A photocleavable affinity tag for the enrichment of alkyne-modified biomolecules†
Timo
Koopmans
a,
Frank J.
Dekker
b and
Nathaniel I.
Martin
*a
aDepartment of Medicinal Chemistry & Chemical Biology, Utrecht Institute for Pharmaceutical Sciences, University of Utrecht, David de Wied Building, Office: 5.64, Universiteitsweg 99, 3584 CG Utrecht, The Netherlands. E-mail: n.i.martin@uu.nl; Tel: +31 (0)6 1878 5274
bDepartment of Pharmaceutical Gene Modulation, Groningen Research Institute of Pharmacy, University of Groningen, Antonius Deusinglaan 1, 9713 AV, Groningen, The Netherlands
First published on 18th January 2012
Abstract
A new photocleavable affinity tag for use in the enrichment of alkyne-labelled biomolecules is reported. The tag is prepared via a concise synthetic route using readily available materials. The photolytic conditions employed for cleavage of the tag provide for a clean release of enriched biomolecules from streptavidin resin.
The ability to selectively probe and characterize subsets of proteins within complex mixtures is a cornerstone of modern proteomics. Activity based protein profiling (ABPP) has emerged as a robust means by which the unique reactivities of specific enzyme families can be rapidly assessed within biological samples.1 While ABPP provides for the selective labelling of target proteins, subsequent enrichment steps are often essential for proper characterization of the labelled target. The most common approach for enriching labelled biomolecules relies upon the high affinity biotin–streptavidin interaction. Activity based probes can be synthesized to contain a biotin moiety, or as is becoming increasingly common, a biotin tag can be introduced after the covalent attachment of the probe to its target biomolecule. To do so, a bioorthogonal alkyne “handle” can be incorporated into the probe molecule serving as a convenient site for biotin attachment at a later stage via the Cu(I)-catalyzed azide-alkyne “click” cycloaddition (CuAAC).2,3 Biomolecules modified in this way can then be enriched by passage over a streptavidin resin followed by washing and elution, after which SDS-PAGE analysis and mass spectrometry are commonly used to characterise the enriched species (Fig. 1).
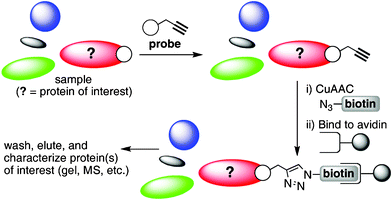 | ||
| Fig. 1 Isolation of alkyne-labelled proteinsviaABPP coupled with CuAAC and biotin-streptavidin enrichment. | ||
A recognized limitation when using biotin-based enrichment tags is the difficulty encountered in obtaining a clean and efficient release from the streptavidin resin. Harsh conditions are required to disrupt the biotin–streptavidin interaction (∼KD 10−15 M) and can lead to degradation of the labelled biomolecule of interest along with introduction of impurities deriving from the streptavidin itself. In addition, endogenously biotinylated proteins present in the original mixture are also released along with the desired target protein. To circumvent these issues, selectively cleavable linkers suitable for use in CuAAC-based protein enrichments have been developed. Among the most notable are Cravatt's cleavable peptide linker based upon a recognition sequence for the tobacco etch virus (TEV) protease2,4 (Fig. 2a) and the dithionite-sensitive azobenzene linker (Fig. 2b) pioneered by Verhelst and Bogyo5 and further developed for use in CuAAC approaches by the groups of Hang6–8 and Hulme.9 In addition to these two well established techniques, cleavable enrichment tags based upon other chemical approaches, including disulfide bond reduction10 and pH-sensitive cleavage,11 have also recently been reported.
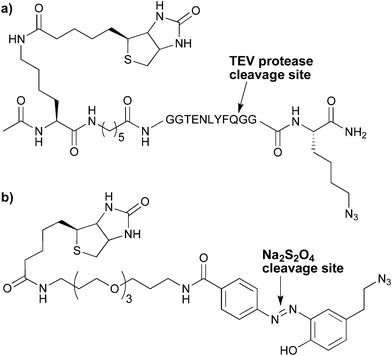 | ||
| Fig. 2 a) TEV protease-cleavable4 and b)dithionite-cleavable7 linkers for use in the CuAAC-based, biotin-streptavidin enrichment of alkyne-modified biomolecules. | ||
In each of the approaches described above, release of the enriched biomolecule is achieved by the addition of a specific cleavage reagent. While such approaches have shown much success, the requisite addition of a cleavage reagent does introduce a new component to the mixture. By comparison, the use of a photocleavable linker circumvents the need for exogenous reagents in releasing the enriched biomolecule. Previous work by the groups of Rothschild12,13 and Tirrell14 has demonstrated that the photosensitive 1-(2-nitrophenyl)-ethyl moiety can serve as a photocleavable linker in biotin–streptavidin enrichments. In such an experiment a protein is labelled with a biotin tag via a linker containing the photocleavable moiety. After binding to the streptavidin resin and washing, the protein of interest is eluted upon cleavage of the linker by irradiation at the appropriate wavelength. Building upon these approaches, we here report a new photocleavable affinity tag for use in the enrichment of alkyne-modified biomolecules.
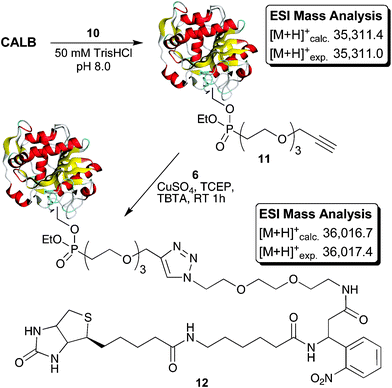
In preparing this new enrichment tag, commercially available β-amino acid 3-amino-3-(2-nitrophenyl)-propionic acid was employed. This building block has previously been utilized in other applications requiring photolytic peptide bond cleavage.15,16 As illustrated in Scheme 1, synthesis of the enrichment tag began with the conversion of triethylene glycol to the required azido amine spacer 2via a modified literature procedure.17,18 Coupling of spacer 2 with Boc-protected 3-amino-3-(2-nitrophenyl)-propionic acid provided intermediate 5. After Boc group removal, condensation with biotinamidohexanoic acid N-hydroxy-succinimide ester (NHS-LC-Biotin) led to formation of the target compound. Via this route, multi-hundred-milligram quantities of photocleavable enrichment tag 6 were readily prepared.
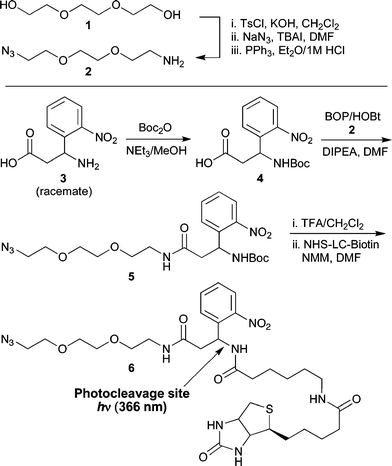 | ||
| Scheme 1 Synthesis of photocleavable tag 6 (note: 3-amino-3-(2-nitrophenyl)-propionic acid is obtained commercially as the racemate resulting in the production of 6 as a diastereomeric mixture). | ||
With photocleavable linker 6 in hand, its performance in the enrichment of alkyne labelled proteins was evaluated. For this purpose we chose a model system based upon the well-characterized family of fluorophosphonate probes frequently employed to characterize serine hydrolase activity.19,20 The fluorophosphonate-alkyne probe 1021 was prepared as shown in Scheme 2 and subsequently used to covalently label Candida antarcticalipase B (CALB) which served as a model hydrolase enzyme (Scheme 3). Labelling of CALB by probe 10 was confirmed by ESI-MS (Fig. S1, ESI†) after which linker 6 was appended to the alkyne tag using standard CuAAC conditions.22,23 Incorporation of 6 was essentially quantitative, as indicated by the ESI-MS analysis of the ligation reaction mixture (Fig. S2, ESI†). The modified CALB species 12 was then incubated with streptavidin resin followed by extensive washing. Binding to the resin by 12 was confirmed by boiling a sample of the treated resin with a sodium dodecyl sulfate (SDS) solution, indicating release of 12 (lane D, Fig. 3). Also detected in the same elution sample was a significant amount of low MW contaminant derived from the streptavidin resin itself as a result of the harsh SDS-boil conditions. By comparison, when a sample of the same loaded resin was exposed to 366 nm UV light at 0 °C, the tagged CALB species was cleanly eluted with no low MW contamination detected (lane E, Fig. 3).
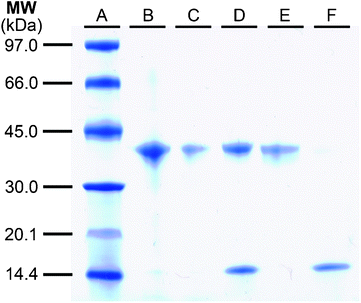 | ||
| Fig. 3 Performance assessment of photocleavable enrichment tag 6 by SDS-PAGE analysis. Lane A) MW ladder; B) solution of 12 before resin binding; C) solution post resin binding; D) elution by SDS boil; E) elution by irradiation at 366 nm; F) control lane (fresh resin treated with non-biotinylated CALB species 11 followed by SDS boil). | ||
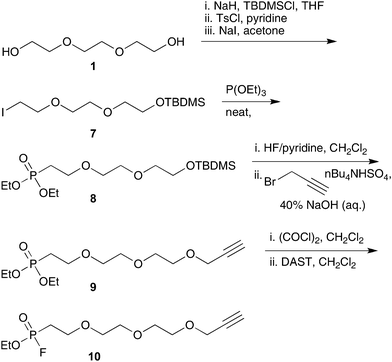 | ||
| Scheme 2 Synthesis of model fluorophosphonate probe 10. | ||
As demonstrated above, elution of the modified CALB species 12 from streptavidin resin upon exposure to UV light provides for a cleaner recovery relative to that obtained with an SDS boil. In this regard, the use of photocleavable linker 6 clearly offers a superior alternative to the harsh conditions associated with SDS cleavage protocols. Also, unlike the other cleavable azido-biotin tags commonly used for the enrichment of alkyne-modified proteins (Fig. 2), photocleavable linker 6 requires the addition of no external cleavage reagents that can complicate subsequent analysis. Furthermore, the concise synthetic route to 6 presents a notable advantage relative to the preparation of other cleavable enrichment tags. Studies aimed at utilizing photocleavable linker 6 to enrich proteins from more complex biological mixtures are underway. Preliminary results involving the enrichment of modified CALB 12 from E. coli lysates are encouraging (Fig. S3, ESI†) and indicate that linker 6 will be of value in proteomic analyses requiring the enrichment of alkyne-modified biomolecules.
In summary, we have developed a new photocleavable linker for use in the enrichment of alkyne-modified proteins. The straightforward synthesis of 6 as well as the reagent-free photolytic conditions required for its cleavage are particularly appealing. Access to such enrichment tags serves to expand the repertoire of available tools for use in the proteomics-based characterization of proteins central to ABPP. In this regard, photocleavable linker 6 is made freely available to interested parties upon request (by contacting the corresponding author).
This work was supported by The Netherlands Organization for Scientific Research (NWO VENI/VIDI grants to N.I.M.). Dr Katherine Barglow is thanked for valued discussion and helpful suggestions. Kees Versluis is kindly acknowledged for providing assistance with MS analysis.
References
- B. F. Cravatt, A. T. Wright and J. W. Kozarich, Annu. Rev. Biochem., 2008, 77, 383–414 CrossRef CAS.
- A. E. Speers and B. F. Cravatt, J. Am. Chem. Soc., 2005, 127, 10018–10019 CrossRef CAS.
- G. Charron, M. M. Zhang, J. S. Yount, J. Wilson, A. S. Raghavan, E. Shamir and H. C. Hang, J. Am. Chem. Soc., 2009, 131, 4967–4975 CrossRef CAS.
- E. Weerapana, A. E. Speers and B. F. Cravatt, Nat. Protoc., 2007, 2, 1414–1425 CrossRef CAS.
- S. H. Verhelst, M. Fonovic and M. Bogyo, Angew. Chem., Int. Ed., 2007, 46, 1284–1286 CrossRef CAS.
- Y. Y. Yang, J. M. Ascano and H. C. Hang, J. Am. Chem. Soc., 2010, 132, 3640–3641 CrossRef CAS.
- Y. Y. Yang, M. Grammel, A. S. Raghavan, G. Charron and H. C. Hang, Chem. Biol., 2010, 17, 1212–1222 CrossRef CAS.
- M. Grammel, M. M. Zhang and H. C. Hang, Angew. Chem. Int. Ed. Engl., 2010, 49, 5970–5974 CAS.
- F. Landi, C. M. Johansson, D. J. Campopiano and A. N. Hulme, Org. Biomol. Chem., 2010, 8, 56–59 CAS.
- M. A. Nessen, G. Kramer, J. Back, J. M. Baskin, L. E. Smeenk, L. J. de Koning, J. H. van Maarseveen, L. de Jong, C. R. Bertozzi, H. Hiemstra and C. G. de Koster, J. Proteome Res., 2009, 8, 3702–3711 CrossRef CAS.
- P. van der Veken, E. H. Dirksen, E. Ruijter, R. C. Elgersma, A. J. Heck, D. T. Rijkers, M. Slijper and R. M. Liskamp, ChemBioChem, 2005, 6, 2271–2280 CrossRef CAS.
- J. Olejnik, S. Sonar, E. Krzymanska-Olejnik and K. J. Rothschild, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 7590–7594 CrossRef CAS.
- M. Lim and K. J. Rothschild, Anal. Biochem., 2008, 383, 103–115 CrossRef CAS.
- J. Szychowski, A. Mahdavi, J. J. Hodas, J. D. Bagert, J. T. Ngo, P. Landgraf, D. C. Dieterich, E. M. Schuman and D. A. Tirrell, J. Am. Chem. Soc., 2010, 132, 18351–18360 CrossRef CAS.
- C. J. Bosques and B. Imperiali, J. Am. Chem. Soc., 2003, 125, 7530–7531 CrossRef CAS.
- M. Toebes, M. Coccoris, A. Bins, B. Rodenko, R. Gomez, N. J. Nieuwkoop, W. van de Kasteele, G. F. Rimmelzwaan, J. B. Haanen, H. Ovaa and T. N. Schumacher, Nat. Med., 2006, 12, 246–251 CrossRef CAS.
- E. Klein, S. DeBonis, B. Thiede, D. A. Skoufias, F. Kozielski and L. Lebeau, Bioorg. Med. Chem., 2007, 15, 6474–6488 CrossRef CAS.
- K. M. Bonger, R. J. van den Berg, L. H. Heitman, I. J. AP, J. Oosterom, C. M. Timmers, H. S. Overkleeft and G. A. van der Marel, Bioorg. Med. Chem., 2007, 15, 4841–4856 CrossRef CAS.
- Y. Liu, M. P. Patricelli and B. F. Cravatt, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 14694–14699 CrossRef CAS.
- D. Kidd, Y. Liu and B. F. Cravatt, Biochemistry, 2001, 40, 4005–4015 CrossRef CAS.
- L. C. Gillet, K. Namoto, A. Ruchti, S. Hoving, D. Boesch, B. Inverardi, D. Mueller, M. Coulot, P. Schindler, P. Schweigler, A. Bernardi and S. Gil-Parrado, Mol. Cell. Proteomics, 2008, 7, 1241–1253 CAS.
- Q. Wang, T. R. Chan, R. Hilgraf, V. V. Fokin, K. B. Sharpless and M. G. Finn, J. Am. Chem. Soc., 2003, 125, 3192–3193 CrossRef CAS.
- S. R. Hanson, T. L. Hsu, E. Weerapana, K. Kishikawa, G. M. Simon, B. F. Cravatt and C. H. Wong, J. Am. Chem. Soc., 2007, 129, 7266–7267 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental protocols and product characterization. See DOI: 10.1039/c2ra20082a |
| This journal is © The Royal Society of Chemistry 2012 |