | Carboxylic acid | |
| 1574 | N–H | Bending | Primary amine | |
| 1485 | N–H | Bending | Secondary amine | |
| 1386 | C–H | Bending | Aldehyde | |
| 1299 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
Bending | Aromatic ester | |
| 1167 | C–O | Stretching | Alcohol | |
| 1098 | C–O | Stretching | Alcohol | |
| 959 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C C |
Bending | Alkene | |
| 776 | C–CL | Bending | Halo compound | |
| 698 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C C |
Stretching | Alkene |
The elemental analysis of APTES-CQDs was conducted by XPS demonstrating four peaks at 533.25, 286.04, 401.73, and 103.91 eV referring to O 1s, C 1s, N 1s, and Si 2p, respectively (Fig. 1D). The results of XPS further confirmed that successfulness of the surface functionalization of CQDs with APTES. The optical properties of APTES-CQDs were examined with UV-vis absorption spectrum showing a strong absorption peak located at 300 nm (Fig. 1B). Furthermore, APTES-CQDs exhibited highest fluorescence at excitation and emission wavelengths of 485/20 nm and 528/20 nm, respectively using a filter-based microplate reader with different Excitation/Emission wavelengths (360/40–460/40, 360/40–528/20, 360/40–590/20, 485/20–528/20, 485/20–590/20), respectively.
The occurred hydrothermal reaction involved chemical interactions between various carbonaceous precursors under high temperature and pressure. This technique is well recognized as an environmentally friendly, regulated, and cost-effective solution.48,49 The synthesis of CQDs typically comprises four processes: breakdown, polymerization/aromatization, nucleation, and growth. Initially, the carbonaceous precursors undergo intermolecular dehydration and polymerization, forming the carbon skeleton and aromatic clusters. When clusters achieve critical super saturation, CQD nucleation occurs. During this process, aromatic clusters move towards the surface of the particle, resulting in the functionalization of the surface of CQDs. As the reaction progresses, intermediates are gradually transforms into fully formed CQDs with a narrow particle size distribution.50 The size of the formed CQDs produced depends on temperature, time, and reactants used in the process.51 Surface functionalization of CQDs is accomplished simultaneously during the hydrothermal process. This phase involves modifying the fluorophore, passivating the surface, and oxidizing the surface of all synthesized CQDs. Most CQDs exhibit a strong hydrophilicity and their surfaces are typically treated with oxygen-containing functional groups.52 Usually, CQDs hydrophilicity is influenced by both the reactant and the synthesis process. For instance, CQDs derived from soluble precursors typically display significant hydrophilicity due to the retention of functional groups from the precursors on the CQD surface during their fabrication. Besides, the choice of reactants affects CQDs optical characteristics. Moreover, elemental analysis showed that the reaction time and reactant type had a direct impact on the amount of nitrogen atoms doped into the carbon core. When comparing CQDs synthesized from secondary and tertiary amine to those made from citric acid and primary amine, the latter presented the highest nitrogen doping degree and the maximum photoluminescence intensity.52 Herein, surface functionalization of CQDs with APTES was confirmed through FTIR and XPS analysis.
