MC-LR@HSA: non-covalent interaction and effect†
Chao
Song
a,
Yan-Qin
Zi
b and
Hong-Wen
Gao
*a
aState Key Laboratory of Pollution Control and Resource Reuse, College of Environmental Science and Engineering, Tongji University, Shanghai, 200092, China. E-mail: EMSL@tongji.edu.cn; Fax: (+)86-21-6598-8598
bSchool of Chemistry & Materials, Huaibei Normal University, Anhui, 235000, China
First published on 26th June 2012
Abstract
Three microcystin-LR molecules binding to subdomains IA, IIA, IIIA and IIB of human serum albumin (HSA) via hydrophobic interaction and hydrogen bonding induced the HSA conformation to transfer from α-helix to β-pleated sheet and random coils so that HSA transport of vitamin B2 was inhibited.
Intermolecular interactions exist extensively, including non-covalent interactions in nature.1 Microcystins (MCs) are widespread in lakes with many toxic algal blooms and coastal waters with red tides. For example, a large bloom of blue-green algae in Taihu Lake caused water quality to deteriorate severely in 2007.2 MC-LR (CAS no. 101043-37-2), the chemical structure of which is shown in Fig. 1A, is the most abundant and most toxic worldwide,3 produced by several species of cyanobacteria.4 It is recognized as an inducer of potent environmental stress in aquatic ecosystems and as a potential threat to human health.5 A few studies have suggested a relationship between liver and colorectal cancers and the occurrence of cyanobacteria in drinking water.6 Once MCs are ingested, most travel to the liver via the bile acid transport system and are stored. However, some remain in the blood stream and bind to human serum albumin (HSA). HSA is the major transport protein in the circulation system. It is a globular protein composed of 585 amino acid residues in three homologous α-helical domains (I, II and III) where each contains 10 helices, and is divided into six antiparallel helices and two subdomains (A and B).7 As a multifunctional plasma carrier, it can bind a wide variety of ligands, including amino acids, vitamins, fatty acids, bilirubin, and bile acids.8 Investigating the interaction of a chemical with HSA can elucidate the properties of a chemical–protein complex9 and provide useful information about the structural features that determine the therapeutic effectiveness of the chemical. As a much more complicated cyclic seven nonribosomal peptide than common chemicals, MC-LR contains many hydrophobic groups in addition to a few hydrophilic groups. There have been many studies about the toxicity-causing of MC-LR, most of which have investigated dose–effect relationships on exposing model animals by observation of physiological behavior.10 The aim of this work is to understand the toxicological interaction of a harmful peptide at the molecular level by investigating the interaction of MC-LR with HSA.
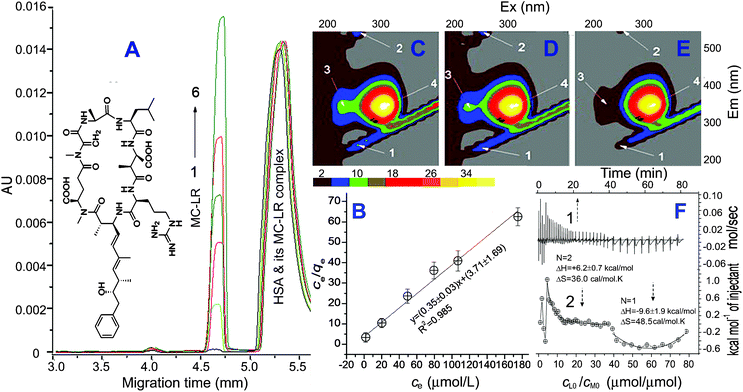 | ||
| Fig. 1 Electropherogram of 5.0 μM HSA mixed with MC-LR (1 to 6: 5, 30, 60, 90, 120 and 190 μM) (A) and plot of ce/qevs. ce (B). 3D-fluorescence of the MC-LR/HSA solutions (C to E: 0, 10, 50 and 90 μM MC-LR, all 5 μM HSA) and ITC curves (F) (1-time effect and 2-effect of cLo/cMo. The experiment was conducted by injecting 2 μL of 2.0 mM MC-LR every time into 200 μL of the ITC cell containing 10.0 μM HSA at pH 7.4 and 26 °C.). | ||
From capillary electrophoresis (CE) of the MC-LR/HSA interaction solutions, the mobility of the MC-LR@HSA complex approached that of HSA because the mass of MC-LR is much less than that of HSA. Thus, MC-LR free in solution (ce) was determined at equilibrium from the peak area (Fig. S1, ESI†, Fig. 1A). A plot of ce/qevs. ce (the symbol qe is the number of MC-LR binding to one HSA) indicated that the binding of MC-LR obeyed a Langmuir adsorption isotherm (Fig. 1B). The saturation mole number (N) of MC-LR binding to HSA was calculated to be 2.8 ± 0.1 and the adsorption constant (Ka) to be (1.6 ± 0.6) × 105 M−1. Three MC-LR molecules bound to one HSA. The high Ka value indicated that a strong adsorption occurred between MC-LR and HSA.
3D-fluorescence is often used to investigate a change of protein structure on interacting with small molecules.11 From Fig. 1C–E, peaks 1 and 2 as the Rayleigh scattering (λEx/λEm at 250 nm/250 nm) and the second-ordered one (250 nm/500 nm) decreased with an increase of MC-LR. This may be attributed to the fact that insertion of MC-LR emaciated HSA. Peak 3 (230 nm/330 nm) provided information on the π→π* transition of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O backbone in HSA.12 The disappearance of peak 3 (Fig. 1E) indicated that an obvious alteration of the peptide chain structure occurred in the presence of MC-LR.
O backbone in HSA.12 The disappearance of peak 3 (Fig. 1E) indicated that an obvious alteration of the peptide chain structure occurred in the presence of MC-LR.
Isothermal titration calorimetry (ITC) is one effective instrumentation for determining the thermodynamic factors of an intermolecular interaction.13 MC-LR consists of seven hydrophobic groups, two carboxyl groups and one amino group, and thus shows good hydrophobicity (Fig. 1).4 The interaction of MC-LR with HSA is endothermic when their initial mole ratio (cL0/cM0) is less than 20 μmol μmol−1 (Fig. 1F). In this process, two MC-LRs bound to HSA and the enthalpy change (ΔH) and entropy change (ΔS) were calculated to be +6.2 kcal mol−1 and +36 cal mol−1 K. Thus, non-covalent interaction occurred between MC-LR and HSA owing to the low ΔH.14 The HSA fluorescence quenched by 80% at 20 of cL0/cM0 (Fig. S2, ESI† and peak 4 in Fig. 1C–E), which reflected a variation of the microenvironment around the Trp and Tyr residues. One MC-LR inserts in the hydrophobic cavity of HSA from the opening (Fig. 2A, arrow 1). It bound around Try21415via hydrophobic interactions e.g. π–π stacking, where the hydrophobic amino acid residues (AARs) are concentrated. The possible connecting side-groups of HSA included Phe326, Leu327, Gly328, Phe330, Leu331, Val343, Val344, Leu345, Leu346, Leu347, Arg348 and Ala350 in subdomain IIB (Fig. 2B). In addition, carboxyl groups (–COOH) of MC-LR may be further fixed by connecting to the adjacent side-chain (–NHR) of alkaline AARs e.g. Lys351 in IIB and Lys212 in IIA and the amino group (–NH2) of MC-LR to Arg218 in IIA via charge attraction and hydrogen bonding. The other MC-LR may insert in HSA according to arrow 2 and the possible connecting side-groups included Leu430, Gly431, Gly434, Tyr452, Leu453, Ser454, Val456, Leu457 and Asn458 in IIIA (Fig. 2C). Similarly, the carboxyl and amino groups of MC-LR were further fixed by Arg428, Arg485 and Glu425. During the first process, the free energy change (ΔG) was calculated to be −4.4 kcal mol−1. The binding of MC-LR to HSA is spontaneous, driven by the entropy increase. With cL0/cM0 increasing to >40 μmol μmol−1, the MC-LR/HSA interaction turned into an exothermic reaction (Fig. 1F). In this process, ΔH, ΔS and ΔG were calculated to be −9.6 kcal mol−1, +48.5 cal mol−1 K and −21.7 kcal mol−1. The binding of MC-LR is spontaneous, driven by the combination of the entropy increase and enthalpy. It is possible for the previous two MC-LRs to destroy the conformation of HSA so that the third MC-LR bound readily to HSA according to arrow 3, confirmed below. The possible connecting groups included Leu22, Val23, Leu24, Ile25, Ala26, Phe27, Ala28, Ala158, Phe157, Phe156, Leu155, Leu154, Pre152 and Ala151 in IA (Fig. 2D). Similarly, carboxyl and amino groups of MC-LR were further fixed by Glu16, Lys20 and Arg145. As a result, the combination of some non-covalent interactions, including hydrophobic stacking, hydrogen bonding and charge attraction, made MC-LR firmly bind to HSA so that MC-LR@HSA owned a high Ka value.
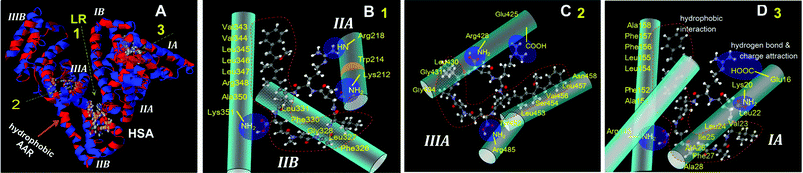 | ||
| Fig. 2 Cartoon illustration of the possible sites of MC-LR binding to HSA (A). The first MC-LR binding to subdomain IIA and IIB according to arrow 1 (B), the second one to IIIA according to arrow 2 (C) and the third one to IA according to arrow 3 (D). | ||
Circular dichroism (CD) is often used to evaluate the secondary structure of a protein.14 The percentage fractions of the conformational factors e.g. α-helix, β-pleated sheet, β-turn and random coils were calculated from CD curves (Fig.S3, ESI†), where two negative bands appeared at 208 and 222 nm. With the addition of MC-LR, the α-helix fraction of HSA reduced from 42.6% in the absence of MC-LR, 24.8% at 1 of cL0/cM0 down to 0 at 4 of cL0/cM0 (Fig. 3). On the other hand, the β-pleated sheet and random coils increased from 36.6 to 44.5 and 55.2%, and 20.8 to 30.7 and 44.8%. During insertion of the first MC-LR (Fig. 2A, arrow 1), the hydrophobic interactions with large numbers of hydrophobic groups in the cavity pulled the peptide chain which shifted. The original hydrogen bonds on helices were destroyed and the cavity narrowed, confirmed from the 3D-fluorescence analysis above. The great entropy increase found above is due to the transition of α-helix into β-pleated sheet and random coils. Thus, the 3rd MC-LR is more accessible for binding to HSA.
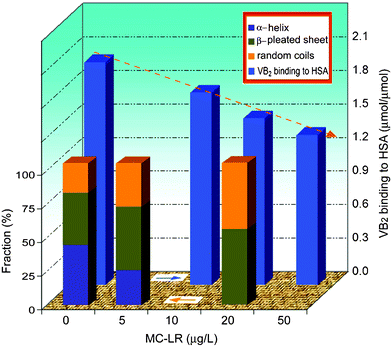 | ||
| Fig. 3 Fraction of α-helix, β-pleated sheet, β-turn and random coils of HSA (5.0 μM) in the presence of MC-LR at pH 7.4. Effect of MC-LR on HSA transporting VB2. All solutions contained 0.02 mM HSA and 0.08 mM VB2 at pH 7.4 and 26 °C. | ||
The number of MC-LR binding to HSA is much lower than that of small organic compounds14 but the potential site number of MC-LR connecting to HSA is enough. The similarity–compatibility law occurring between a cyclic seven nonribosomal peptide MC-LR and the peptide chain of HSA led to a subversive alteration of the HSA conformation. Certainly, a subversive conformation alteration will influence the physiological function and activity of a protein.15 The effect of MC-LR on HSA transport of vitamin B2 (VB2) is illustrated in Fig. 3 (Fig. S4, ESI†). With an increase of MC-LR, the binding number of VB2 to HSA decreases obviously under the normally physiological conditions. For example, the binding number of VB2e.g. approximately 2, decreased by 13% in 10 μM MC-LR up to one-third in 50 μM MC-LR. One reason is that MC-LR occupied competitively the original sites of VB2 binding to HSA. The other is due to the disintegration of α-helices in HSA decreasing the binding of VB2.
Conclusion
At present, eutrophication tends towards acceleration in rivers, lakes and coastal waters. Various algae breed abundantly and water blooms and red tides appear frequently. The entry of persistent and highly toxic MCs into water bodies is unstoppable. The already tense global shortage of fresh water is further aggravated. This work has demonstrated the toxicological interaction mechanism of MC-LR by various methods including CE, fluorometry, CD, ITC and equilibrium dialysis. The non-covalent interactions of MC-LR with HSA were determined to fit a Langmuir adsorption isotherm. The binding site of MC-LR was recognized and the thermodynamic characterization indicated the binding force and interaction type, in which hydrophobic interactions play the primary role. The binding of MC-LR causes a great alteration of HSA conformation and thus the physiological function of HSA transport of VB2 may be inhibited.Acknowledgements
We thank the Foundation of State Key Laboratory of Pollution Control and Resource Reuse (Tongji University, China) (No. PCRRK11003) for financial support of this work.Notes and references
- (a) M. E. Alberto, G. Mazzone, N. Russo and E. Sicilia, Chem. Commun., 2010, 46, 5894 RSC; (b) R. Cabot and C. A. Hunter, Chem. Commun., 2009, 2005 RSC; (c) I. K. Mati and S. L. Cockroft, Chem. Soc. Rev., 2010, 39, 4195 RSC.
- M. Yang, J. Yu, Z. Li, Z. Guo, M. Burch and T. F. Lin, Science, 2008, 319, 158a CrossRef.
- (a) J. Fastner, G. A. Codd, J. S. Metcalf, P. Woitke, C. Wiedner and H. Utkilen, Anal. Bioanal. Chem., 2002, 374, 437 CrossRef CAS; (b) J. Zhang, J. Lei, R. Pan, C. Leng, Z. Hu and H. Ju, Chem. Commun., 2011, 47, 668 RSC.
- (a) C. Djediat, M. Malecot, A. de Luze, C. Bernard, S. Puiseux-Dao and M. Edery, Toxicon, 2010, 55, 531 CrossRef CAS; (b) J. A. Kalaitzis, F. M. Lauro and B. A. Neilan, Nat. Prod. Rep., 2009, 26, 1447 RSC.
- (a) M. Tachi, S. Y. Imanishi and K. Harada, Environ. Toxicol., 2007, 22, 620 CrossRef CAS; (b) N. Gan, X. Sun and L. Song, Chem. Res. Toxicol., 2010, 23, 1477 CrossRef CAS; (c) O. A. Sadik and F. Yan, Chem. Commun., 2004, 1136 RSC.
- (a) L. Zhou, H. Yu and K. Chen, Biomed. Environ. Sci., 2002, 15, 166 Search PubMed; (b) B. H. Kim, S. J. Hwang, M. H. Park and Y. J. Kim, Bull. Environ. Contam. Toxicol., 2010, 85, 457 Search PubMed.
- X. M. He and D. C. Carter, Nature, 1992, 358, 209 CrossRef.
- (a) J. A. Hamilton, S. Era, S. P. Bhamidipati and R. G. Reed, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 2051 CAS; (b) A. A. Bhattacharya, S. Curry and N. P. Franks, J. Biol. Chem., 2000, 275, 38731 CrossRef CAS; (c) X. Wang, X. Wang, Y. Wang and Z. Guo, Chem. Commun., 2011, 47, 8127 RSC.
- (a) I. Petitpas, A. A. Bhattacharya, S. Twine, M. East and S. Curry, J. Biol. Chem., 2001, 276, 22804 CrossRef CAS; (b) S. L. Cockroft and C. A. Hunter, Chem. Soc. Rev., 2007, 36, 172 RSC; (c) V. M. Hernández-Rocamora, S. W. A. Reulen, B. Waal, E. W. Meijer, J. M. Sanz and M. Merkx, Chem. Commun., 2011, 47, 5997 RSC; (d) J. Hamblin, N. Abboyi and M. P. Lowe, Chem. Commun., 2005, 657 RSC.
- (a) A. Schwarzenberger, C. Courts and E. von Elert, BMC Genomics, 2009, 10, 527 Search PubMed; (b) R. Jayaraj, T. Anand and P. V. Rao, Toxicology, 2006, 220, 136 CrossRef CAS.
- U. S. Mote, S. H. Han, S. R. Patil and G. B. Kolekar, J. Lumin., 2010, 130, 2059 Search PubMed.
- A. N. Glazer and E. L. Smith, J. Biol. Chem., 1961, 236, 2942 Search PubMed.
- A. Shval and Y. Mastai, Chem. Commun., 2011, 47, 5735 RSC.
- F. F. Chen, Y. N. Tang, S. L. Wang and H. W. Gao, Amino Acids, 2009, 36, 399 CrossRef CAS.
- A. Desai, C. Lee, L. Sharma and A. Sharma, Biochimie, 2006, 88, 1435 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details and Fig. S1–S3. See DOI: 10.1039/c2ra20874a |
| This journal is © The Royal Society of Chemistry 2012 |
