Open Access Article
Open Access ArticleDetermination of the uptake of lanthanide doped-carbon dots by human cells using single cell ICP-ToF-MS†
Guillermo
Redondo-Fernandez
 ab,
Kharmen
Billimoria
ab,
Kharmen
Billimoria
 b,
Simon
Cowen
b,
David
Ojeda
b,
Simon
Cowen
b,
David
Ojeda
 b,
Dorota
Bartczak
b,
Dorota
Bartczak
 b,
Ana
Soldado
b,
Ana
Soldado
 a,
Jose M.
Costa-Fernandez
a,
Jose M.
Costa-Fernandez
 a and
Heidi
Goenaga-Infante
a and
Heidi
Goenaga-Infante
 *b
*b
aDepartment of Physical and Analytical Chemistry, University of Oviedo, Avda. Julián Claveria 8, 33006, Oviedo, Spain
bNational Measurement Laboratory, LGC Limited, Queens Road, Teddington, TW11 0LY, UK. E-mail: Heidi.Goenaga-Infante@LGCGroup.com
First published on 28th April 2025
Abstract
As the application of engineered nanoparticles in medical fields grows, there is a rising demand for techniques capable of determining how they interact with biological entities, including cells. Single Cell Inductively Coupled Plasma Time-of-Flight Mass Spectrometry (sc-ICP-ToF-MS) has been shown potential to assess the mass of nanoparticle tags uptaken by individual cells. However, this approach suffers from challenges related to inconsistencies in transport efficiency (TE) determination and data processing protocols, which greatly impact the quality of the data. To address these issues, a novel metrological approach for the identification of cell events and determination of their TE is presented here for the first time. It is based on using the ratio of Eu to Yb signals in a single event to distinguish cellular events from background noise when analyzing HeLa cells tagged with lanthanide-doped carbon dots (Ln-CDs). To achieve this, Ln-CDs with Eu (4.3%) and Yb (3.4%) were synthesized, characterized and cytotoxicity assays were performed to confirm their biocompatibility. HeLa cells were exposed to these multielement Ln-CDs at varying concentrations to evaluate their cellular uptake. Laser ablation (LA)-ICP-MS analysis of individual isolated cells (n = 1578) confirmed cell tagging efficiency of 99.87%. Using this approach and CytoNeb–CytoSpray interface, TE values between 40–50% were achieved. The results showed a dosage dependent uptake of Ln-CDs by cells, with final concentrations ranging from 6.8 to 5115 fg Ln-CDs per cell.
Introduction
During the last two decades, advances in the development of novel nanomaterials (NM) for use in clinical fields has increased exponentially. Among the different type of existing NM, Carbon Dots (CDs) were discovered in 2004,1 and researchers have been exploiting their unique and tuneable properties for imaging, sensing, and even theragnostic applications.2,3 Depending on their chemical composition or size, CDs can present different properties and characteristics which make them ideal tags for bioanalytical research.4,5 Some highlighted properties of this NM are high biocompatibility, stability, low-cost synthetic procedures, highly luminescent emission and easy functionalization. Moreover, the introduction of heteroatoms, a strategy known as doping, has increased the potential use of CDs as this introduces multifunctionality to the NM.6,7 Metal doping with elements such as Gd or Yb potentially provide an alternative means for producing robust contrast agents for fluorescence/MRI/CT bioimaging.8,9 Whilst there is increasing research interest in this type of multifunctional NM, their potential use in biomedical applications require further biocompatibility testing and would greatly benefit from cell studies to improve understanding of their interactions with biological samples.In this regard, single cell analysis is crucial as individual cells, even those from the same population and type behave completely different.10 This cellular heterogeneity has led to demand for new approaches to enable the study and characterization of biological processes or mechanisms at a single cell level, promoting the development of single-cell analysis methodologies.11,12
Amongst all developed methodologies, single-cell (sc) ICP-MS has been demonstrated to provide quantitative data for the largest range of elements.13,14 sc-ICP-MS follows the same principle as single-particle (sp) ICP-MS and was first used in 2005.15 For sc-ICP-MS analysis, a sufficiently dilute suspension of cells is introduced such that a cell is nebulized and then, the ions generated in the plasma from a single cell are individually detected by utilising high time resolution.