3.2 Characterization of the CQDs-MIPs-NCM
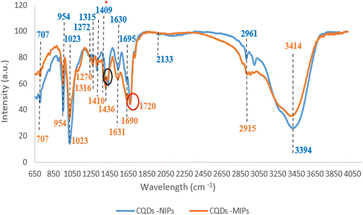
Molecular imprinting involves the synthesis of polymers that possess molecular recognition sites that precisely correspond to the size, shape, and functional group of template molecules. Due to their low selectivity, CQDs should be paired with other technologies viz MIPs to enhance their selectivity while maintaining cost-effectiveness.53,54After successfully synthesizing the CQDs-MIPs, chemical composition and involved functional groups in the binding of LSDV with CQDs -MIPs were evaluated using ATR-FTIR at scanning range of 4000–400 cm−1.48 Hence, the FTIR spectra of CQDs -MIPs were compared before and after binding with LSDV (Fig. 2 and Table 2) to inspect the altered functional groups upon binding. For CQDs -MIPs before LSDV binding, the FTIR revealed peaks located at 3414, 2915, 2133, and 1690 cm−1 that are referred to the stretching vibration of N–H, O–H, N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N
N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, C
N, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups, respectively. Also, peaks were detected at 1630, 1542, 1436 and 1409 cm−1 indicate the bending vibration of N–H, N–H, O–H and O–H groups, respectively. While the peaks detected at 1315, 1272 and 1023 cm−1 correspond to the stretching vibration of –NO2, C–O and C–O groups, respectively. In addition, peaks detected at 954 and 707 cm−1 refer to the bending vibration of C
O groups, respectively. Also, peaks were detected at 1630, 1542, 1436 and 1409 cm−1 indicate the bending vibration of N–H, N–H, O–H and O–H groups, respectively. While the peaks detected at 1315, 1272 and 1023 cm−1 correspond to the stretching vibration of –NO2, C–O and C–O groups, respectively. In addition, peaks detected at 954 and 707 cm−1 refer to the bending vibration of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C group and the stretching vibration peak of C-CL, respectively. FTIR spectra of the CQDs -MIPs after rebinding with LSDV revealed shifting of the functional groups N–H, O–H and C
C group and the stretching vibration peak of C-CL, respectively. FTIR spectra of the CQDs -MIPs after rebinding with LSDV revealed shifting of the functional groups N–H, O–H and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O represented by peaks at 3394, 2961 and 1695 cm−1 with stretching mode, respectively. Also, shifting of two N–H groups were detected at 1631 and 1540 cm−1. Likewise, O–H, –NO2 and C–O groups shifting were detected at 1410, 1316 and 1270 cm−1, respectively. Furthermore, new bonds were recorded at 1720 cm−1 which referred to C
O represented by peaks at 3394, 2961 and 1695 cm−1 with stretching mode, respectively. Also, shifting of two N–H groups were detected at 1631 and 1540 cm−1. Likewise, O–H, –NO2 and C–O groups shifting were detected at 1410, 1316 and 1270 cm−1, respectively. Furthermore, new bonds were recorded at 1720 cm−1 which referred to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups with stretching mode. Additionally, the two functional groups, N
O groups with stretching mode. Additionally, the two functional groups, N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N
N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and O–H, represented previously by peaks at 2133 and 1436 cm−1 Disappeared. The occurred changes in the FTIR spectra peaks approve the successfulness of the formation of LSDV-CQDs-MIPs complex and the involvement of multiple functional groups indicates the presence of strong efficient binding of between the synthesized CQDs-MIPs and LSDV.
N and O–H, represented previously by peaks at 2133 and 1436 cm−1 Disappeared. The occurred changes in the FTIR spectra peaks approve the successfulness of the formation of LSDV-CQDs-MIPs complex and the involvement of multiple functional groups indicates the presence of strong efficient binding of between the synthesized CQDs-MIPs and LSDV.
