New dictyodendrins as BACE inhibitors from a southern Australian marine sponge, Ianthella sp.†
Hua
Zhang‡
,
Melissa M.
Conte
,
Zeinab
Khalil
,
Xiao-Cong
Huang
and
Robert J.
Capon
*
Division of Chemistry and Structural Biology, Institute for Molecular Bioscience, The University of Queensland, St. Lucia, Queensland, 4072, Australia. E-mail: r.capon@uq.edu.au; Tel: +61 7 3346 2979; Fax: +61 7 3346 2090
First published on 22nd February 2012
Abstract
Chemical analysis of a southern Australian marine sponge, an Ianthella sp., yielded dictyodendrins F–J (1–5) as new examples of a rare class of marine alkaloid. Structures were assigned on the basis of detailed spectroscopic analysis, while biosynthetic considerations suggested a relationship between the dictyodendrins and co-metabolites belonging to the lamellarin and ianthellidone structure classes. The dictyodendrins 1 and 3–5 exhibited significant BACE inhibitory activity (IC50 1–2 μM), with the differential cytotoxicity displayed by 1–4 towards two human colon cancer cell lines (IC50 2–16 μM) marking them as both cytotoxins and probable substrates for the multi-drug resistance efflux pump P-glycoprotein. The dictyodendrins 1–5 did not inhibit growth of Gram −ve bacteria or fungi, but 1, 3, and 4 were selective Gram +ve antibacterials (IC50 1–3 μM). Dictyodendrin J (5), with its unique seco-carbon skeleton and unusual 1,2-diketone functionality, exhibited a promising non-cytotoxic biological activity profile, inclusive of significant BACE inhibitory activity (IC50 2 μM), supportive of further investigation.
Introduction
Neurodegenerative diseases present a very significant challenge to modern healthcare, none more so than Alzheimer's disease (AD) where patients are confronted with progressive and irreversible deterioration of cognitive ability, and where medical intervention is largely limited to palliative care.1 Since its discovery late last century the protease β-secretase (BACE) has been recognized for its role in processing the essential neuronal transmembrane amyloid precursor protein (APP), releasing the peptide beta-amyloid (Aβ) that in turn drives the formation of amyloid plaques.1 In addition to being a dominant AD pathology, amyloid plaques are believed to be responsible for AD symptoms and disease progression.1 Discovery of the key role played by BACE raises the hope that chemical inhibitors could lower Aβ levels, reducing amyloid plaque pathology and mediating the onset and severity of AD. Although an attractive hypothesis, clinically useful BACE inhibitors have proved elusive.In an attempt to contribute to the discovery of novel small molecule BACE inhibitors we set out to test the proposition that marine natural products might deliver such inhibitors. This was achieved by screening a collection of ∼2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 southern Australian and Antarctic marine invertebrates and algae, to detect 27 extracts (1%) that exhibited BACE inhibitory properties. Prioritization of these extracts by chemical (HPLC-DAD-MS) and spectroscopic (1H NMR) profiling drew our attention to a particularly noteworthy marine sponge, Ianthella sp. (CMB-01245), collected during scientific trawling operations in Bass Strait, Australia. A portion of the aqueous EtOH extract from this specimen was concentrated in vacuo and the residue subjected to fractionation by solvent partitioning and sequential trituration, followed by reverse phase chromatography (SPE and HPLC). In a preliminary report2 we described sixteen metabolites from this Ianthella sp., consisting of six examples of a new class of pyrrolidone, ianthellidones A–F, two examples of a new class of furanone, ianthellidones G–H, two new and two known examples of the lamellarin class of pyrrole alkaloid, lamellarins O1, O2, O and Q, plus the known aromatics 4-hydroxybenzaldehyde, 4-hydroxybenzoic acid, 4-methoxybenzoic acid and ethyl 4-hydroxybenzoate.
600 southern Australian and Antarctic marine invertebrates and algae, to detect 27 extracts (1%) that exhibited BACE inhibitory properties. Prioritization of these extracts by chemical (HPLC-DAD-MS) and spectroscopic (1H NMR) profiling drew our attention to a particularly noteworthy marine sponge, Ianthella sp. (CMB-01245), collected during scientific trawling operations in Bass Strait, Australia. A portion of the aqueous EtOH extract from this specimen was concentrated in vacuo and the residue subjected to fractionation by solvent partitioning and sequential trituration, followed by reverse phase chromatography (SPE and HPLC). In a preliminary report2 we described sixteen metabolites from this Ianthella sp., consisting of six examples of a new class of pyrrolidone, ianthellidones A–F, two examples of a new class of furanone, ianthellidones G–H, two new and two known examples of the lamellarin class of pyrrole alkaloid, lamellarins O1, O2, O and Q, plus the known aromatics 4-hydroxybenzaldehyde, 4-hydroxybenzoic acid, 4-methoxybenzoic acid and ethyl 4-hydroxybenzoate.