In this sense, sc-ICP-ToF-MS allows fast, multiple, and simultaneous analysis of different elements and isotopes within the same acquisition.16,17 Whilst the principle of single-cell can be elucidated from single particle methodologies, aspects such as the transport efficiency (TE) determination, data treatment and accurate element/tag quantification are remaining challenges. Along the last years, several sc-ICP-MS approaches have been applied to obtain elemental information or to study the uptake of NM in cells.18–20 Although there is an improvement in sample introduction systems that exploit the capabilities of direct cell infusion21 and/or total consumption spray chambers,12 there is still a lack of consensus on approaches used to determine TE and define thresholds, which ultimately impact quality and comparability of data quality of the data. Despite having similar principles to spICP-MS, it is important to note that cells do not behave like NM, their bigger sizes (micron scale) and their complex composition make their density different to engineered NM (average cell density is of approximately 1.1 g cm−3).22
Moreover, both spICP-MS and sc-ICP-MS acquisition modes generate large data sets, making manual analysis almost impossible. To address that issue, commercial software packages which are often integrated in the main instrument software, enable processing and visualization of sp and sc data. Nonetheless, it is not completely clear how these algorithms process the data, limiting the capabilities of users to modify critical parameters. As a result, some researchers have developed different data processing approaches and platforms to analyze sp and sc data efficiently.23–25 These novel platforms allow the control of individual processing parameters by the user and moreover, they have been tested along with the commercial software packages. Again, it is worth stressing that sc data can often differ from typical sp analysis due to the dynamic background arising from the complex cell matrix.
In this work, we present a novel multi-modal metrological approach for the quantification of lanthanide tags (Eu and Yb) present in carbon nanoparticles uptaken by HeLa cells using sc-ICP-ToF-MS. It is based on utilizing the ratio between Eu and Yb signals detected in a single event to identify true cell events from the background and determine their TE. To achieve this, nitrogen and lanthanide-doped carbon dots (Ln-CDs) containing Eu (4.3%) and Yb (3.4%) were synthesized, characterized and cytotoxicity assays were used to confirm their biocompatibility. HeLa cells were exposed to these multielement CDs at varying concentrations to find an optimal concentration at which sufficient signal could be detected and discriminated by sc-ICP-ToF-MS and for which the majority of the cells were tagged. Laser ablation (LA)-ICP-MS analysis of individual cells isolated and deposited on a substrate was used to determine cell tagging efficiency. An in-house data processing approach was developed for an all-in-one R-based platform for threshold application and event filtering, quantification, and plotting of large data sets.
Materials and methods
Chemicals
EuCl3·6H2O (99.9% trace metals basis, Sigma-Aldrich), YbCl3·6H2O (99.9% trace metals basis, Sigma-Aldrich), citric acid monohydrate (99.5–100.5%, Sigma-Aldrich) and ammonium citrate dibasic (≥98%, Sigma-Aldrich) were employed as precursors for the synthesis of Ln-CDs. Pur-A-Lyzer cassettes (Sigma-Aldrich) with 1 kDa pore size and 0.22 μm PVDF membrane filters were employed for Ln-CDs purification.HeLa and NIH/3T3 cells (ATCC) were cultured in Corning® cell culture flasks and plates. Cells grew in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% (v/v) inactivated fetal bovine serum (Gibco, Thermo-Fischer Scientific). Phosphate buffer saline (PBS) 10 mM pH 7.4 (Gibco, Thermo-Fischer Scientific) was employed as working buffer. Cells were detached from the flasks and plates using Accutase (Gibco, Thermo-Fischer Scientific) and fixated with paraformaldehyde 4% (PFA; Agar Scientific Ltd). Cell counting kit-8 (CCK-8; Sigma-Aldrich) was employed to carry out cell proliferation studies. ICP calibrations were performed with ICP standards (1000 μg mL−1; CPI International, Sigma-Aldrich and ROMIL Ltd) in 1–2% ultrapure HNO3 (ROMIL Ltd). Ultrapure water (18 MΩ, 25 °C) from and Elga water purification unit (Elga) was employed throughout.