| Material | Absorption (cm−1) | Chemical bond | Vibration mode | Functional group |
|---|---|---|---|---|
| CQDs-MIPs before LSDV binding | 3414 | N–H | Stretching | Amine |
| 2915 | O–H | Stretching | Carboxylic acid | |
| 2133 | N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N N |
Stretching | Azide | |
| 1690 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
Stretching | Primary amide | |
| 1630 | N–H | Bending | Primary amine | |
| 1542 | N–H | Bending | Secondary amine | |
| 1436 | O–H | Bending | Carboxylic acid | |
| 1409 | O–H | Bending | Carboxylic acid | |
| 1315 | –NO2 | Stretching | Aliphatic nitro | |
| 1272 | C–O | Stretching | Aromatic ester | |
| 1023 | C–O | Stretching | Alcohol | |
| 954 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C C |
Bending | Alkene | |
| 707 | C–CL | Bending | Halo compound | |
| CQDs-MIPs after LSDV rebinding | 3394 | N–H | Stretching | Amine |
| 2961 | O–H | Stretching | Carboxylic acid | |
| 1720 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
Stretching | Carboxylic acid | |
| 1695 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
Stretching | Primary amide | |
| 1631 | N–H | Bending | Primary amine | |
| 1540 | N–H | Bending | Secondary amine | |
| 1410 | O–H | Bending | Carboxylic acid | |
| 1316 | –NO2 | Stretching | Carboxylic acid | |
| 1270 | C–O | Stretching | Aliphatic nitro | |
| 1023 | C–O | Stretching | Aromatic ester | |
| 954 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C C |
Bending | Alcohol | |
| 707 | C–CL | Bending | Alkene |
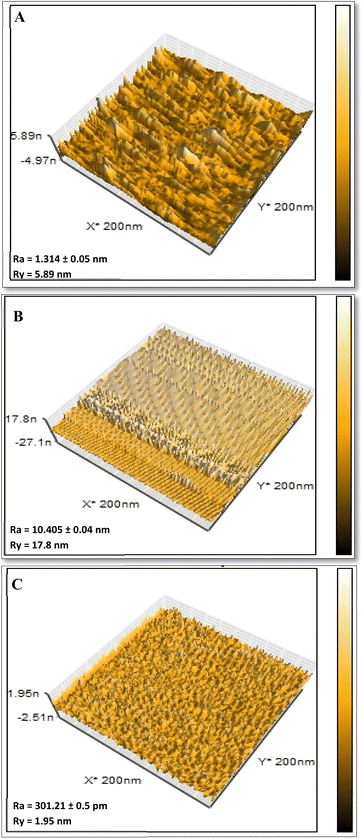
Using AFM, the topographical properties of CQDs-NIPs, CQDs-MIPs, and CQDs-MIPs-LSDV complex were observed and documented. These features included structure, size of pores, and degree of roughness and arrangement of imprinted sites on the imprinted polymeric layer. AFM (3D) images were recorded for the imprinted bare CQDs-MIPs following polymerization (Fig. 3A), CQDs-MIPs-LSDV complex after template removal (Fig. 3B) and CQDs-NIPs (Fig. 3C). Significantly, imprinted bare CQDs-MIPs layers demonstrated homogenous distribution across the surface, exhibiting height of 5.89 nm and a roughness of 1.314 ± 0.05 nm (Fig. 3A) with N grade of (01). The rebind of LSDV to its specific empty sites on the bare MIP resulted in an increase in both; surface height from 5.89 to 17.8 nm and roughness from 1.314 ± 0.05 to 10.405 ± 0.04 nm with N grade of (0) demonstrating successful rebinding (Fig. 3B). Whereas, CQDs-NIPs exhibited a smooth surface with 1.95 nm height and 301.21 ± 0.5 pm roughness (Fig. 3C). FE-SEM images demonstrate the topographical morphology of the CQDs-MIPs empty cavities after template removal versus LSDV filled cavities and CQDs-NIPs (Fig. 4A–C). CQDs-NIPs thin film arranged uniformly on the surface of NCM (Fig. 4A1 and A2) with a range size of 500.8–650.7 nm. CQDs-MIPs directly after washing showed well distributed holes on the MIPs film with the range size of 850 nm–2.45 μm (Fig. 4B1 and B2) which may indicate the agglomeration of virus clusters in some imprinting sites. In CQDs-MIPs-LSDV complex images, the holes appeared to be uniformly sealed with viral particles, particularly in binding sites, following the rebinding of LSDV template (Fig. 4C1 and C2). AFM and FE-SEM images showed compatible results approving the successfulness of the imprinting process.
In this study, APTES-CQDs was copolymerized with other functional monomers as acrylamide, methacrylic acid, methyl methacrylate, N-vinyl pyrrolidone in the presence of bisacrylamide as a cross linker to imprint LSDV template forming recognition cavities inside the polymer structure. The results of ATR-FTIR, AFM, and FE-SEM were in accordance proving the successfulness of the imprinting process on the NCM surface.
