Catalytic capsids: the art of confinement†
Inge J.
Minten
a,
Victor I.
Claessen
a,
Kerstin
Blank
a,
Alan E.
Rowan
a,
Roeland J. M.
Nolte
a and
Jeroen J. L. M.
Cornelissen
*b
aRadboud University Nijmegen, Institute for Molecules and Materials, Heyendaalseweg 135, 6525 AJ, Nijmegen, the Netherlands
bLaboratory for Biomolecular Nanotechnology, MESA+ Institute, University of Twente, P.O. Box 217, 7500 AE, Enschede, the Netherlands. E-mail: J.J.L.M.Cornelissen@tnw.utwente.nl; Fax: +31 534894645; Tel: +31 53489 4380
First published on 9th November 2010
Abstract
In the cell, enzymes are almost always spatially confined in crowded and tightly controlled cellular compartments. The entrapment of enzymes in artificial nanoreactors as biomimetic systems can be expected to contribute to the understanding of the activity and the interactions of enzymes in confined spaces. The capsid of the Cowpea Chlorotic Mottle virus (CCMV) represents such an artificial nanoreactor that can be used to encapsulate multiple proteins in its interior. Employing a controlled encapsulation process we are able to load a precise number of proteins (Pseudozyma antarctica lipase B and EGFP) into the CCMV capsid and to study their activity. In the case of the enzyme, our results indicate that the apparent overall reaction rate increases upon encapsulation and is almost independent of the number of enzymes in the capsid. These observation are the result of the extremely high confinement molarity of the enzyme inside the capsid (Mconf = ∼ 1 mM) leading to very rapid formation of the enzyme–substrate complex. These results highlight the importance of small volumes for efficient multi-enzyme cascade catalysis.
Introduction
To better understand the complex workings of the cell, it is important to appreciate the interplay between the enzyme and its environment. In the cell, enzymes are invariably spatially confined in crowded and tightly controlled cellular organelles. Even in bacteria, which do not have organelles, enzyme-loaded proteinaceous microcompartments (MCPs) have been recently discovered, which have a similar function. It is therefore important to study enzymes in an environment which mimics these confined conditions where normal solution reaction kinetics is no longer valid. It is, however, challenging to find suitable biomimetic containers and to entrap enzymes within these artificial systems in a controlled way. Polymeric vesicles,1,2 mimicking lipid vesicles, are a commonly employed system for the encapsulation of enzymes as these are robust and easy to use. Despite their potential, their relatively large size (diameter (D) = ∼ 100 nm) combined with a high polydispersity results in non-uniform reaction conditions in each vesicle hindering kinetic analysis.In contrast, protein cages, like virus capsids, can have diameters of only tens of nanometres and are exceptionally monodisperse. Viruses are the most abundant biological entities on earth,3 and are mostly known for their infectious properties. They often consist of only one or a few different protein components and nucleic acids, presented by Nature in numerous variations.
Their simple composition makes them very useful as versatile nanobuilding blocks, e.g. for the creation of conductive nanowires,4MRI contrast agents5 and drug delivery systems.6 One example of a frequently used virus is the Cowpea Chlorotic Mottle virus (CCMV). It is an icosahedral plant virus of a diameter D = 28 nm, which is composed of 180 identical capsid proteins surrounding single-stranded RNA. A unique property of CCMV is the ability to reassemble into the native capsid structure after removal of the viral RNA. At pH 7.5, the capsid disassembles into 90 capsid protein dimers allowing for extraction of its RNA. The original D = 28 nm capsid structure is reformed when the pH is lowered to 5.0. This process is completely reversible, and has been used to encapsulate inorganic materials,5,7 negatively charged polymers,8,9 or even enzymes10 by adding these molecules during the assembly process.