Synthesis and characterization of Ln-CDs
Ln-CDs doped with N, Eu, and Yb were synthesized via one-step hydrothermal treatment of citric acid and ammonium citrate, used as carbon and nitrogen precursors respectively, in presence of EuCl3 and YbCl3, used as lanthanide doping agents. 1.2206 g of citric acid, 1.0044 g of ammonium citrate, 0.1671 g of EuCl3·6H2O, and 0.1686 g YbCl3·6H2O were dissolved in 10 mL of deionized water, transferred into a 50 mL Teflon-lined stainless steel autoclave reactor and heated at 170 °C for 8 h. After cooling down to room temperature, a brown solution was observed. The obtained solution was purified by dialysis against Milli-Q water using a 1 kDa cutoff dialysis cassette for 24 h. Finally, the resultant solution was filtered through a 0.22 μm PVDF membrane filter to remove potential bigger particles remaining in the solution. After purification steps, the light brown aqueous solution was stored at room temperature and dark conditions prior to its use. To know the Ln-CDs concentration, a lyophilization of the purified solution was carried out.In order to characterize the prepared Ln-CDs, several techniques were used. Scanning transmission electron microscopy (STEM) imaging was carried out in a probe-corrected Titan (Thermofisher, formerly FEI) operated at 300 kV and equipped with a high brightness X-FEG and a spherical aberration Cs-corrector (CEOS) for the condenser system to provide sub-angstrom probe size. To analyze the chemical composition of the materials, X-ray Energy Dispersive Spectra (EDS) were obtained with an Ultim Max detector (Oxford Instruments). Images were further processed using ImageJ. X-ray photoelectron spectroscopy (XPS) experiments were carried out in a SPECS spectrometer with an Al X-ray source (1486.74 eV), using for high resolution spectra 0.05 eV and 30 eV as energy step and energy pass, respectively and 1 eV and 90 eV for survey spectra. Infrared spectra were taken in a Varian 670 FTIR spectrometer using 4 cm−1 at 2000 cm−1 resolution and 32 scans per sample. Photoluminescence experiments were carried out using a Varian Cary Eclipse spectrophotometer, with both excitation and emission slits width of 5 nm. UV-vis measurements were performed in a Genesys 10S spectrophotometer. Quantum yield of the Ln-CDs (φS) was calculated by using the following equation (eqn (1)):
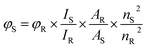 | (1) |
Cell culture procedures and Ln-CDs exposure
Cytotoxic activity of Ln-CDs was tested against a human tumor cell line (HeLa). Mouse embryonic fibroblast cell line NIH/3T3 was used as control to evaluate cytotoxicity against non-malignant cells. Firstly, 3000 cells were seeded per well, in 96-well plates using the CCK-8 and they were grown for 24 h in DMEM supplemented with 10% fetal bovine serum and an antibiotic antimycotic commercial solution (A5955, from Sigma-Aldrich). After such incubation at 37 °C under 5% CO2, 10 μL of Ln-CDs were added to each well by triplicate (assayed concentrations: 0, 4, 8, 16, 31, 63, 125, 250 and 500 mg kg−1). Cells were incubated for another 24 h a 37 °C under 5% CO2. Lastly, 10 μL of CCK-8 reagent were added, left incubating for 2 h at the same conditions, and finally measured at 450 nm using a Plate Reader Synergy LX (BioTek). Viability of treated cells was expressed as the percentage of the absorbance measured in untreated cells. The value for each sample is the mean of triplicate determinations (±SD).To carry out the cellular uptake assays, HeLa cells were seeded in 6-well plates and allowed to attach for 24 h in DMEM supplemented with 10% fetal bovine serum at 37 °C under 5% CO2. Cells were then incubated in presence of different concentrations of Ln-CDs (0, 6, 15, 30, 60 and 120 mg kg−1) for 24 h, at the same conditions. After that, media was removed, and each well was washed with sterile PBS 10 mM pH 7.4. Cells were detached from the well with accutase and fixated with PFA at 4% in PBS 10 mM pH 7.4 for 10 min. Fixed cells were kept in PBS 10 mM pH 7.4 at 4 °C until use.
Fixed cell number concentration measurements in cell suspensions were performed using a Vi-Cell XR (Beckman) instrument, prior to the single-cell experiments. Cell integrity was assessed by nebulizing a cell solution of 105 cells per mL at different working conditions and collecting the spray from the nebulizer. Obtained solutions were studied using an optical microscope (Leica DMi1).