3.3 LSDV-CQDs-MIP sensor validation
The change in fluorescence intensity in response to the addition of various LSDV concentrations (101–106 log10 TCID50 per ml) was evaluated in order to examine the sensitivity and recognition ability of the LSDV-CQDs-MIP sensor. Comparing its result to the negative control, a considerable increase in the fluorescence intensity was detected (Fig. 5A). Furthermore, the results showed direct correlation between measured fluorescence signal and LSDV concentration. When operating under optimum conditions, LSDV-CQDs-MIP sensor's detection limit was found to be 1 log10 copies per ml, which is comparable to the CT value of 37 of qPCR findings. In this study, we successfully performed a novel, rapid, accurate, sensitive, and specific tool for LSDV compatible with PCR findings and can be applied to clinical samples at a large scale in a short time, which will be helpful in the diagnosis of clinical samples in case of outbreaks. Within 15 minutes, our assay could identify LSDV with high fluorescence intensity. The presence of sufficient empty cavities for LSDV rebind in a specific manner results in its rapid detection. Moreover, our sensor demonstrated the same sensitivity in detecting LSDV in complex sample matrixes, spiked or real (Fig. 5C). Fluorescence intensities just before (F0) and after (F) addition of LSDV template are shown in (Fig. 5D). Additionally, the fluorescence intensity changes in response to the presence of SPV, FMDV, and BVDV at a concentration of 105 log10 TCID50 per ml was used to assess the specificity of the LSDV-CQDs-MIP sensor (Fig. 5B). Following the addition of samples, a significant reduction in the recorded fluorescence intensity was observed indicating a turn-off activity. This behaviour might be the consequence of these viruses' incompetence to fit as precisely into the empty imprinted cavities as LSDV due to the lack of the essential functional groups required to bind to the imprinted surface selective recognition site resulted in weaker or no attachment. Hence, it can distinguish between members of the Capripoxviridae family that attain identical morphological characteristics as well as other possible viral contaminants could be present within the sample. These findings support the developed sensor's eligibility for application as an effective tool for LSDV detection in a quantitative manner. It is the first sensor incorporating fluorescent CQDs and MIPs loaded on NCM, and there are few studies that design rapid diagnostic tools for LSDV. Although we mentioned all recent studies designed rapid diagnostic tools for LSDV as a Real-Time Recombinase Polymerase Amplification Assay55 (RPA), a colorimetric sandwich-type lateral flow immunoassay (LFIA),56 gold nanoparticles-lateral flow test57 and Loop-mediated isothermal amplification (LAMP).58 The advantages and disadvantage of these assays are in details in Table 3. Sensors and biosensors based on CQDs can work with variable mechanisms such as fluorescence static and dynamic quenching, fluorescence-resonance energy transfer (FRET), inner-filter effect (IFE), and photo-induced electron transfer (PET).39 In FRET, the interaction between two fluorescent components results in the transfer of energy from a greater energy donor to a lower energy acceptor. Typically, the distance between the two fluorescent components should be around 10 nm.59 Our proposed LSDV-CQDs-MIPs sensor depends on the FRET mechanism. In a previous study,38 it was noticed that LSDV has a fluorescence intensity at Excitation/Emission wavelength (360/40–460/40). Since both LSDV and CQDs have fluorescence intensity, when they come in close contact due to the fitting of LSDV in the specific imprinted groove of CQDs labeled MIPs, their emission spectra overlapped. Hence, the high fluorescent signal. When the other viruses could not fit into the groove, a low fluorescent signal was detected. This proves the ability of the sensor to specifically detect LSDV. Our assay provides high specificity and sensitivity with LOD of 1 log10 copies per ml. Moreover, it is cost effective due to low cost of required reagents and easy fabrication. Also, samples don't require extraction or extensive pre-processing and results can be detected in a short time within 15 min. Besides, it can be easily used to detect a large number of samples simultaneously. All these, gives our developed test the age over the other rapid diagnostic test. Further studies should be conducted to determine the other factors that could affect the accuracy of the proposed sensor.| Assay | LOD | Advantages | Disadvantages | Reference |
|---|---|---|---|---|
| Real-time recombinase polymerase amplification (RPA) | 15 copies per μL | High sensitivity | Specific primer design | 55 |
| High specificity | High price of reagents | |||
| Short detection time | Require specific instruments | |||
| A colorimetric sandwich-type lateral flow immunoassay (LFIA) | 103.4 TCID50 per mL | High inter-and intra-assay repeatability | Qualitative nature not quantitative | 56 |
| Specificity | Critical antibody preparation step | |||
| Time consuming in case of large scale of samples | ||||
| Gold nanoparticles-lateral flow test | — | Accurate | Low sensitivity | 57 |
| Rapid | Monoclonal and polyclonal antibody preparation | |||
| Loop-mediated isothermal amplification (LAMP) | 8 copies per μL | Simple | Complex primer design | 58 |
| Specific | Require expensive kits | |||
| Accuracy similar to PCR |
4. Conclusion
LSDV-CQDs-MIP is a novel cost-effective paper-based turn-on fluorescence sensor designed for accurate detection of LSDV within few minutes. Our sensor exhibited high specificity and sensitivity for LSDV, with a lower low of detection reaching 101 log10 TCID50 per ml with high accuracy and reproducibility. Moreover, it detected LSDV in various real blood, skin nodules, and serum samples successfully, so it can be applied to clinical samples on a large scale and considered a local product. The findings of this study highlighted the approach for developing a novel, facile, and low-cost sensor that can be fabricated for efficient detection of other microorganisms.Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.Data availability
All the data and materials are accessible on request.Conflicts of interest
Authors declared no conflict of interest.Acknowledgements
The experimental section was supported by the Science, Technology & Innovation Funding Authority (STDF), Innovation Grant-cycle 8, Egypt (Grant number: IG-43570).References
- E. Orlova, A. Shcherbakov, V. Diev and V. Zakharov, Differentiation of capripox virus species and strains by polymerase chain reaction, J. Appl. Mol. Cell Biol., 2006, 40(1), 139–145 CAS.