Although our preliminary study established lamellarin O1 as a BACE inhibitor (IC50 ∼10 μM), we nevertheless concluded that the dominant BACE inhibitory metabolites were neither ianthellidones nor lamellarins. In this report we continue our investigations into this Ianthella sp., describing the isolation, characterization, structure elucidation and biological properties of the principle BACE inhibitory agents as five new examples of a rare class of marine alkaloid, dictyodendrins F–J (1–5) (Fig. 1).
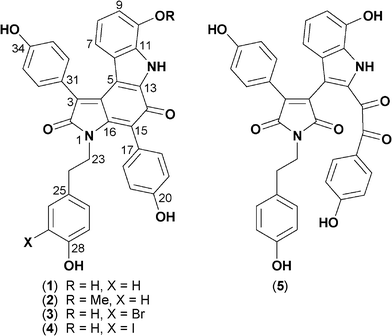 | ||
| Fig. 1 Structures of dictyodendrins F–J (1–5). | ||
Dictyodendrin nomenclature was first coined in 2003 by Fusetani et al. who described dictyodendrins A–E (6–10) as telomerase inhibitors from a Japanese marine sponge, Dictyodendrilla verongiformis,3 albeit acknowledging two structurally related alkaloids, 11, 12, described a decade earlier by Sato et al. as aldose reductase inhibitors from another Japanese Dictyodendrilla sp.4 Although the structural novelty and biological properties of the rare marine alkaloids 6–12 (Fig. 2) attracted the attention of synthetic and medicinal chemists,5 to date there have been no further reports of natural products belonging to the “dictyodendrin” family, impeding access to and exploration of this unusual pharmacophore.
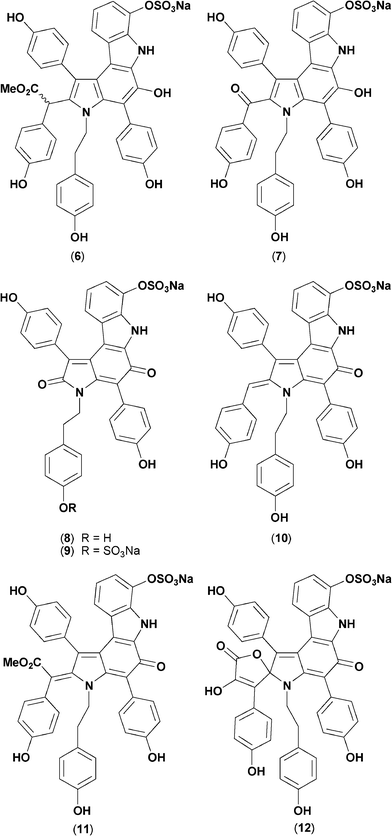 | ||
| Fig. 2 Structures of known “dictyodendrins” 6–12. | ||
Results and discussion
The structures for dictyodendrins F–J (1–5) were determined by spectroscopic analysis, with a detailed account of the structure elucidation shown below. The 1H and 13C NMR data for 1–5 are summarized in Table 1 and 2 respectively, with more detailed documentation of 1D and 2D NMR data provided in the ESI.†| Pos. | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| a Measured in pyridine-d5. b Measured in methanol-d4. c (d, J 8.2). d (d, J 8.3). e (dd, J 8.1, 0.7). f (dd, J 8.2., 7.5). g (dd, J 8.3, 7.7). h (dd, J 8.2, 7.6). i (dd, J 8.1, 7.5). j (d, J 7.5). k (d, J 7.7). l (d, J 7.6). m (dd, J 7.5, 0.7). n (d, J 8.5). o (d, J 8.9). p (t, J 8.1). q (t, J 8.2). r (t, J 7.6). s (br). t (d, J 2.1). u (d, J 1.9). v (dd, J 8.2, 2.1). w (dd, J 8.2, 1.9). x (d, J 8.4). y (d, J 8.6). z (s). | |||||
| 7 | 6.66c | 6.64d | 6.64c | 6.65c | 6.53e |
| 8 | 6.96f | 6.91g | 6.94h | 6.93h | 6.82i |
| 9 | 7.05j | 6.76k | 7.06l | 7.06l | 6.71m |
| 18/22 | 7.65d | 7.62c | 7.63n | 7.62d | 7.76o |
| 19/21 | 7.36d | 7.35c | 7.38n | 7.38d | 6.79o |
| 23 | 3.86p | 3.83p | 3.81q | 3.81p | 3.57r |
| 24 | 2.76p | 2.73p | 2.69q | 2.68p | 2.58s |
| 26 | 7.05d | 7.03d | 7.37t | 7.66u | 6.96n |
| 27 | 7.09d | 7.09d | 6.70n | ||
| 29 | 7.09d | 7.09d | 7.08c | 7.02c | 6.70n |
| 30 | 7.05d | 7.03d | 6.92v | 6.95w | 6.96n |
| 32/36 | 7.92d | 7.88x | 7.91y | 7.92n | 7.35o |
| 33/35 | 7.38d | 7.39x | 7.39y | 7.39n | 6.59o |
| 10-OMe | 3.86z | ||||
| Pos. | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| a Measured in pyridine-d5. b Measured in methanol-d4. c Interchangeable signals within the column. d Interchangeable signals within the column. e Interchangeable signals within the column. f Interchangeable signals within the column. g Signals not detected. | |||||
| 2 | 172.3 | 172.2 | 172.1 | 172.3 | 172.3 |
| 3 | 128.9 | 129.2 | 128.8 | 128.8 | 138.5 |
| 4 | 134.9c | 134.6d | 134.9e | 129.6f | |
| 5 | 114.0 | 113.9 | 113.8 | 113.9 | 114.8 |
| 6 | 126.7 | 126.0 | 126.6 | 126.6 | 129.4 |
| 7 | 116.4 | 117.7 | 116.3 | 116.4 | 114.2 |
| 8 | 122.9 | 122.4 | 122.9 | 123.0 | 123.7 |
| 9 | 109.9 | 105.5 | 109.9 | 109.9 | 111.2 |
| 10 | 147.1 | 148.4 | 146.9 | 147.0 | 145.9 |
| 11 | 131.7 | 131.0 | 131.5 | 131.6 | 130.6 |
| 13 | 134.2c | 134.2d | 134.1e | ,f | |
| 14 | 180.4 | 180.2 | 180.3 | 187.0 | |
| 15 | 118.5 | 118.5 | 118.4 | 118.5 | 192.4 |
| 16 | 149.6 | 149.4 | 149.2 | 149.4 | 172.3 |
| 17 | 124.2 | 124.0 | 124.0 | 124.0 | 126.0 |
| 18/22 | 133.9 | 133.8 | 133.8 | 133.8 | 134.3 |
| 19/21 | 116.3 | 116.3 | 116.3 | 116.3 | 116.8 |
| 20 | 159.8 | 159.7 | 159.7 | 159.8 | 165.5 |
| 23 | 44.0 | 43.9 | 43.6 | 43.7 | 41.0 |
| 24 | 35.0 | 35.0 | 34.5 | 34.3 | 34.8 |
| 25 | 129.6 | 129.4 | 131.1 | 131.8 | 130.5 |
| 26 | 130.9 | 130.8 | 134.1 | 140.3 | 131.0 |
| 27 | 116.6 | 116.5 | 111.3 | 86.3 | 116.4 |
| 28 | 157.9 | 157.8 | 154.4 | 157.2 | 157.2 |
| 29 | 116.6 | 116.5 | 117.0 | 115.8 | 116.4 |
| 30 | 130.9 | 130.8 | 129.8 | 130.8 | 131.0 |
| 31 | 123.7 | 123.5 | 123.6 | 123.6 | 121.9 |
| 32/36 | 133.8 | 133.7 | 133.8 | 133.8 | 132.9 |
| 33/35 | 116.6 | 116.6 | 116.6 | 116.5 | 116.4 |
| 34 | 160.8 | 160.8 | 160.7 | 160.7 | 160.8 |
| 10-OMe | 55.8 | ||||
High resolution ESI(+)MS analysis of 1 returned a pseudo molecular ion [M + Na]+ consistent with a molecular formula (C34H24N2O6, Δmmu −1.0) requiring twenty-four double bond equivalents (DBE). Analysis of the NMR (pyridine-d5) data for 1 revealed resonances attributed to four aromatic sub-structure fragments A–D (Fig. 3), with deshielded carbon resonances requiring that fragment C incorporate an amide/lactam residue (δC 172.3, C-2), and that all fragments bear phenolic residues (δC 147.1, C-10; 159.8, C-20; 157.9, C-28; 160.8, C-34). The remaining NMR resonances comprised three fully substituted sp2 carbons, attributed to two olefinic carbons (δC 134.9, C-4; 134.2, C-13) and a ketone (δC 180.4, C-14), together with five deshielded exchangeable 1H NMR resonances. These observations (Fig. 3) accounted for the molecular formula and required that 1 was heptacyclic. On reviewing the literature we noted that 1 was isomeric with and possessed identical 1H NMR (methanol-d4) data to a known acid hydrolysis product of 6–10.3 As 6–10 were not detected in the Ianthella extract we propose dictyodendrin F (1) as a natural product.
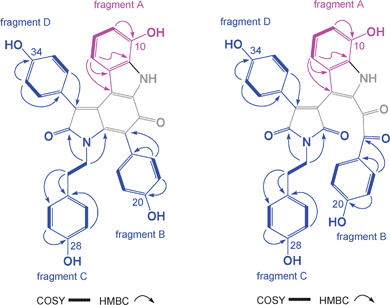 | ||
| Fig. 3 Key 2D NMR correlations and structure fragments for 1 and 5. | ||
High resolution ESI(+)MS analysis of 2 returned a pseudo molecular ion [M + Na]+ consistent with a molecular formula (C35H26N2O6, Δmmu −1.4) for a methylated analogue of 1. Comparison of the NMR (pyridine-d5) data for 2 with 1 revealed a high level of similarity with the only significant difference being attributed to resonances (δH 3.86, s; δC 55.8) and diagnostic shielding (H-9, ΔδH −0.29; C-9, ΔδC −4.4) for a 10-OMe moiety. These considerations permitted assignment of the structure for dictyodendrin G (2) as shown.
High resolution ESI(+)MS analysis of 3 returned a pseudo molecular ion [M − H]− consistent with a molecular formula (C34H23BrN2O6, Δmmu 1.9) for a mono brominated analogue of 1. Comparison of the NMR (pyridine-d5) data for 3 with 1 revealed a high level of similarity with the only significant difference being attributed to a C-27 bromo substituent (δH 7.37, d, J = 2.1 Hz, H-26; 7.08, d, J = 8.2 Hz, H-29; 6.92, dd, J = 8.2, 2.1 Hz, H-30; with HMBC correlations from H2-24 to C-25 and C-26/30). These considerations permitted assignment of the structure for dictyodendrin H (3) as shown.
High resolution ESI(+)MS analysis of 4 returned a pseudo molecular ion [M − H]− consistent with a molecular formula (C34H23IN2O6, Δmmu −1.8) for a mono iodinated analogue of 1. Comparison of the NMR (pyridine-d5) data for 4 with 3 revealed a high level of similarity with the only significant difference being a diagnostic shielding of C-27 (ΔδC–45) with HMBC correlations from H2-24 to C-25 and C-26/30. These considerations permitted assignment of the structure for dictyodendrin I (4) as shown.
High resolution ESI(+)MS analysis of 5 returned a pseudo molecular ion [M − H]− consistent with a molecular formula (C34H24N2O8, Δmmu 0.3) for an oxidized (+O2) analogue of 1.
Analysis of the NMR (methanol-d4) data for 5 revealed resonances consistent with structure fragments A–D described above for 1 (Fig. 3), albeit with significantly deshielded chemical shifts for C-15 (δC 192.4) and C-16 (δC 172.3), consistent with oxidative cleavage of Δ15,16 in 1 to yield a C-15 ketone and a C-16 lactam in 5. On the basis of these observations we propose the structure for dictyodendrin G (5) as shown.
As multiple examples of the dictyodendrin, lamellarin and ianthellidone structure classes were determined to be co-metabolites in Ianthella sp. (CMB-01245), this raises the prospect that these marine alkaloids share a common (or at least related) biosynthetic origin. In our earlier report2 we proposed that the ianthellidones were oxygen addition adducts of lamellarins, as illustrated in Fig. 4 with the relationship between ianthellidone F and lamellarin O. As an extension of that earlier hypothesis we now draw attention to the biosynthetic similarities between lamellarins and dictyodendrins. The biosynthesis of lamellarins can be viewed as a condensation between tyrosine and one or more substituted 4-hydroxyphenyl residues, further elaborated by a limited repertoire of cyclizations, oxidations and methylations. Likewise, the biosynthesis of dictyodendrins can be viewed in a similar fashion, as a condensation between tryptophan and one or more substituted 4-hydroxyphenyl residues. In this hypothesis the principle difference between the biosynthesis of lamellarins and dictydendrins is the choice of amino acid, tyrosine versus tryptophan, and in the case of the latter the appending of a substituted 4-hydroxyphenyl residue to C-2 of the tryptophan which then engages in an intramolecular decarboxylation and ring closure. The conserved nature of the proposed dictyodendrin biosynthesis is illustrated in Fig. 4, where dictyodendrin F (1) can be viewed as the biosynthetic precursor to dictyodendrin G (2) via methylation at the 10-OH, dictyodendrins H (3) and I (4) by bromination and iodination at C-27, and dictyodendrin J (5) by oxidative cleavage of Δ15,16. This biosynthetic hypothesis can also be extended to 6–12. The proposed biosynthetic relationship between lamellarins, ianthellidones and dictyodendrins has the potential to inform future biomimetic synthetic strategies.
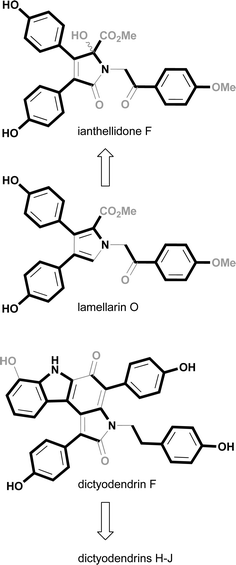 | ||
| Fig. 4 Plausible biosynthetic links between lamellarins, ianthellidones and dictyodendrins. | ||
The alkaloids 1–5 did not inhibit growth of the fungus Candida albicans (ATCC 90028), or the Gram −ve bacteria Escherichia coli (ATCC 11775) and Pseudomonas aeruginosa (ATCC 10145), or the Gram +ve bacteria Staphylococcus aureus (ATCC 9144 and ATCC 25923). Selected dictyodendrins did however inhibit growth of the Gram +ve bacteria Bacillus subtilis (ATCC 6051 and ATCC 6633); 1 (IC50 2.7 and 2.3 μM), 3 (IC50 1.2 and 3.1 μM) and 4 (IC50 2.5 and 2.8 μM).
On assessing the BACE inhibitory activity of dictyodendrins F–J (1–5) we noted that 1 and 3–5 were significant inhibitors (IC50 1–2 μM), while the 10-OMe analogue 2 was inactive (Table 3). This observation was noteworthy in that all the prior published members of this structure class (6–12) incorporate a 10-sulfate moiety. The BACE inhibitory results for 1–5 contrast sharply with cytotoxicity data as measured against the human colon cancer cell line SW620, and the P-glycoprotein (P-gp) over-expressing multi-drug resistant variant SW620 Ad300 (Table 3). Dictyodendrins 1–4 were cytotoxic to SW620, with the 10-OMe analogue 2 being the most cytotoxic, while the ring opened analogue 5 was not cytotoxic to either cell line. That the cytotoxicity displayed by 1–4 towards SW620 did not translate to SW620 Ad300 strongly suggested that these alkaloids were P-gp substrates, and as such were subject to efficient P-gp mediated efflux. The latter observation is significant as P-gp is expressed at high levels in endothelial cells at the blood brain barrier (BBB), where its broad substrate recognition and high transport capacity provide cellular protection against many endogenous and exogenous toxins. Clinically useful BACE inhibitors need to penetrate the BBB and not be subject to P-gp mediated efflux.
| BACE | SW620 | SW620 Ad300 | |
|---|---|---|---|
| a = not active | |||
| (1) | 1.5 | 8.5 | >30 |
| (2) | 2.0 | >30 | |
| (3) | 1.0 | 16 | >30 |
| (4) | 2.0 | 10 | >30 |
| (5) | 2.0 | >30 | >30 |
Conclusions
In summary, the Bass Strait sponge Ianthella sp. (CMB-01245) has proved to be a rich source of marine alkaloids spanning three structure classes, lamellarins,2 ianthellidones2 and dictyodendrins. In this report we describe the isolation, characterization and structure elucidation of dictyodendrins F–J (1–5), as new examples of a very rare class of alkaloid for which the only known natural examples were reported in 2003 (6–10)3 and 1993 (11,12)4 from Japanese sponges of the genus Dictyodendrilla. We propose a plausible biosynthetic relationship between all Ianthella sp. (CMB-01245) alkaloids, and in doing so highlight the future potential for convergent biomimetic syntheses. Our biological investigations have established that 1 and 3–5 are the dominant BACE inhibitory metabolites (IC50 1–2 μM), and highlight the critical structure activity significance of the 10-OH moiety. Extending these studies we determined that 1–4 are cytotoxic to a human colon cancer cell line, and are likely substrates for the multi-drug resistance efflux pump P-glycoprotein. We also determined that 1, 3 and 4 are selective Gram +ve antibacterials (IC50 1–3 μM). Our studies reveal that the non-cytotoxic, non-antibacterial dictyodendrin J (5), with its unique seco-carbon skeleton and unusual 1,2-diketone functionality, is a promising BACE inhibitor scaffold deserving further investigation.Experimental
General experimental procedures
Optical rotations were measured on a JASCO P-1010 polarimeter with a 10 cm length cell. UV spectra were obtained on a Cary 50 UV-visible spectrophotometer with 1 cm pathway cell. NMR experiments were performed on a Bruker Avance DRX600 spectrometer and are referenced to residual 1H signals in the deuterated solvents. ESIMS experiments were carried out on an Agilent 1100 series LC/MSD instrument. HR-ESIMS data were acquired on a Bruker micrOTOF mass spectrometer by direct infusion in MeCN at 3 μL min−1 using sodium formate clusters as an internal calibrant. All HPLC analyses and purifications were performed on Agilent 1100 series LC instruments with corresponding detectors, collectors and software. All chemicals were purchased from Merck, Sigma-Aldrich or Fluka. Solvents used for general purposes were of at least analytical grade, and solvents used for HPLC were of HPLC grade. Agilent Zorbax SB-C18 (5μm, 4.6 × 150 mm (analytical) and 9.4 × 250 mm (semi-preparative)) columns were used for HPLC analyses and separations. An Alltech Extract-Clean 5 g C18 cartridge was used for crude fractionation.Animal material (collection and taxonomy)
The sponge CMB-01245 was collected in May 1991 during a scientific trawling expedition in Bass Strait, Australia, aboard the RV Franklin. The description of the sponge is as follows: growth form – stalked, biplanar, anastomosing branches 5–10 mm thick; colour on deck and in EtOH – dark green–black, probable aerophobic colour change; texture – soft and flexible in life, light but tough and fibrous once preserved in EtOH; oscules – 1–2 mm diameter, sunken between conules; surface – opaque, membranous, conulose, fibre reticulation visible at the surface; spicules – none; ectosome – collagenous with fibre tips protruding; choanosome – a rectangular reticulation of thick spongin fibres 50–100 μm in diameter, branched and anastomosing, laminated, secondary connecting fibres slightly smaller. Light collagen in the mesohyl filled with green pigment granules. Choanocyte chambers eurypylous.Based on this analysis the sponge CMB-01245 was identified as an Ianthella sp. (Order Verongida, Family Ianthellidae; Museum Victoria Registry No. MVF167496).
Extraction, fractionation and isolation
The initial extraction and partition procedures were as previously reported.2 SPE and HPLC fractionation on the CH2Cl2 soluble fraction afforded lamellarin O, ianthellidones A, C, D, E–H, along with 4-hydroxybenzaldehyde, 4-hydroxybenzoic acid, 4-methoxybenzoic acid and ethyl 4-hydroxybenzoate.2A portion of the MeOH solubles (116 mg) was eluted through an Alltech C18 SPE cartridge using a stepwise gradient elution (as above) to return five fractions. HPLC separation on the second fraction yielded lamellarins O, Q, O1 and O2, as well as ianthellidones B and F.2 The third fraction (5.7 mg) was separated by a Zorbax SB-C18 semi-preparative column (4.0 mL min−1, 15 min 60–40% H2O–MeCN gradient elution) to yield dictyodendrin F (1, 0.6 mg) and dictyodendrin J (5, 0.4 mg). The fourth fraction (34.5 mg) was separated by a Zorbax SB-C18 semi-preparative column (4.0 mL min−1, 15 min 60–40% H2O–MeCN gradient elution) to yield (in order of elution) dictyodendrin F (1, 10.5 mg), a mixture (1.6 mg), dictyodendrin H (3, 0.7 mg), dictyodendrin I (4, 1.2 mg) and dictyodendrin G (2, 3.2 mg). The 1.6 mg of mixture noted above was further purified by a Prep-C18 Scalar column (1.0 mL min−1, 15 min 90–60% H2O–MeOH gradient elution, with 0.01% constant TFA) to return another portion of dictyodenrin J (5, 0.6 mg).
An isolation scheme is provided in the ESI illustrating the whole fractionation process including yields for all co-metabolites.†
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) [M + H]+, ESI(−)MS m/z 633/635 (1
1) [M + H]+, ESI(−)MS m/z 633/635 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) [M − H]−; HR-ESI(−)MS m/z 633.0686 [M − H]− (calcd for C34H2279BrN2O6, 633.0667).
1) [M − H]−; HR-ESI(−)MS m/z 633.0686 [M − H]− (calcd for C34H2279BrN2O6, 633.0667).
Bioassays
BACE1 assay
The TruPoint Beta-Secretase Assay was conducted according to manufacturer's instructions (Perkin Elmer, AD0258). Briefly, compounds of interest were dispensed in duplicate into a 384 well plate at the desired concentrations (7% DMSO). BACE (0.3 mU μL−1; Invitrogen, P2947) was added before incubating the plate at room temperature for 30 min. The reaction was started by the addition of substrate (200 nmol L−1) to a final assay volume of 30 μL. The assay plate was incubated for 24 h at room temperature, protected from light and covered with parafilm to prevent evaporation. The assay was read at 340/615 nm using an Envision plate reader. Concentrations given are for final assay conditions.Antibacterial assay
The bacterium to be tested was streaked onto a tryptic soy agar plate and was incubated at 37 °C for 24 h. One colony was then transferred to fresh tryptic soy broth (15 mL) and the cell density was adjusted to 104–105 cfu/mL. Test compounds were dissolved in DMSO and diluted with H2O to give a 300 μM stock solution (10% DMSO). The stock solution was then serially diluted with 10% DMSO to give final concentrations of 30 μM to 0.01 μM in 1% DMSO. An aliquot (20 μL) of each dilution was transferred to a 96-well microtitre plate and freshly prepared microbial broth (180 μL) was added to each well. The plates were incubated at 37 °C for 24 h and the optical density of each well was measured spectrophotometrically at 600 nm. Each test compound was screened against the Gram-negative bacterium Escherichia coli (ATCC 11775) and Pseudomonas aeruginosa (ATCC 10145), and the Gram-positive bacteria Staphylococcus aureus (ATCC 9144 and ATCC 25923) and Bacillus subtilis (ATCC 6633 and ATCC 6051).Antifungal assay
The fungus to be tested was streaked onto a Sabouraud agar plate and was incubated at 26.5 °C for 48 h. One colony was then transferred to fresh Sabouraud broth (15 mL) and the cell density was adjusted to 104–105 cfu/mL. Test compounds were dissolved in DMSO and diluted with H2O to give a 300 μM stock solution (10% DMSO). The stock solution was then serially diluted with 10% DMSO to give final concentrations of 30 μM to 0.01 μM in 1% DMSO. An aliquot (20 μL) of each dilution was transferred to a 96-well microtitre plate and freshly prepared microbial broth (180 μL) was added to each well. The plates were incubated at 26.5 °C for 48 h and the optical density of each well was measured spectrophotometrically at 600 nm. Each test compound was screened against the fungus Candida albicans (ATCC 90028).Cell lines and cell culture
The parental cell line SW620 (American Type Culture Collection, Manassasm VA, CCL-227), is a human colon cell line that was originated from a lymph node metastasis in the patient with primary adenocarcinoma of the colon. The multi-drug resistant (MDR) cell line, SW620 Ad300, which over-expresses Permeable-glycoprotein (P-gp), was selected from SW620 by growth in the presence of increasing concentrations of doxorubicin. SW620 and SW620 Ad300 were grown in RPMI medium 1640 as adherent mono-layer in flasks supplemented with 10% foetal bovine serum, 2 mM L-glutamine, 100 unit/mL penicillin and 100 μg mL−1 streptomycin in a humidified incubator containing of 5% CO2 at 37 °C. After SW620 Ad300 exhibited stable phenotype of P-gp, the cells were maintained in 300 ng mL−1 doxorubicin.MTT cytotoxicity assay
The MTT assay was used to evaluate the cytotoxicity of compounds against cancer cell. This assay was modified slightly from that previously described.6 Briefly, cells (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000/well in 180 μL of RPMI 1640 supplemented with 10% FBS) were seeded evenly in a 96-well micro-plate, and the plate was incubated for 18 h (37 °C; 5% CO2) to allow cells to attach. Compounds to be tested were dissolved in 5% DMSO (v/v) and diluted from 300 μM–300 nM. Aliquots (20 μL) of each dilution (or of 5% aqueous DMSO for control wells) were added to the plate in duplicate. After 68 h of incubation (37 °C; 5% CO2), a solution of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma, USA) in PBS was added to each well to a final concentration of 0.4 mg mL−1 and the plate was incubated for a further 4 h (37 °C; 5% CO2). After that, the medium was carefully aspirated and precipitated formazan crystals were dissolved in DMSO (100 μL/well). Finally, the absorbance of each well at 580 nm was measured with a PowerWave XS Microplate Reader from Bio-Tek Instruments Inc. (Vinooski, VT). The IC50 value was calculated as the concentration of the compound required for 50% inhibition of the cancer cells using Prism 5.0 from GraphPad Software Inc. (La Jolla, CA).
000/well in 180 μL of RPMI 1640 supplemented with 10% FBS) were seeded evenly in a 96-well micro-plate, and the plate was incubated for 18 h (37 °C; 5% CO2) to allow cells to attach. Compounds to be tested were dissolved in 5% DMSO (v/v) and diluted from 300 μM–300 nM. Aliquots (20 μL) of each dilution (or of 5% aqueous DMSO for control wells) were added to the plate in duplicate. After 68 h of incubation (37 °C; 5% CO2), a solution of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma, USA) in PBS was added to each well to a final concentration of 0.4 mg mL−1 and the plate was incubated for a further 4 h (37 °C; 5% CO2). After that, the medium was carefully aspirated and precipitated formazan crystals were dissolved in DMSO (100 μL/well). Finally, the absorbance of each well at 580 nm was measured with a PowerWave XS Microplate Reader from Bio-Tek Instruments Inc. (Vinooski, VT). The IC50 value was calculated as the concentration of the compound required for 50% inhibition of the cancer cells using Prism 5.0 from GraphPad Software Inc. (La Jolla, CA).
Acknowledgements
The authors would like to thank the CSIRO Division of Oceanography and crew of the RV Franklin for the collection of Ianthella sp. (CMB-01245), L. Goudie (Museum Victoria) for sponge taxonomy, Noscira (Madrid, Spain) for BACE inhibitory screening of crude marine extracts, S. Bates and R. Robey (NCI) for providing the SW620 and SW620 Ad300 cell lines, and A. M. Piggott (IMB, UQ) for the acquisition of HRESIMS data. X.-C. Huang and Z. Khalil acknowledge the provision of International Postgraduate Scholarships from The University of Queensland. This work was funded partially by the Institute for Molecular Bioscience, the University of Queensland, the Australian Research Council (ARC LP0775547) and Noscira (Madrid, Spain).References
- (a) M. Goedert and M. G. Spillantini, Science, 2006, 314, 777–781 CrossRef CAS; (b) E. D. Roberson and L. Mucke, Science, 2006, 314, 781–784 CrossRef.
- H. Zhang, M. M. Conte, X.-C. Huang, Z. Khalil and R. J. Capon, Org. Biomol. Chem., 2012, 10, 2656–2663 CAS.
- K. Warabi, S. Matsunaga, R. W. M. van Soest and N. Fusetani, J. Org. Chem., 2003, 68, 2765–2770 CrossRef CAS.
- A. Sato, T. Morishita, T. Shiraki, S. Yoshioka, H. Horikoshi, H. Kuwano, H. Hanzawa and T. Hata, J. Org. Chem., 1993, 58, 7632–7634 CrossRef CAS.
- (a) H. Tokuyama, K. Okano, H. Fujiwara, T. Noji and T. Fukuyama, Chem.–Asian J., 2011, 6, 560–572 CrossRef CAS; (b) K. Okano, H. Fujiwara, T. Noji, T. Fukuyama and H. Tokuyama, Angew. Chem., Int. Ed., 2010, 49, 5925–5929 CAS; (c) S. Hirao, Y. Yoshinaga, M. Iwao and F. Ishibashi, Tetrahedron Lett., 2010, 51, 533–536 CrossRef CAS; (d) S. Hirao, Y. Sugiyama, M. Iwao and F. Ishibashi, Biosci., Biotechnol., Biochem., 2009, 73, 1764–1772 CrossRef CAS; (e) P. Buchgraber, M. M. Domostoj, B. Scheiper, C. Wirtz, R. Mynott, J. Rust and A. Fürstner, Tetrahedron, 2009, 65, 6519–6534 CrossRef CAS; (f) C. Ayats, R. Soley, F. Albericio and M. Álvarez, Org. Biomol. Chem., 2009, 7, 860–862 RSC; (g) A. Frstner, M. M. Domostoj and B. Scheiper, J. Am. Chem. Soc., 2006, 128, 8087–8094 CrossRef; (h) A. Frstner, M. M. Domostoj and B. Scheiper, J. Am. Chem. Soc., 2005, 127, 11620–11621 CrossRef.
- J. Carmichael, W. G. DeGraff, A. F. Gazdar, J. D. Minna and J. B. Mitchell, Cancer Res., 1987, 47, 943–946 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Tabulated 1D and 2D NMR data for new compounds, along with 1H NMR spectra for all compounds. See DOI: 10.1039/c2ra20322g |
| ‡ Current address: Department of Natural Medicinal Chemsitry, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, 201203, P. R. China |
| This journal is © The Royal Society of Chemistry 2012 |