Single-cell analysis and tagging efficiency
Fixed cell suspensions obtained from the Ln-CDs exposure were washed and resuspended in D. I. water to get a concentration number close to 105 cells per mL prior to their use. Single-cell analysis was performed by using an ICP-ToF-MS 2R (Tofwerk) with a sample introduction system consisting of a CytoNeb 50 nebulizer, a CytoSpray chamber, and a one-piece ICP-MS torch (Elemental Scientific, Inc.). Solutions were introduced at 15 μL min−1 using the main peristaltic pump of the instrument and measurements were performed using a dwell time of 3 ms. Instrumental parameters for data acquisition are listed in Table S1.† Cell tagging efficiency was evaluated by single-cell analysis by using a NWRImageBio (ESL) 266 nm laser with Two Volume 3 sample chamber and dual concentric injector coupled to the ICP-ToF-MS 2R (Tofwerk). Laser operating conditions were: 5 μm laser spot, 26% laser energy (∼3 J cm−2 fluence), 100 Hz rep rate, 100 μm s−1 scan speed. Cells were deposited onto a standard microscope slide for imaging studies using a single cell dispensing system (cellenONE®, Cellenion) which deposited individual cells in a 20 × 10 grid, allowing the ablation and acquisition of data from single cells. Transport efficiency was evaluated by using the lanthanide ratio of the tagged cells themselves. The detected cell events were compared with the number of cells pumped into the ICP, being the number of cells in suspension determined by coulter particle counting analysis. This was assisted by improved data processing as detailed in the following section.The determination of the mass of tag per cell was derived from a calibration curve of multi-elemental ionic standard solutions that were prepared daily within a concentration range from 0 to 200 mg kg−1.
Data processing
All data treatment from sc-ICP-ToF-MS analysis was carried out by applying an algorithm developed in R (R Statistical Software, v4.3.1; R Core Team 2021, R Foundation for Statistical Computing, http://www.r-project.org), a programming language for statistical computing and graphics, open-source, and free software environment. RStudio (RStudio Desktop v.2023.06.1+524; Posit PBC) is an integrated development environment for R that was used along this work. This designed algorithm uses the exported csv files obtained from the ToF instrument in which, at least, two elements that are present in the cells (in this case, coming from the Ln-CDs that are tagging each cell) are measured simultaneously in single-cell mode. Firstly, the data is uploaded to the algorithm, moving the high intensity events (peaks above 5 times the mean value of the element along the spectrum) into a temporary datasheet, to avoid their contribution in the next step. Then, the algorithm computes a mathematical treatment based on a running median window calculation to define the background of the selected element. A running median was chosen as it reflects local changes in the spectrum, whilst being less influenced by true events. To carry out this calculation, an optimized value of 101 points is taken to calculate each point of such baseline (k = 101; where k is the window size parameter). Once the baseline is defined, it applies a 3.29σ threshold above that baseline, to discriminate potential false events from the potential true and detectable events. This function will define the detection threshold (DT) for the measurement. A 3.29σ was chosen as in a further step the events above this first threshold will be evaluated again to check if they meet an elemental ratio condition (the second threshold), in order to be considered true events. After these steps, the temporarily removed events are returned to the main data set, and the script finds the events above the defined DT and filters the split-events. Then, it applies a second threshold in which another criterion has to be met, in this case, a lanthanide ratio. For this example, if a peak of 153Eu is found in the absence of 172Yb or vice versa, then it is considered as a false event or an artifact. Additionally, if there are two peaks (153Eu and 172Yb) but the relationship between their intensities does not meet the empirical cumulative distribution function range for the measurement (calculated for each acquisition), then it is not reliable enough to be considered a real event. Most of the rejected peaks or events do not meet the first condition as expected due to the natural absence of lanthanides in human cells. Finally, the script determines a value of transport efficiency for each measurement by comparing the number of peaks after the thresholding and filtering process to the expected number of cell events based on the cell number concentration, acquisition time and uptake flow. These calculations allow the later quantification of elements in each cell as their masses are quantified at single-cell level by using the eqn (2): | (2) |
 | (3) |
Results and discussion
Synthesis and characterization of Ln-CDs
Full details about the characterization of the Ln-CDs using different analytical techniques are shown and discussed in the ESI (Fig. S1–S5†). Their doping percentage was found to be of Eu (4.28%) and Yb (3.38%) by ICP-MS analysis.Cell exposure, integrity, and tagging efficiency
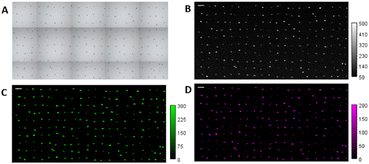
Potential cytotoxic effects of Ln-CDs were evaluated prior to their use in biological experiments. To achieve this, Ln-CDs cytotoxicity was determined using HeLa and NIH-3T3 cells. Both cell lines were incubated in absence of the NM (employed as control) and in presence of several Ln-CDs concentrations, ranged between 4 and 500 mg kg−1, for 24 h. Results revealed that at lower concentrations of Ln-CDs (<125 mg kg−1), no cytotoxicity was observed for either cell line, as it can be seen in Fig. S6.† At higher concentrations no cytotoxic effects were observed for the NIH/3T3 line. For HeLa cells a small decrease in cell viability was observed when the concentration of Ln-CDs exceeded 125 mg kg−1. These results reveal that there is a need for a comprehensive understanding of such NM-cell interactions to know the potential cytotoxic effects.Cellular tagging efficiency was evaluated by LA-ICP-ToF-MS. For this purpose, HeLa cells from a culture batch were incubated with 60 mg kg−1 of Ln-CDs, and fixed. After fixation these tagged cells were washed with D. I. water and arrayed using the cellenONE on a microscope slide, achieving enough spacing between them, to ablate individual cells and determine NM uptake. As can be seen in Fig. 1, the 23Na signal was used to identify the fixed cells, whilst the 153Eu and 172Yb signals are from the Ln-CDs tags. These elemental maps also overlap with the pre-ablation microscope image of the cell samples and as a result tagging efficiency was calculated as 99.87% (n = 1578 cells), confirming the quantitative cellular uptake of Ln-CDs. This result confirms the feasibility of using such multi-elemental NM as a tag for cells to determine the cellular TE, since a true cell event must show both lanthanides and meeting the elemental ratio criterium.
Finally, cell integrity during the sample introduction phase was evaluated using suspensions of fixed HeLa cells (105 cells per mL). The fixed cells were nebulized using different conditions to assess their integrity and stability in water before entering the plasma (see Fig. S7†). Pictures taken by optical microscopy showed that cell integrity was kept after nebulisation. The tested conditions allow whole cells to enter the plasma intact with the sample introduction system used throughout this work. Furthermore, aggregated cells were not found after their nebulization which, in accordance with the capillary size of the nebulizer, reduces the possibility of finding double events in single-cell measurements.
These results demonstrate the use of biocompatible and non-toxic metal-doped CDs as an alternative approach for cell labeling strategies in comparison with traditional labeling techniques, that often incorporate antibodies with metallic elements as tags (NM or chelates).26,27
Quantitative analysis of CDs uptake and data processing
For sc-ICP-ToF-MS three critical experimental parameters were highlighted as having the greatest impact on TE for the single cell sample introduction system used. They are: (1) nebulizer and make up gas flow rates, (2) cell number and (3) sample uptake rate. Therefore, in order to maximize cell TE all three parameters were optimized carefully. Nebulizer and additional gas flows were modified for maximum TE whilst also keeping the cells intact during introduction into the ICP-MS. Optimal conditions were 0.3 L min−1 nebulizer gas and 0.5 L min−1 additional gas (Tables S2 and S3†). An optimal transport efficiency value (of approximately 40%) was achieved at a sample uptake flow of 20 μL min−1 and a cell concentration of 105 cells per mL (see Fig. S8†). Sample uptake was later reduced from 20 to 15 μL min−1 to obtain higher transport efficiency values. The number of detected events could have potentially been increased further if lower uptake flows were used, however lower flows resulted in peristaltic pump instability resulting in pulsations and clogging of the nebulizer. sc-ICP-ToF-MS was used to quantify the uptake of Ln-CDs in HeLa cells and to evaluate its potential use as transport efficiency tool in single-cell studies. To do so, cells were incubated in presence of different concentrations of Ln-CDs (6, 15, 30, 60 and 120 mg kg−1) for 24 h and later fixed. Prior to their use, cells were washed and resuspended in D. I. water, and counted to know the number of cells in each solution before being measured by sc-ICP-ToF-MS. Also, control cells were incubated under the same conditions but in absence of Ln-CDs (see Fig. S9†). The resulting LOD was found to be 0.106 ± 0.003 fg per cell and 0.226 ± 0.004 fg per cell for 153Eu and 172Yb, respectively, with the LOD for the sum of both lanthanides 0.332 ± 0.007 fg per cell. The raw data obtained after acquisitions corresponding to the Ln-CDs exposed cells (see example, Fig. S10†) was treated by applying the R algorithm previously described. A threshold was calculated for each measurement to discriminate the cell events from the background and then, the split events were corrected. After checking the lanthanide relationships (a condition that has to be met, as every cell contains particles with these elements in the corresponding ratios) and removing the false events (see representative example in Fig. 2), true cell events were determined, and transport efficiency values were assigned to each measurement taking into account the corresponding sample flow, time, and cell number concentration values.In order to validate the algorithm's performance, the window size was carefully studied. In this sense, several values of k (window size) were evaluated as shown in Fig. S11.† For low k values, the background is strongly influenced by true events and as a result the obtained baseline would not fit the spectral background. As there are more values corresponding to the background noise than true events, the higher is the k value, the closer the calculation of such baseline will be, receiving a minimum influence from the background and true events. However, as the k value increases (1) the number of filtered events is almost constant, as the curve flattens and (2) the baseline values will be influenced by further points that do not correspond to the behaviour of the local background. For such reason, a k value of 101 was consider as the optimum window size. Experimental results showed that transport efficiency values were ranged between 40 and 50% (see Table S4†). This confirmed the reliability of the approach to be used for transport efficiency determination of the employed system. To calculate the amount of Ln-CDs inside each cell, 153Eu and 172Yb signals were transformed into masses of Eu and Yb using eqn (2). The lanthanide mass distribution per cell and the average cell diameter information for each cell batch is shown in Table 1 and Fig. 3, where it can be seen that there is a clear increase in the cellular uptake when cells are exposed to higher concentrations of Ln-CDs.
| [Ln-CDs] (mg kg−1) | Average cell diameter (μm) | Lanthanide mass per cell (fg per cell) |
|---|---|---|
| 6 | 17.93 | 0.52–197.96 |
| 15 | 17.71 | 0.70–304.42 |
| 30 | 17.52 | 1.28–248.22 |
| 60 | 19.22 | 2.09–296.13 |
| 120 | 18.51 | 6.14–391.82 |
However, Fig. 3 also shows that single cell distributions do not present a unique population around the median but rather the distributions have long tails after the main population, potentially due to cellular heterogeneity within the population. In addition, with the box plot related with mass distribution of lanthanides in cells (see Fig. 4), the following observations can be noticed: the median values of mass increases along with increased exposure to Ln-CDs, whereas, comparable when using the highest one.
Moreover, the dispersion above the median was larger than below it for all Ln-CDs concentrations showing that there are some cells that are able to uptake higher amounts of Ln-CDs even at low concentration. It is also worth noting that for all concentrations of CDs exposed to the cells a maximum mass of lanthanide within the cells was reached, suggesting that there is a e number of CDs the cells can uptake irrespective of the initial spiking amount, and that there are cells able to uptake a large number of particles even at low concentration levels. Such behaviour could be caused not only by differences in cell metabolism activities or interaction with the Ln-CDs but also by cell size differences. A future study correlating cell size and NM uptake by monitoring endogenous elements or other tags would be of interest.
To conclude such study, and to have a better understanding about the interaction cell-NM, statistical analysis was carried out to evaluate the relationship between the mean and median uptake values vs. the Ln-CDs concentration added to the cell culture medium, being all data summarized in the ESI.† Firstly, a linear fitting was set to both datasets (see Table S6†), obtaining good statistic values for both the mean and median sets. It must be highlighted that the p value is lower than 0.05, suggesting a clear relationship between the uptake values and the concentration of Ln-CDs added to the medium. Moreover, to test and study the potential curvature of such values, a quadratic fitting was also carried out to compare the linear fitting of the data with a quadratic one. Both fittings are also represented in Fig. S12,† where both approaches show a clear relationship between the studied parameters. The statistical analysis showed evidence that both models are likely to fit the data, following the statistical values obtained from this analysis for both mean and median datasets, as it can be seeing in Tables S5 and S6.† The final statistical comparison was made comparing the models. Although the R2 is better for the quadratic fitting, following the rest statistical values, it cannot be concluded which is the model that better fits the data as the quadratic fitting did not provide a significantly better fit to the data than the linear and it could represent a model overfitting.
Conclusions
The strategy proposed in this work outlines methodology for the identification of true cell events in sc-ICP-MS to determine their TE. This is achieved through exploring for the first time the potential of elemental ratios detected by ICP-TOFMS in single cell transient events using HeLa cells tagged with multielement tags (Ln-CDs) as a model. LA-ICP-MS imaging of individual cells confirmed that Ln-CDs are a suitable option for cell tagging with a tagging efficiency of 99.87%. Due to the small size of the nanotag (2.3 nm), a single event with both doping elements (Eu and Yb) present at the correct ratio was considered a true cell event, as it was not possible to detect single Ln-CDs above the background with this experimental setup. TE values ranging from 40 to 50% were obtained with a CytoNeb–CytoSpray interface. The cellular uptake of Ln-CDs in HeLa cells followed a distribution ranging from 6.8 to 5115 fg CDs per cell. The in-house R-based platform developed for single-cell data treatment enabled the analysis of large ICP-ToF-MS data sets and showed reliable performance for simultaneous TE determination, elemental mass quantification and data visualization.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Guillermo Redondo-Fernandez: conceptualization, experimental, methodology, formal analysis, software, writing – original draft, writing – review & editing. Kharmen Billimoria: conceptualization, methodology, experimental, writing – original draft, writing – review & editing. David Ojeda: conceptualization, experimental, writing – review & editing. Dorota Bartczak: conceptualization, writing – review & editing. Simon Cowen: formal analysis, software, writing – review & editing. Ana Soldado: conceptualization, resources, writing – review & editing, supervision, funding acquisition. J. M. Costa-Fernandez: conceptualization, resources, writing – review & editing, supervision, funding acquisition. Heidi Goenaga-Infante: conceptualization, resources, supervision, paper review and editing, funding acquisition.Conflicts of interest
Authors declare that the reported experimental results are not influenced by competing financial interests or personal relationships.Acknowledgements
Guillermo Redondo-Fernandez acknowledges his PhD grant (BP20-050) from the Asturias Regional Government (Spain). This research was funded by Spanish Ministry of Science and Innovation PID2020-117282RB-I00 and PID2022-142323NB-I00, funded by MCIN/AEI/10.13039/501100011033/and by “ERDF A way of making Europe”. Funding from the UK Department for Science, Innovation and Technology (DSIT) is also acknowledged. Authors would like to thank to Marcos García Ocaña from Biotechnology Preparative Unit of University of Oviedo for his advice. Authors acknowledge the use of instrumentation as well as the technical advice provided by the National Facility ELECMI ICTS, node “Laboratorio de Microscopias Avanzadas (LMA)” at “Universidad de Zaragoza”.References
- X. Xu, R. Ray, Y. Gu, H. J. Ploehn, L. Gearheart, K. Raker and W. A. Scrivens, J. Am. Chem. Soc., 2004, 126, 12736–12737 CrossRef CAS PubMed.
- M. Lan, L. Guo, S. Zhao, Z. Zhang, Q. Jia, L. Yan, J. Xia, H. Zhang, P. Wang and W. Zhang, Adv. Ther., 2018, 1, 1800077 CrossRef.
- Y. Zhang, M. Wu, M. Wu, J. Zhu and X. Zhang, ACS Omega, 2018, 3(8), 9126–9145 CrossRef CAS PubMed.
- S. Zhu, X. Zhao, Y. Song, S. Lu and B. Yang, Nano Today, 2016, 11, 128–132 CrossRef CAS.
- S. Li, L. Li, H. Tu, H. Zhang, D. S. Silvester, C. E. Banks, G. Zou, H. Hou and X. Ji, Mater. Today, 2021, 51, 188–207 CrossRef CAS.
- G. Redondo-Fernandez, J. C. Canga, A. Soldado, J. Ruiz Encinar and J. M. Costa-Fernandez, Anal. Chim. Acta, 2023, 1284, 341874 CrossRef CAS PubMed.
- L. Ansari, S. Hallaj, T. Hallaj and M. Amjadi, Colloids Surf., B, 2021, 203, 111743 CrossRef CAS.
- Y. Zhao, X. Hao, W. Lu, R. Wang, X. Shan, Q. Chen, G. Sun and J. Liu, ACS Appl. Nano Mater., 2018, 1, 2544–2551 CrossRef CAS.
- D. Bouzas-Ramos, J. C. Canga, J. C. Mayo, R. M. Sainz, J. Ruiz Encinar and J. M. Costa-Fernandez, Adv. Funct. Mater., 2019, 29, 1903884 CrossRef.
- L. Mueller, H. Traub, N. Jakubowski, D. Drescher, V. I. Baranov and J. Kneipp, Anal. Bioanal. Chem., 2014, 406, 6963–6977 CrossRef CAS PubMed.
- M. Corte-Rodríguez, R. Álvarez-Fernández, P. García-Cancela, M. Montes-Bayón and J. Bettmer, TrAC, Trends Anal. Chem., 2020, 132, 116042 CrossRef.
- A. B. Santos da Silva and M. A. Zezzi Arruda, J. Trace Elem. Med. Biol., 2023, 75, 127086 CrossRef.
- S. Theiner, K. Loehr, G. Koellensperger, L. Mueller and N. Jakubowski, J. Anal. At. Spectrom., 2020, 35, 1784–1813 RSC.
- L. Yin, Z. Zhang, Y. Liu, Y. Gao and J. Gu, Analyst, 2019, 144, 824–845 RSC.
- F. Li, D. W. Armstrong and R. S. Houk, Anal. Chem., 2005, 77, 1407–1413 CrossRef CAS PubMed.
- A. Schoeberl, M. Gutmann, S. Theiner, M. Schaier, A. Schweikert, W. Berger and G. Koellensperger, Anal. Chem., 2021, 93(49), 16456–16465 CrossRef CAS.
- W. Qin, H.-J. Stärk, S. Müller and T. Reemtsma, Front. Microbiol., 2022, 13, 870931 CrossRef.
- L. Hendriks, V. M. Kissling, T. Buerki-Thurnherr and D. M. Mitrano, Environ. Sci.: Nano, 2023, 10, 3439–3449 RSC.
- S. Hellmann, P. García-Cancela, S. Alonso-Fernández, M. Corte-Rodríguez, J. Bettmer, A. Manteca, D. Merten, T. Gil-Díaz, T. Schäfer and M. Montes-Bayón, Chemosphere, 2024, 347, 140633 CrossRef CAS.
- J. Tsz-Shan Lum and K. Sze-Yin Leung, Anal. Chim. Acta, 2019, 1061, 50–59 CrossRef PubMed.
- Y. Zhou, Z. Chen, J. Zeng, J. Zhang, D. Yu, B. Zhang, X. Yan, L. Yang and Q. Wang, Anal. Chem., 2005, 77, 1407–1413 CrossRef.
- M. Godin, A. K. Bryan, T. P. Burg, K. Babcock and S. R. Manalis, Appl. Phys. Lett., 2007, 91, 123121 CrossRef.
- M. Elinkmann, S. Reuter, M. Holtkamp, S. Heuckeroth, A. Köhrer, K. Kronenberg, M. Sperling, O. Rubner, C. D. Quarles, M. Hipplerb and U. Karst, J. Anal. At. Spectrom., 2023, 38, 2607–2618 RSC.
- T. E. Lockwood, R. Gonzalez de Vega and D. Clases, J. Anal. At. Spectrom., 2021, 36, 2536–2544 RSC.
- M. I. Chronakis, M. von der Au and B. Meermann, J. Anal. At. Spectrom., 2022, 37, 2691–2700 Search PubMed.
- Y. Liang, Q. Liu, Y. Zhou, S. Chen, L. Yang, M. Zhu and Q. Wang, Anal. Chem., 2019, 91, 8341–8349 CrossRef CAS.
- B. Yang, B. Chen, M. He and B. Hu, Anal. Chem., 2017, 89, 1879–1886 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ja00075k |
| This journal is © The Royal Society of Chemistry 2025 |