- B. Moss, Poxviridae: the viruses and their replication, Fundamental virology, 4th edn, 2001, pp. 1249–1283 Search PubMed.
- E. R. Tulman, C. L. Afonso and Z. Lu, et al, The genome of sheep pox and goat pox viruses, J. Virol., 2002, 76(12), 6054–6061 CrossRef CAS PubMed.
- T. C. Oguzoglu, F. Alkan, A. Ozkul, S. Atalay- Vural, A. B. Gungor and I. Burgu, A sheep pox virus outbreak in central turkey in 2003: Isolation and Identification of capripox virus, Vet. Res. Commun., 2006, 30, 965–971 CrossRef CAS PubMed.
- M. R. Benzigar, R. Bhattacharjee, M. Baharfar and G. Liu, Current methods for diagnosis of human coronaviruses: pros and cons, Anal. Bioanal. Chem., 2021, 413, 2311–2330 CrossRef CAS PubMed.
- J. Vidic, M. Manzano, C. M. Chang and N. Jaffrezic-Renault, Advanced biosensors for detection of pathogens related to livestock and poultry, Vet. Res., 2017, 48, 1–22 CrossRef PubMed.
- Z. Tang, X. Zhang, Y. Shu, M. Guo, H. Zhang and W. Tao, Insights from nanotechnology in COVID-19 treatment, Nano Today, 2021, 36, 101019 CrossRef CAS PubMed.
- F. Yuan, S. Li, Z. Fan, X. Meng, L. Fan and S. Yang, Shining carbon dots: synthesis and biomedical and optoelectronic applications, Nano Today, 2016, 11(5), 565–586 CrossRef CAS.
- M. J. Molaei, Carbon quantum dots and their biomedical and therapeutic applications: a review, RSC Adv., 2019, 9(12), 6460–6481 RSC.
- B. Rezaei, H. Lotfi-Forushani and A. A. Ensafi, Modified Au nanoparticles-imprinted sol–gel, multiwall carbon nanotubes pencil graphite electrode used as a sensor for ranitidine determination, Mater. Sci. Eng., C, 2014, 37, 113–119 CrossRef CAS PubMed.
- Z. Peng, X. Han, S. Li, A. O. Al-Youbi, A. S. Bashammakh, M. S. El-Shahawi and R. M. Leblanc, Carbon dots: Biomacromolecule interaction, bioimaging and nanomedicine, Coord. Chem. Rev., 2017, 343, 256–277 CrossRef CAS.
- X. Xu, G. Xu, F. Wei, Y. Cen, M. Shi, X. Cheng and Q. Hu, Carbon dots coated with molecularly imprinted polymers: A facile bio probe for fluorescent determination of caffeic acid, J. Colloid Interface Sci., 2018, 529, 568–574 CrossRef CAS PubMed.
- Y. Du and S. Guo, Chemically doped fluorescent carbon and graphene quantum dots for bioimaging, sensor, catalytic and photoelectronic applications, Nanoscale, 2016, 8, 2532 RSC.