The encapsulation of single enzyme molecules inside the CCMV capsid by a statistical procedure has already been demonstrated.10 Following up on this work, we now encapsulate multiple enzymes in the confined space of the CCMV capsid. The encapsulation of multiple enzymes in a precisely controlled fashion cannot be achieved by statistical encapsulation. We have recently overcome this problem by using directed assembly based on a heterodimeric coiled-coil linker (vide infra).11 With this method the enzyme to be encapsulated is attached to the capsid protein dimers before assembly using the coiled-coil motif. The number of encapsulated enzymes is then controlled by varying the ratio between wild-type capsid protein and the capsid protein–enzyme complex. This method significantly increases the encapsulation efficiency and, most importantly, provides control over the average number of encapsulated enzymes. This directed assembly process closely matches the strategy used by Nature to pack enzymes into MCPsvia short N-terminal peptide sequences.12 In the work presented here, we aim to use this method to encapsulate the enzymePseudozyma (formerly Candida) antartica lipase B (PalB) and to investigate the overall reaction rate as a function of the number of encapsulated PalB molecules. In addition we co-encapsulated EGFP as a non-catalytic protein to investigate the reaction rate as a function of the total protein concentration inside the capsid.
Experimental
Sample preparation
Fusion proteins consisting of monomers of the coiled coil motif and PalB (E-coil), EGFP (E-coil) or the capsid protein (K-coil) were expressed in E. coli and purified by Ni2+-NTA chromatography using standard protocols. The PalB fusion protein was fluorescently labeled with NHS-Alexa Fluor 568. Prior to capsid assembly, coiled coil complexes were formed between PalB and capsid protein as well as between EGFP and capsid protein. Complexes were mixed with wildtype capsid protein in the desired ratios in pH 7.5 buffer. Lowering the pH to 5.0 by dialysis induced capsid assembly. Characterization of the coiled coil complexes as well as of the assembled capsids were performed using size exclusion chromatography. The absorption at 280 or 586 nm was used to determine the concentration of capsid protein or labeled PalB, respectively, neglecting the small contribution of PalB to the absorption at 280 nm (see supporting information for details).Enzyme activity measurements
For the enzyme activity measurements with 6,8-difluoro-4-methylumbelliferyl octanoate (DiFMU-octanoate) either the concentration of PalB or the concentration of capsid was kept constant for all measured samples. Reactions were performed in triplicate using a microplate reader (λex = 355 nm and λem = 460 nm). The reaction was started with the addition of DiFMU-octanoate. The initial reaction velocities were calculated from the obtained data by determining the slope of the initial part of the curves. These values were then normalized by dividing all initial slopes by the same value (i.e. the obtained initial velocity for 1.3 PalB/capsid were the concentration of PalB was kept constant throughout the experiment).Dual-color fluorescence cross-correlation spectroscopy (FCCS)
Laser light of 488 nm (Spectra-Physics 2080 argon ion laser) and 568 nm (Coherent CR-599 dye laser, pumped by 532 nm Spectra-Physics Millennia Nd:YAG laser) was coupled into a single-mode optical fiber, reflected by a dichroic beam splitter (Chroma z488/568rpc) and focused onto the sample by an oil immersion 100× objective (Carl Zeiss, NA = 1.30), which was mounted on a Carl Zeiss Axiovert 200 inverted microscope. Fluorescent light coming from the focal volume was collected by the same objective, passed through the dichroic beamsplitter, focused through a 100 μm pinhole, split by a second dichroic mirror (Chroma 560dcxr), filtered by either a ‘red’ (Chroma D630/60m) or ‘green’ filter (Chroma HQ525/50m) and focused onto avalanche photodiodes (PerkinElmer SPCM-AQR-14). The photon count signals were recorded using a Picoquant PicoHarp 300E TCSPC module.The focal volume was positioned in a solution of CCMV capsids containing EGFP, PalB or both. EGFP is autofluorescent and is excited at 488 nm, while the PalB was labeled with an Alexa568 dye which is excited at 568 nm. Emission of both fluorophores was filtered and recorded by separate photodetectors. The photon correlation functions were calculated using matlab scripts developed in-house. Curves were normalized on the G(0) parameter and then multiplied by the r-square of the fit for clarity.
Results and discussion
PalB (EC 3.1.1.3), a widely used biocatalyst, was chosen as the model enzyme since it retains most of its activity at pH 5.0.13 PalB was genetically modified with the E-coil monomer of the heterodimeric coiled coil, while the capsid protein was modified with the complementary K-coil (see ESI†). To enable the encapsulation efficiency to be determined, PalB was labeled with the fluorescent dye Alexa Fluor 568. Coiled-coil formation was achieved by simply mixing capsid protein with PalB. The resulting complex was purified by size exclusion chromatography and subsequently mixed with wildtype capsid protein in various ratios to form capsids with different amounts of encapsulated PalB. This capsid assembly was induced by lowering the pH to 5.0 (Fig. 1). Following assembly, the capsids were purified and analyzed by size exclusion chromatography using a Superose 6 column (Fig. S1, ESI†). The UV-Vis absorption of the protein and the dye was monitored during the run and the ratio between the dye-specific and protein-specific absorption of the non-encapsulated PalB was used to determine the amount of PalB encapsulated per capsid (see ESI†). Four different batches of capsid were made, with on average 1.3, 2.0, 3.5 and 4.0 PalB enzymes per capsid.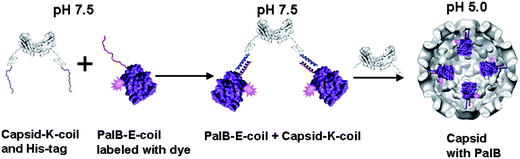 | ||
| Fig. 1 Schematic representation of PalB encapsulation. Capsid with K-coil and His-tag will be referred to as capsid protein in the text, PalB with E-coil will be referred to as PalB in the text. The PalB-capsid protein complex is mixed with wild-type capsid at pH 7.5 and subsequently dialyzed to pH 5.0 to induce capsid formation. | ||
In steady-state enzyme kinetic measurements, when the concentration of the substrate is well above KM, the enzyme functions at its maximum rate (kcat). The reaction velocity is then directly proportional to the enzyme concentration in solution. To test if this is also true for our confined system, the four samples containing different amounts of encapsulated PalB and a sample with non-encapsulated PalB were diluted such that the concentration of PalB was the same for all samples (Fig. 2a). To determine the overall reaction rate of these different samples, the pro-fluorescent substrate 6,8-difluoro-4-methylumbelliferyl octanoate (DiFMU-octanoate) was added to give a final concentration of 40 μM, which is well above the KM values of 8.8 μM determined in this study (ESI†) and 2.6 μM reported for PalB and DiFMU octanoate in the literature.14,15 The initial reaction rates were calculated and normalized with respect to the sample with the lowest encapsulated enzyme concentration (see Fig. 2b and Experimental section).
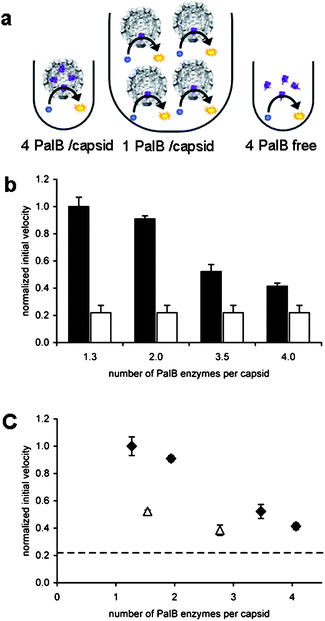 | ||
| Fig. 2 Kinetics of PalB encapsulation. The concentration of PalB is the same in all samples. (A) Schematic representation of experimental setup. (B) Normalized initial velocities with respect to 1.3 PalB/capsid. Black bars represent initial velocities of samples with encapsulated PalB. White bars indicate normalized initial velocity of non-encapsulated PalB (one experiment was plotted four times for clarity. Error bars indicate SD of triplicate experiments. (C) Normalized initial velocities plotted against the number of encapsulated PalB enzymes per capsid. (◆ represent capsid with encapsulated PalB; △ represent capsids with PalB and 7 EGFP proteins. Error bars indicate SD of triplicate experiments. The dashed line indicates the initial velocity of non-encapsulated PalB. | ||
Since the overall enzyme concentration is the same in all five samples, the reaction velocities should also be the same, if these were to follow standard Michaelis–Menten reaction kinetics. We observe, however, that the reaction velocity is inversely proportional to the number of encapsulated enzymes (Fig. 2c) and the encapsulated enzymes appear to outperform the non-encapsulated PalB.
It needs to be considered that the overall amount of capsids is higher in the samples with less PalB per capsid. In order to eliminate any effect of the capsid on the catalytic reaction, four samples containing different amounts of PalB were diluted such that all samples contained the same amount of capsids. Since the concentration of PalB is now different for each sample, four different control experiments were performed, in which the concentration of non-encapsulated PalB was the same as for the corresponding encapsulated PalB samples (Fig. 3a). As expected, the initial reaction rates of non-encapsulated PalB increase with increasing enzyme concentrations (Fig. 3b). The initial reaction velocities of the encapsulated PalB samples, however, are relatively constant within experimental error.
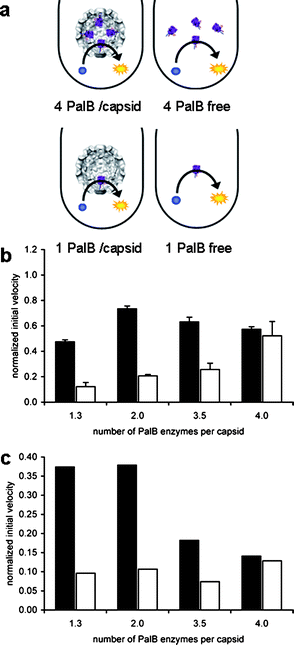 | ||
| Fig. 3 Kinetics of PalB encapsulation. The concentration of capsid is the same in all samples. (A) Schematic representation of the experimental setup. (B) Normalized initial rates with respect to 1.3 PalB/capsid experiment reported in Fig. 2. The reduced normalized velocity is a result from the absolute dilution in order to obtain the same concentration of capsid. Black bars represent initial rates of samples with encapsulated PalB. White bars indicate the normalized initial rate of non-encapsulated PalB. Error bars indicate standard deviation of triplicate experiments. (C) Normalized initial rates per PalB and per capsid, values obtained by dividing values of the initial rates of graph B by the number of PalB enzymes per capsid. | ||
Dividing the activity of the non-encapsulated PalB by its concentration (Fig. 3c), the calculated activity should be the same for the four different samples. Within experimental margins this seems to be true. This calculated activity of the encapsulated PalB, however, appears to be the largest when only one or two PalB molecules are encapsulated per capsid, suggesting that an increase in the protein concentration inside the capsid might have a negative effect on the reaction velocity originating from some form of crowding effect.
In summary, these results show two remarkable effects of the encapsulation of PalB in the very confined space of the CCMV capsid, i.e. (i) encapsulated PalB seems to outperform the non-encapsulated PalB, and (ii) the presence of more PalBs per capsid does not lead to an increase in the overall reaction rate. The former of the two effects could be the result of a change in kcat upon complexation and encapsulation inside the virus. It is known that PalB is more active when immobilized, and one could argue that it is immobilized within the capsids. Further, Chen et al.16 have shown that in vesicle systems the turnover number (kcat) of entrapped enzymes increases when the vesicle size becomes smaller. They observed a two-fold increase in turnover number going from non-encapsulated enzymes to enzymes spatially confined to vesicles with an internal volume of 5 attoliter, and an additional two-fold increase going from these vesicles to vesicles with an internal volume of 0.05 attoliter. In contrast, the second effect (ii) could be explained by crowding inside the virus by the presence of additional protein. This potential contribution of crowding will be discussed in more detail below.
Both effects together can be readily explained by a change in the effective molarity of the enzyme in the confined space of the capsid, which we define as confinement molarity Mconf. The internal volume of the CCMV capsid is approximately 5 zeptoliter, a factor 10 smaller than the smallest vesicles tested in the paper by Chen et al. In this small volume Mconf for the encapsulated enzyme is ∼1 mM, which is a factor 104 higher than the bulk enzyme concentration (30–100 nM)
Assuming that the rate of diffusion through the capsid pores is significantly larger than the rate of catalysis and that the concentration of substrate inside the capsid and outside the capsid is equal (steady state situation) we can calculate that the enzyme concentration inside the capsid is approximately 25 times higher than the substrate concentration of 40 μM. Considering that KM is not altered as a result of the encapsulation, the high Mconf can therefore be expected to shift the equilibrium towards the formation of the enzyme–substrate complex thereby increasing the reaction velocity. The observed results clearly highlight that the advantage of confinement is the shifting of the enzyme substrate equilibrium completely in the direction of the enzyme–substrate complex. Faster formation of the enzyme substrate complex might be further assisted by an increased number of collisions between the enzyme and the substrate, resulting from the confinement, as has been shown by Chen et al.16
Having explained the overall increase in reaction rate, we can now look at the number of molecules inside the capsid in more detail to explain why the reaction velocity does not increase with an increasing number of enzymes in the capsid. Using a Poisson distribution the chance that two or more substrates are present in the capsid at the same time can be calculated. This chance is 0.64%, which is negligibly low. In fact, only ∼10% of the capsids contain a substrate molecule at all. It therefore makes no difference whether a capsid contains one or more enzymes, as there is maximally only one substrate molecule present. In other words, a consequence of the high Mconf is that upon going from one to four enzymes in the capsid, the change in Mconf is so small that it has a minimal effect on the overall reaction velocity. It should be noted than in solution the substrate concentration is in excess whereas in the capsid the enzyme concentration is in excess.
The effect of crowding within the capsid upon the increase in the number of enzymes cannot be fully discarded. To investigate a potential crowding effect, EGFP was co-encapsulated with PalB. This protein does not contribute to product conversion. Two samples were prepared, one containing 1 PalB and 7 EGFP per capsid and the other containing 3 PalB and 7 EGFP per capsid. Co-encapsulation of the enzyme and the protein was demonstrated by dual-color fluorescence cross-correlation spectroscopy (FCCS). Control experiments of capsid containing exclusively PalB or EGFP showed only auto-correlation in the appropriate channel and little cross-correlation, while analysis of the samples containing both proteins does clearly show cross-correlation (Fig. 4) proving that indeed both biomacromolecules are present in the same capsid.
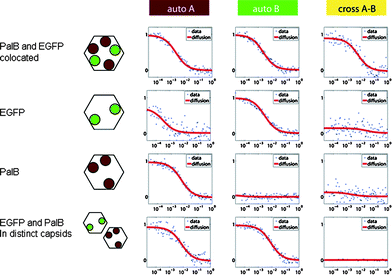 | ||
| Fig. 4 Dual-color FCCS correlation curves of CCMV capsids containing EGFP (grey circles), PalB (black circles) or both. When EGFP is encapsulated, there is correlation in the green channel B, when PalB is encapsulated there is correlation in the red channel A, but only when both are present in the same capsid the cross-correlation arises. Due to spectral overlap of EGFP and the Alexa Fluor 568 dye at higher wavelengths some of the emission of EGFP is recorded in the red channel, thereby causing some red-channel auto-correlation even when the red dye Alexa Fluor 568 is absent. | ||
Co-encapsulation of EGFP did indeed result in a small effect in the reaction rate (Fig. 2c), but since the total protein concentration was approximately two times higher than the highest amount of PalB encapsulated, the effect should have been more pronounced if the high local enzyme concentration would really have been the only cause for the decrease in activity. Instead the observed lower reaction velocity in Fig. 2c might be attributed to a decreased diffusion speed of the substrate and product molecules over the capsid shell. The EGFP and PalB molecules are attached to the N-termini of the capsid proteins and thus located in the vicinity of the pores in the capsid shell. There are 20 pores of D = 2 nm in the capsid shell, together covering roughly 1% of the solvent-exposed outer surface of the capsid. Therefore diffusion over the capsid wall is likely slower than diffusion in solution.8 If diffusion over the capsid wall was slower than the substrate conversion, diffusion could be the rate-limiting step. This is unlikely however, since substrate conversion of native PalB is about nine orders of magnitudes slower than unhindered diffusion of a substrate molecule over the capsid wall. This diffusion rate however could be affected by an increasing number of proteins close to the pores, which will then result in a reduced overall reaction velocity, as is observed (Fig. 2c).
Conclusions
In conclusion, we have studied the effects of encapsulation of multiple PalB enzymes in the spatial confinement of the CCMV capsid. The spatially confined PalB seems to have a higher activity than non-encapsulated PalB. Although this might be caused by the immobilization of PalB in the capsid viz. a change in kcat, there is no evidence for this. It is more likely that the observed effect is caused by Mconf of the enzyme combined with an increased collision chance due to the spatial confinement. Since a capsid rarely contains more than one substrate molecule, one enzyme is sufficient to convert the substrate, and the presence of additional enzymes has no effect on the reaction velocity. Further research will be needed to confirm this theory by varying the substrate concentration, which in the described system, however, is hindered by the limited solubility of the DiFMU.We have shown here that it is indeed possible to encapsulate two different proteins inside the CCMV capsid, i.e.EGFP and PalB. In the future, multiple different enzymes, capable of performing cascade reactions might be encapsulated in the CCMV capsid using this method. When multiple different enzymes are encapsulated in the CCMV capsid it can readily predicted, that due to Mconf, the reaction speed of the cascade reactions will be considerably enhanced. The converted substrate of one encapsulated enzyme would more likely collide and bind with another enzyme in the capsid than diffuse through the capsid wall. Thus the converted substrate would likely react with a different enzyme present in the same capsid at a much faster speed than if the enzymes were not encapsulated. The study of cascade reactions in confinement could not only potentially provide insight into enzymatic cascade reactions inside cellular organelles, but could also lead to the creation of a highly efficient nano-sized reaction vessel with potential industrial applications.
Acknowledgements
The Chemical Council of the Netherlands' National Science Foundation NWO-CW), the European Science Foundation (ESF) and the Royal Netherlands Academy of Arts and Sciences (KNAW) are acknowledged for financial support. A plasmid encoding PalB was a generous gift of S. Schoffelen.Notes and references
- S. F. M. van Dongen, M. Nallani, J. J. L. M. Cornelissen, R. J. M. Nolte and J. C. M. van Hest, Chem.–Eur. J., 2009, 15, 1107–1114 CAS.
- F. Caruso, D. Trau, H. Möhwald and R. Renneberg, Langmuir, 2000, 16, 1485–1488 CrossRef CAS.
- C. A. Suttle, Nature, 2005, 437, 356–361 CrossRef CAS.
- Y. J. Lee, H. Yi, W.-J. Kim, K. Kang, D. S. Yun, M. S. Strano, G. Ceder and A. M. Belcher, Science, 2009, 324, 1051–1055 CAS.
- L. Liepold, S. Anderson, D. Willits, L. Oltrogge, J. A. Frank, T. Douglas and M. Young, Magn. Reson. Med., 2007, 58, 871–879 CrossRef CAS.
- W. Wu, S. C. Hsiao, Z. M. Carrico and M. B. Francis, Angew. Chem., Int. Ed., 2009, 48, 9493–9497 CrossRef CAS.
- T. Douglas and M. Young, Nature, 1998, 393, 152–155 CrossRef CAS.
- I. J. Minten, Y. J. Ma, M. A. Hempenius, G. J. Vancso, R. J. M. Nolte and J. J. L. M. Cornelissen, Org. Biomol. Chem., 2009, 7, 4685–4688 RSC.
- F. D. Sikkema, M. Comellas-Aragones, R. G. Fokkink, B. J. M. Verduin, J. J. L. M. Cornelissen and R. J. M. Nolte, Org. Biomol. Chem., 2007, 5, 54–57 RSC.
- M. Comellas-Aragones, H. Engelkamp, V. I. Claessen, N. Sommerdijk, A. E. Rowan, P. C. M. Christianen, J. C. Maan, B. J. M. Verduin, J. J. L. M. Cornelissen and R. J. M. Nolte, Nat. Nanotechnol., 2007, 2, 635–639 CrossRef CAS.
- I. J. Minten, L. J. A. Hendriks, R. J. M. Nolte and J. J. L. M. Cornelissen, J. Am. Chem. Soc., 2009, 131, 17771–17773 CrossRef CAS.
- C. G. Fan, S. Q. Cheng, Y. Liu, C. M. Escobar, C. S. Crowley, R. E. Jefferson, T. O. Yeates and T. A. Bobik, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 7509–7514 CrossRef CAS.
- N. Miletic, V. Abetz, K. Ebert and K. Loos, Macromol. Rapid Commun., 2010, 31, 71–74 CrossRef CAS.
- N. Zhang, W.-C. Suen, W. Windsor, L. Xiao, V. Madison and A. Zaks, Protein Eng., 2003, 16, 599–605 CrossRef CAS.
- Y. Ying and L. Stefan, Biotechnol. Bioeng., 2009, 105, 44–50.
- Q. Chen, H. Schonherr and G. J. Vancso, Small, 2009, 5, 1436–1445 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Additional experimental details and figures. See DOI: 10.1039/c0sc00407c |
| This journal is © The Royal Society of Chemistry 2011 |