- J. Zhang and S. H. Yu, Carbon dots: large-scale synthesis, sensing and bioimaging, Mater. Today, 2016, 19(7), 382–393 CrossRef CAS.
- Y. Guo and W. Zhao, In situ formed nanomaterials for colorimetric and fluorescent sensing, Coord. Chem. Rev., 2019, 387, 249–261 CrossRef CAS.
- Q. M. Zhong, X. H. Huang, Q. M. Qin, A. M. SU, Y. Y. Chen, X. Y. Liu and Y. L. Wang, Determination of glucose based on carbon quantum dots as peroxidase mimetic enzyme. CHINESE, J. Anal. Chem., 2018, 1062–1068 CAS.
- H. C. Zhang and Y.-M. GUO, Advances of carbon quantum dots for fluorescence turn-on detection of reductive small biomolecules. CHINESE, J. Anal. Chem., 2021, 49(1), 14–23 Search PubMed.
- Y. Guo and W. Zhao, Hydrothermal synthesis of highly fluorescent nitrogen-doped carbon quantum dots with good biocompatibility and the application for sensing ellagic acid, Spectrochim. Acta, Part A, 2020, 240, 118580 CrossRef CAS PubMed.
- F. Yuan, S. Li, Z. Fan, X. Meng, L. Fan and S. Yang, Shining carbon dots: synthesis and biomedical and optoelectronic applications, Nano Today, 2016, 11(5), 565–586 CrossRef CAS.
- R. Wang, K. Q. Lu, Z. R. Tang and Y. J. Xu, Recent progress in carbon quantum dots: synthesis, properties and applications in photocatalysis, J. Mater. Chem. A, 2017, 5(8), 3717–3734 RSC.
- S. S. Liang, L. Qi, R. L. Zhang, M. Jin and Z. Q. Zhang, Ratiometric fluorescence biosensor based on CdTe quantum and carbon dots for double strand DNA detection, Sens. Actuators, B, 2017, 244, 585–590 CrossRef CAS.
- O. J. Achadu, K. Takemura, I. M. Khoris and E. Y. Park, Plasmonic/magnetic molybdenum trioxide and graphitic carbon nitride quantum dotsbased fluoro immune sensing system for influenza virus, Sens. Actuators, B, 2020, 321, 128494 CrossRef CAS PubMed.
- A. Kurdekar, L. Chunduri, E. P. Bulagonda, M. K. Haleyurgirisetty, V. Kamisetti and I. K. Hewlett, Comparative performance evaluation of carbon dot-based paper immunoassay on Whatman filter paper and nitrocellulose paper in the detection of HIV infection, Microfluid. Nanofluid., 2016, 20, 1 CrossRef CAS.
- B. Demir, M. Lemberger, P. X. Panagiotopoulou, P. X. Medina Rangel, S. Timur and T. Hirsch, etal, Tracking Hyaluronan: Molecularly Imprinted Polymer Coated Carbon Dots for Cancer Cell Targeting and Imaging, ACS Appl. Mater. Interfaces, 2018, 10, 3305–3313 CrossRef CAS PubMed.
- F. Wei, G. Xu, Y. Wu, X. Wang, J. Yang, L. Liu, P. Zhou and Q. Hu, Molecularly imprinted polymers on dual-color quantum dots for simultaneous detection of norepinephrine and epinephrine, Sens. Actuators, B, 2016, 229, 38–46 CrossRef CAS.
- M. S. Amorim, M. G. F. Sales and M. F. Frasco, Recent advances in virus imprinted polymers, Biosens. Bioelectron., 2022, 10, 100131 CAS.
- A. A. Malik, C. Nantasenamat and T. Piacham, Molecularly imprinted polymer for human viral pathogen detection, Mater. Sci. Eng., C, 2017, 77, 1341–1348 CrossRef CAS PubMed.
- L. Uzun and A. P. F. Turner, Molecularly-imprinted polymer sensors: realising their potential, Biosens. Bioelectron., 2016, 76, 131–144 CrossRef CAS PubMed.
- Y. Saylan, F. Yilmaz, E. Ozgur, A. Derazshamshir, H. Yavuz and A. Denizli, Molecular Imprinting of Macromolecules for Sensor Applications, Sensors, 2017, 17, 898 CrossRef PubMed.
- S. Suryana, Y. Mutakin Rosandi and A. N. Hasanah, An update on molecularly imprinted polymer design through a computational approach to produce molecular recognition material with enhanced analytical performance, Molecules, 2021, 26, 1891 CrossRef CAS PubMed.
- C. Liu, L. Shang, H. T. Yoshioka, B. Chen and K. Hayashi, Preparation of molecularly imprinted polymer nanobeads for selective sensing of carboxylic acid vapors, Anal. Chim. Acta, 2018, 1010, 1–10 CrossRef CAS PubMed.
- Y. Liu, N. Cao, W. Gui and Q. Ma, Nitrogen-doped graphene quantum dots-based fluorescence molecularly imprinted sensor for thiacloprid detection, Talanta, 2018, 183, 339–344 CrossRef CAS PubMed.
- M. M. Samarin, F. Faridbod, A. Shiralizadeh Dezfuli and M. R. Ganjali, A novel metronidazole fluorescent nanosensor based on graphene quantum dots embedded silica molecularly imprinted polymer, Biosens. Bioelectron., 2017, 92, 618–623 CrossRef PubMed.
- H. Liu, G. Fang and S. Wang, Molecularly imprinted optosensing material based on hydrophobic CdSe quantum dots via a reverse microemulsion for specific recognition of ractopamine, Biosens. Bioelectron., 2014, 55, 127–132 CrossRef CAS PubMed.
- A. A. Ensafi, N. Kazemifard and B. Rezaei, Development of a selective prilocaine optical sensor based on molecularly imprinted shell on CdTe quantum dots, Sens. Actuators, B, 2017, 242, 835–884 CrossRef CAS.
- B. Rezaei, H. Lotfi-Forushani and A. A. Ensafi, Modified Au nanoparticles-imprinted sol–gel, multiwall carbon nanotubes pencil graphite electrode used as a sensor for ranitidine determination, Mater. Sci. Eng., C, 2014, 37, 113–119 CrossRef CAS PubMed.
- S. Kassem, M. E. Hamdy, K. M. Selim, D. M. A. Elmasry, M. A. Shahein and D. M. El-Husseini, Development of Paper-Based Fluorescent Molecularly Imprinted Polymer Sensor for Rapid Detection of Lumpy Skin Disease Virus, Molecules, 2024, 29(7), 1676 CrossRef CAS PubMed.
- M. J. Molaei, Principles, mechanisms, and application of carbon quantum dots in sensors: a review, Anal. Methods, 2020, 12(10), 1266–1287 RSC.
- M. P. Ajith, S. Pardhiya and P. Rajamani, Carbon dots: an excellent fluorescent probe for contaminant sensing and remediation, Small, 2022, 18(15), 2105579 CrossRef PubMed.
- Y. Xue, C. Liu, G. Andrews, J. Wang and Y. Ge, Recent advances in carbon quantum dots for virus detection, as well as inhibition and treatment of viral infection, Nano Convergence, 2022, 9(1), 15 CrossRef CAS PubMed.
- Y. Guo, Y. Chen, F. Cao, L. Wang, Z. Wang and Y. Leng, Hydrothermal synthesis of nitrogen and boron doped carbon quantum dots with yellow-green emission for sensing Cr (VI), anti-counterfeiting and cell imaging, RSC Adv., 2017, 7(76), 48386–48393 RSC.
- M. Li, C. Yu, C. Hu, W. Yang, C. Zhao, S. Wang and J. Qiu, Solvothermal conversion of coal into nitrogen doped carbon dots with singlet oxygen generation and high quantum yield, Chem. Eng. J., 2017, 320, 570–575 CrossRef CAS.
- Y. Choi, N. Thongsai, A. Chae, S. Jo, E. B. Kang, P. Paoprasert and I. In, Microwave-assisted synthesis of luminescent and biocompatible lysine-based carbon quantum dots, J. Ind. Eng. Chem., 2017, 47, 329–335 CrossRef CAS.
- H. Li, X. He, Y. Liu, H. Huang, S. Lian, S. T. Lee and Z. Kang, One-step ultrasonic synthesis of water-soluble carbon nanoparticles with excellent photoluminescent properties, Carbon, 2011, 49(2), 605–609 CrossRef CAS.
- A. Rahy, C. Zhou, J. Zheng, S. Y. Park, M. J. Kim, I. Jang and D. J. Yang, Photoluminescent carbon nanoparticles produced by confined combustion of aromatic compounds, Carbon, 2012, 50(3), 1298–1302 CrossRef CAS.
- S. Chahal, N. Yousefi and N. Tufenkji, Green synthesis of high quantum yield carbon dots from phenylalanine and citric acid: Role of stoichiometry and nitrogen doping, ACS Sustain. Chem. Eng., 2020, 8, 5566–5575 CrossRef CAS.
- N. Tejwan, S. K. Saha and J. Das, Multifaceted applications of green carbon dots synthesized from renewable sources, Adv. Colloid Interface Sci., 2020, 275, 102046 CrossRef CAS PubMed.
- B. Wang, J. Yu, L. Sui, S. Zhu, Z. Tang, B. Yang and S. Lu, Rational design of multi-color-emissive carbon dots in a single reaction system by hydrothermal, Adv. Sci., 2021, 8, 2001453 CrossRef CAS PubMed.
- N. Papaioannou, M. M. Titirici and A. Sapelkin, Investigating the effect of reaction time on carbon dot formation, structure, and optical properties, ACS Omega, 2019, 4, 21658 CrossRef CAS PubMed.
- D. Qu, M. Zheng, L. Zhang, H. Zhao, Z. Xie, X. Jing, R. E. Haddad, H. Fan and Z. Sun, Formation mechanism and optimization of highly luminescent N-doped graphene quantum dots, Sci. Rep., 2014, 4, 1 Search PubMed.
- P. Zhao and L. Zhu, Dispersibility of carbon dots in aqueous and/or organic solvents, Chem. Commun., 2018, 54, 5401–5406 RSC.
- L. Wu, Z. Z. Lin, H. P. Zhong, X. M. Chen and Z. Y. Huang, Rapid determination of malachite green in water and fish using a fluorescent probe based on CdTe quantum dots coated with molecularly imprinted polymer, Sens. Actuators, B, 2017, 239, 69–75 CrossRef CAS.
- A. A. Ensafi, P. Nasr-Esfahani and B. Rezaei, Synthesis of molecularly imprinted polymer on carbon quantum dots as an optical sensor for selective fluorescent determination of promethazine hydrochloride, Sens. Actuators, B, 2018, 257, 889–896 CrossRef CAS.
- C. E. Probst, P. Zrazhevskiy, V. Bagalkot and X. Gao, Quantum dots as a platform for nanoparticle drug delivery vehicle design, Adv. Drug Delivery Rev., 2013, 65(5), 703–718 CrossRef CAS PubMed.
- Q. Zhai, X. Zhou, L. Du, N. Yang, Y. Lou, J. Liu and S. Zhai, A Real-Time Recombinase Polymerase Amplification Assay for Specific Detection of Lumpy Skin Disease Virus, Vet. Sci., 2023, 10(10), 625 Search PubMed.
- S. Cavalera, G. Pezzoni, S. Grazioli, E. Brocchi, S. Baselli, D. Lelli and L. Anfossi, Investigation of the “Antigen hook effect” in lateral flow sandwich immunoassay: The case of lumpy skin disease virus detection, Biosensors, 2022, 12(9), 739 CrossRef CAS PubMed.
- A. M. Abdalhamed, S. M. Naser, A. H. Mohamed and G. S. G. Zeedan, Development of gold nanoparticles-lateral flow test as a novel field diagnostic assay for detecting foot-and-mouth disease and lumpy skin disease viruses. Iran, J. Microbiol., 2022, 14(4), 574 Search PubMed.
- J. J. Mwanandota, M. Macharia, C. M. Ngeleja, R. S. Sallu, M. G. Yongolo, C. Mayenga and T. A. Holton, Validation of a diagnostic tool for the diagnosis of lumpy skin disease, Vet. Dermatol., 2018, 29(6), 532–536 CrossRef PubMed.
- G. B. Kim and Y. P. Kim, Analysis of protease activity using quantum dots and resonance energy transfer, Theranostics, 2012, 2(2), 127 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |