Programmable multimetallic linear nanoassemblies of ruthenium–DNA conjugates†
Joris
Irvoas
ab,
Arielle
Noirot
ab,
Nadia
Chouini-Lalanne
ab,
Olivier
Reynes
cd,
Jean-Christophe
Garrigues
ab and
Valerie
Sartor
*ab
aUniversité de Toulouse; Université Paul Sabatier; Laboratoire IMRCP; Bat. II R1, 118 route de Narbonne, F-31062, Toulouse cedex 09, France. E-mail: sartor@chimie.ups-tlse.fr; Fax: +33 (0)5 61 55 81 55; Tel: +33 (0)5 61 55 25 62 74
bCNRS; Laboratoire IMRCP UMR 5623, F-31062, Toulouse cedex 09, France. E-mail: sartor@chimie.ups-tlse.fr; Fax: +33 (0)5 61 55 81 55; Tel: +33 (0)5 61 55 62 74
cUniversité de Toulouse; Université Paul Sabatier; Laboratoire de Genie Chimique; Bat. II R1, 118 route de Narbonne, F-31062, Toulouse cedex 09, France
dCNRS; LCG UMR 5503, F-31062, Toulouse cedex 09, France
First published on 2nd August 2012
Abstract
A new ruthenium–DNA conjugates family was synthesized, made up of a ruthenium complex bound to one or two identical DNA strands of 14–58 nucleotides. The formation of controlled linear nanoassemblies containing one to seven ruthenium complexes is described.
Introduction
Metals are largely used by chemists to make original and varied nanostructures by complexation with organic backbones.1,2 More recently, inorganic complexes were employed to stabilize, to study and to form DNA assemblies.3–5 The nature of the metals used is as diverse as the nature of the assemblies formed. Ruthenium is one of them due to the remarkable photophysical and photochemical properties of ruthenium complexes, which can be easily modulated by the nature of the ligands employed. This allowed researchers to develop a wide range of applications concerning these components, for example, dye-sensitized solar cells,6 cancer treatments and medical diagnosis7,8 and catalysis.9 Despite the potential of these complexes in various fields of scientific research, only few examples of DNA assemblies involving ruthenium compounds are described.10–15It was previously shown that the insertion of DNA backbones on terpyridine ruthenium complexes permitted the formation of linear arrays.11,12 The formation of cyclic structures has also been described. The control of the hybridization process allowed the formation of such constructions bearing one or two ruthenium molecules.13 Another original work reported the assembly of cyclic DNA architectures by using the tris(2,2′-bipyridine) ruthenium complex as a molecular template not being part of the structure, to control the duplex assembly.14 Furthermore, a star building block for future nanoassemblies was achieved. Indeed, a ruthenium tris(bipyridine) centre with six identical oligonucleotide arms was synthesized and the formation of six DNA double strands surrounding the complex was shown.15
Considering the potential of ruthenium complexes on varied applications and the development of DNA hybridization strategies to construct nanodevices, we developed a very simple synthesis pathway to obtain ruthenium–DNA conjugates and a flexible hybridization strategy to control the distance and the position of the functionalized building blocks. The synthetic pathway is only based on two reaction steps and on commercially available modified oligonucleotides and chemical products. This synthetic strategy relies on the use of the carboxylic acid moiety. As it is a commonly used functionalization, it allows us to easily modulate the nature of the molecules inserted into DNA, therefore broadening the range of possible applications. In this paper, we describe the work made with Ru(II)(2,2′-bipyridine)2(4,4′-dicarboxy-2,2′-bipyridine) complex.16 We developed a new family of ruthenium complexes bearing one or two DNA single strands with lengths varying between 14 and 58 nucleic bases. The modular hybridization strategy is based on the choice of the oligonucleotide sequences. They have been wisely designed to self-assemble into linear nanoarrays in a modular fashion to lead to original one-dimensional constructions with one to seven ruthenium complexes at different controlled positions and distances (Fig. 1).
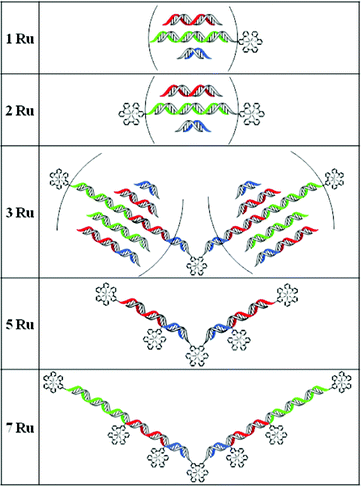 | ||
| Fig. 1 Multimetallic linear nanoassemblies bearing one to seven ruthenium complexes and DNA sequence lengths of 14–58 nucleotides. | ||
Results and discussion
Synthesis of ruthenium–DNA conjugates
This new series of ruthenium–DNA hybrids were synthesized by coupling oligonucleotides substituted by an amino hexyl linker in the 5′ position with a Ru(II)(2,2′-bipyridine)2(4,4′-dicarboxy-2,2′-bipyridine) complex.16 The amide link has been performed with the DNA strand kept on a solid support. The reaction took place using 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride (DMT-MM) in 0.8 M 3-morpholinopropane-1-sulfonic acid (MOPS) buffer (pH = 7) in DMSO/H2O solvents with a large excess of ruthenium complex.17 After deprotection and cleavage of crude DNA products from the solid support, analysis and purification were realized using denaturing polyacrylamide gel electrophoresis (PAGE). The polyacrylamide gels were directly observed under UV light at two wavelengths (254 and 365 nm), which were convenient to discriminate DNA and Ru–DNA conjugates. Indeed, at λ = 365 nm or on a Safe Imager™ 2.0 Blue-light transilluminator, the presence of a Ru centre induces a characteristic orange luminescence. The latter does not appear at λ = 254 nm, but DNA is still visible. Two molecules per reaction were isolated and characterized both carrying a ruthenium complex. The fast-moving orange band in the gel corresponds to the incorporation of one DNA sequence on the ruthenium complex. The slowest moving orange band in the gel was identified as the bis-DNA-functionalized ruthenium complex centre. With this experimental protocol, we obtained a family of ruthenium–DNA hybrids constituted of one 14 (1m and 2m), two 14 (2b), one 20 (3m and 4m), two 20 (4b), one 24 (5m and 6m), two 24 (6b), two 34 (7b) or two 58 (8b) nucleotide arms (Fig. 2).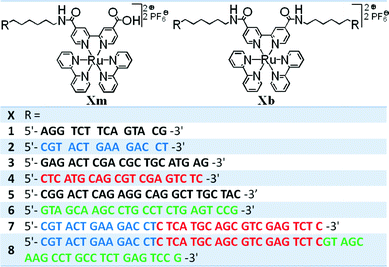 | ||
| Fig. 2 Synthesized mono-DNA-functionalized ruthenium complexes (Xm) and bis-DNA-functionalized ruthenium complexes (Xb) with their corresponding sequences. | ||
The modified oligonucleotides were characterized by PAGE, UV-Vis and fluorescence spectrophotometries and mass spectroscopy.
The polyacrylamide gel electrophoresis of this new family of ruthenium–DNA conjugates was visualized directly on a Safe Imager™ 2.0 Blue-light transilluminator (Fig. 3). The increase of the sequence lengths and the insertion of two DNA single strands on one ruthenium complex consequently showed a decreased migration length of the molecules in the gel.
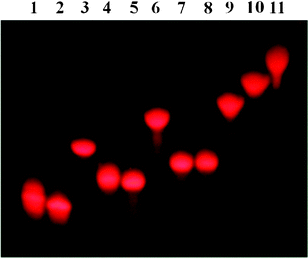 | ||
| Fig. 3 Denaturing PAGE analysis of ruthenium–DNA conjugates on a Safe Imager™ 2.0 Blue-light transilluminator. Lane 1: 1m; lane 2: 2m; lane 3: 2b; lane 4: 3m; lane 5: 4m; lane 6: 4b; lane 7: 5m; lane 8: 6m; lane 9: 6b; lane 10: 7b; lane 11: 8b. | ||
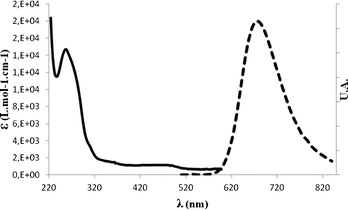
The UV-Vis absorption spectra of these conjugates show at λ = 260 nm the characteristic band of DNA oligomers and at λ = 280 nm and λ = 458 nm, the bipyridine π–π* and the complex MLCT (metal-to-ligand charge-transfer) transitions, respectively. The fluorescence spectra are all identical and characteristic of the ruthenium complex, namely having an emission peak at λ = 650 nm with an excitation wavelength at 458 nm. An example of UV-Vis and fluorescence spectra of a mono DNA-functionalized ruthenium complex is shown in Fig. 4.
High resolution electrospray mass spectra of 1m, 2m, 2b, 3m, 4m, 4b and 5m confirm the structure of these ruthenium–DNA conjugates (see in ESI†). The compounds 6m, 6b, 7b and 8b did not provide HRMS. However, the migration of their bands in PAGE compared to the fully characterized compounds and the presence of a ruthenium signal in UV-Vis and fluorescence spectrophotometries provided convincing evidence and characterizations of theirs structures.
Multimetallic linear nanoassemblies
The selective and specific hybridization of these ruthenium–DNA conjugates afforded a variety of linear assemblies. They provided DNA double helices with a length of 14 to twice 58 base pairs and made up of one to seven ruthenium polypyridine complexes (Fig. 1). They were highlighted by native PAGE (Fig. 5 and 6 and in ESI†).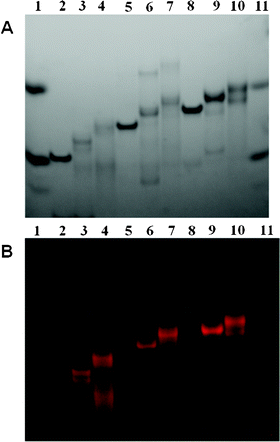 | ||
| Fig. 5 Non-denaturing PAGE analysis of linear assemblies with 0, 1 and 2 ruthenium complexes with λ = 254 nm (A) and on Safe Imager™ (B). Lanes 1 and 11: two products for migration control, upper: xylene cyanol, lower: bromophenol blue; lane 2: 1 + 2; lane 3: 1m + 2; lane 4: 1m + 2m; lane 5: 3 + 4; lane 6: 3m + 4; lane 7: 3m + 4m; lane 8: 5 + 6; lane 9: 5m + 6; lane 10: 5m + 6m. | ||
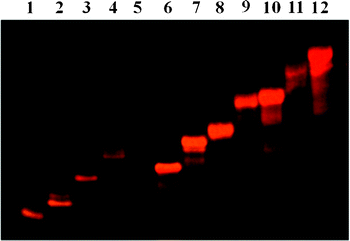 | ||
| Fig. 6 Non-denaturing PAGE analysis of multimetallic linear assemblies on Safe Imager™. Lane 1: 2b; lane 2: 4b; lane 3: 6b; lane 4: 7b; lane 5: 8b; lane 6: 2b + 2 × 1m; lane 7: 4b + 2 × 3m; lane 8: 6b + 2 × 5m; lane 9: 7b + 2 × 1 + 2 × 3m; lane 10: 7b + 2 × 1m + 2 × 3m; lane 11: 8b + 2 × 1 + 2 × 3 + 2 × 5m; lane 12: 8b + 2 × 1m + 2 × 3m + 2 × 5m. | ||
Hybridization of mono DNA-functionalized ruthenium complex 1m with 2 or 2m yields a 14-base double strand with one (Fig. 5, lane 3) or two ruthenium moieties (Fig. 5, lane 4), respectively. Similar assemblies were obtained based on the mono 20 (Fig. 5, lanes 6 and 7) and 24 (Fig. 5, lanes 9 and 10) nucleotide-long DNA-substituted ruthenium conjugates. Three ruthenium complex–DNA duplex structures were built by hybridization of the bis(DNA)-ruthenium hybrids with two equivalents of the sequence complementary mono(DNA)-ruthenium conjugates (Fig. 6). In these three-ruthenium assemblies, the distances between two ruthenium complexes are modulated by the duplexes lengths, which are 14 (lane 6), 20 (lane 7), 24 (lane 8), 34 (lane 9) or 58 (lane 11) base pairs long.
To easily and efficiently modulate the number of ruthenium motifs and their positions in the linear assemblies, the 34 and 58 nucleotide-long DNA oligomers 7 and 8 were designed to be successively composed with the 14- and 20-base sequences (1 and 2) and 14-, 20- and 24-base sequences (1, 2 and 3), respectively. Consequently, longer three-ruthenium linear assemblies were generated by hybridization of the bis(34 bases oligonucleotide)–ruthenium hybrid 7b with two complementary mono(20 bases oligonucleotide)–Ru conjugates 3m and two 14 nucleotide-long single strands 1 (Fig. 6 lane 9). Hybridization of the bis(58 bases oligonucleotide)–ruthenium hybrid 8b with two complementary mono(24 bases oligonucleotide)–Ru conjugates 5m and two 14 and 20 nucleotide-long single strands 1 and 3 led to an even longer three-ruthenium linear structure (Fig. 6 lane 11). This approach allowed us to easily adjust and increase the number of ruthenium moieties in the architecture. Indeed, it was achieved by using Ru–DNA conjugates instead of DNA single strands in the previously formed structures. With this methodology, hybridization of 7b with 2 eq. of 1m and 3m (Fig. 6, lane 10), and 8b with 2 eq. of 1m, 3m and 5m (Fig. 6, lane 12) provided linear DNA assemblies bearing five and seven ruthenium complexes, respectively.
All of these linear assemblies were studied by UV-thermal denaturation experiments and circular dichroism (CD) spectroscopy.
The melting temperature (Tm) analysis shows no significant difference in Tm values for duplexes in presence of no, one, two or three ruthenium complexes (Table 1). Nevertheless, the Tm curves are broadened (Tm curves available in ESI†). The metallic motifs seem to affect the thermal stability of the double helices. It can be watched on the polyacrylamide gel in Fig. 5. The bands for one- and two-ruthenium–DNA conjugate duplexes (lanes 3, 4, 6, 7, 9 and 10) are thicker and more diffuse than duplexes without a ruthenium complex (lanes 2, 5 and 8). For the 34 and 58 base pair duplexes formed with two and three distinct double strands, respectively, two Tm were observed, the values corresponding to an average of the hybridization temperatures of the separate double strands.
| Linear assemblies | T m1 (°C) | T m2 (°C) |
|---|---|---|
| 1 + 2 | 47.3 | |
| 1m + 2 | 48 | |
| 1m + 2m | 48 | |
| 3 + 4 | 68 | |
| 3m + 4 | 67.5 | |
| 3m + 4m | 68.5 | |
| 5 + 6 | 71.1 | |
| 5m + 6 | 72.2 | |
| 5m + 6m | 71.2 | |
| 2b + 2 × 1m | 51 | |
| 4b + 2 × 3m | 67.8 | |
| 6b + 2 × 5m | 72.2 | |
| 7b + 2 × 1 + 2 × 3m | 54 | 66.6 |
| 7b + 2 × 1m + 2 × 3m | 54 | 66.8 |
| 8b + 2 × 1 + 2 × 3 + 2 × 5m | 58.3 | 70.1 |
| 8b + 2 × 1m + 2 × 3m + 2 × 5m | — | 69 |
Circular dichroism (CD) was used to check Ru complex effects on the duplex integrity. The CD spectra are identical for the whole duplex family. They present a characteristic spectrum of DNA B form with a positive band at 282 nm and a negative band at 252 nm (Fig. 7). No CD signal next to the MLCT band of the ruthenium complex was observed. The presence of internal and/or terminal metallic centres did not affect the CD signals and thus the shape of the duplexes.
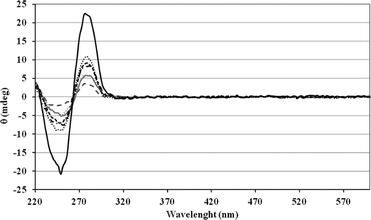 | ||
| Fig. 7 Circular dichroism spectra of 2b + 2 × 1m (dash grey line); 4b + 2 × 3m (grey line); 6b + 2 × 5m (dash dot black line); 7b + 2 × 1 + 2 × 3m (dot grey line); 7b + 2 × 1m + 2 × 3m (dash black line); 8b + 2 × 1 + 2 × 3 + 2 × 5m (black line); 8b + 2 × 1m + 2 × 3m + 2 × 5m (dot black line). | ||
Conclusions
Herein, we have shown the synthesis and the characterization of a new family composed of eleven mono- and bis-DNA-functionalized ruthenium complexes. A simple and efficient route was described to assemble ruthenium complexes in a linear and programmable fashion using the DNA self-assembly property. This strategy allowed us to selectively choose the number and the position of metallic complexes introduced in a DNA backbone. The extension of this methodology could afford symmetrical nanowires of various lengths with other metals.Experimental
Materials and methods
5′-C6 amino-modified oligonucleotides were purchased from Eurogentech. 4,4′-dicarboxy-2,2′-bipyridine, cis-bis(2,2′-bipyridine)dichlororuthenium(II) and 3-morpholinopropane-1-sulfonic acid (MOPS) was purchased at Alfa-Aesar France. 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride (DMT-MM) was purchased at Sigma Aldrich France. [Ru2+(2,2′-bipyridine)2(4,4′-dicarboxy-2,2′-bipyridine](PF6−)2 was synthesized as already reported.16UV-Vis spectra were obtained using Hewlett-Packard 8452A equipment. Fluorescence spectra were acquired on a Photon Technology International modular setup. Circular dichroism was achieved on a Jasco J-815 CD spectrometer. Polyacrylamide gel electrophoresis (PAGE) analysis was carried out on a Hoefer SE40-15-1.5 unit and the gels finally visualized on a Safe Imager™ 2.0 blue light transilluminator, under UV light at λ = 254 nm or at λ = 365 nm. HRMS analyses were realized on a Qtof Ultima API (Waters) mass spectrometer, see ESI†. The capillary, cone and RF Lens tensions were, respectively, 3.5 kV, 130 V and 40 V. The source and desolvation temperatures were 60 °C and 80 °C. The collision energy (Ar) was fixed at 8 eV in Tof ms mode, with nitrogen for the nebulization gas. The solutions were analyzed by infusion (10μL min−1), with an acquisition time of 5 min, realized with the Masslynx 4.1 software (Waters).
General procedure for ruthenium–DNA hybrids synthesis
A 2 mL aqueous buffer solution (pH = 7) of 3-morpholinopropane-1-sulfonic acid (MOPS) (1,046 g, 0.8 mol L−1) and 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride (DMT-MM) (138,4 mg, 0.08 mol L−1) was prepared. 4 mL DMSO solution of [Ru2+(2,2′-bipyridine)2(4,4′-dicarboxy-2,2′-bipyridine)](PF6−)2) (4,7 mg, 5 μmol, 25 eq.) was added, followed by the introduction of 5′-C6 amino-modified oligonucleotide kept on a solid support (200 nmol, 1 eq.). The solution was stirred for 48 h at 25 °C and then filtered. The resulting orange beads were washed once with DMSO and 3 times with water. The beads were put in a concentrated ammonia solution (32%) at 55 °C for 20 h and then filtered. The filtrate was evaporated and redissolved in an aqueous solution. Ruthenium–DNA conjugates were purified by PAGE on 20% denaturing polyacrylamide gels. The bands were cut, crushed in water and incubated at 55 °C. The mixture was filtered to remove the gel. The filtrate was concentrated and washed twice with water on a Amicon® Ultra centrifugal filter 3 K (Millipore Ireland Ltd).Hybridization conditions
A solution with the appropriated number of equivalents of each DNA strand was prepared for a final 50 μM (duplex) concentration in a buffer solution (50 mM Tris, 100 mM NaCl and 10 mM MgCl2 at pH = 7.2). Hybridization was achieved by heating this solution to 90 °C for 3 min and then leaving its temperature to decrease slowly to room temperature. The solution was finally incubated for 12 h at 4 °C.Acknowledgements
Financial support was provided by the French National Agency of Research (ANR) (ANR-08-JCJC-0141).References
- Y. E. Alexeev, B. I. Kharisov, T. C. H. Garcia and A. D. Garnovskii, Coord. Chem. Rev., 2010, 254, 794–831 CrossRef CAS.
- R. Chakrabarty, P. S. Mukherjee and P. J. Stang, Chem. Rev., 2011, 111, 6810–6918 CrossRef CAS.
- H. Yang, K. L. Metera and H. F. Sleiman, Coord. Chem. Rev., 2010, 254, 2403–2415 CrossRef CAS.
- S. Ghosh and E. Defrancq, Chem.–Eur. J., 2010, 16, 12780–12787 CrossRef CAS.
- J. Müller, Eur. J. Inorg. Chem., 2008, 2008, 3749–3763 CrossRef.
- G. C. Vougioukalakis, A. I. Philippopoulos, T. Stergiopoulos and P. Falaras, Coord. Chem. Rev., 2011, 255, 2602–2621 CrossRef CAS.
- O. Lentzen, C. Moucheron and A. K.-D. Mesmaekerin Metallotherapeutic Drugs and Metal-Based Diagnostic Agents, John Wiley & Sons, Ltd, 2005, pp. 359-378 Search PubMed.
- W. H. Ang, A. Casini, G. Sava and P. J. Dyson, J. Organomet. Chem., 2011, 696, 989–998 CrossRef CAS.
- A. L. Noffke, A. Habtemariam, A. M. Pizarro and P. J. Sadler, Chem. Commun., 2012, 48, 5219–5246 RSC.
- K. Wiederholt and L. W. McLaughlin, Nucleic Acids Res., 1999, 27, 2487–2493 CrossRef CAS.
- K. M. Stewart and L. W. McLaughlin, Chem. Commun., 2003, 2934–2935 RSC.
- S. Ghosh, I. Pignot-Paintrand, P. Dumy and E. Defrancq, Org. Biomol. Chem., 2009, 7, 2729–2737 CAS.
- D. Mitra, N. Di Cesare and H. F. Sleiman, Angew. Chem., Int. Ed., 2004, 43, 5804–5808 CrossRef CAS.
- F. A. Aldaye and H. F. Sleiman, J. Am. Chem. Soc., 2007, 129, 10070–10071 CrossRef CAS.
- K. M. Stewart, J. Rojo and L. W. McLaughlin, Angew. Chem., Int. Ed., 2004, 43, 5808–5811 CrossRef CAS.
- E. Terpetschnig, H. Szmacinski, H. Malak and J. R. Lakowicz, Biophys. J., 1995, 68, 342–350 CrossRef CAS.
- X. Li, J. Gartner, B. N. Tse and D. R. Liu, J. Am. Chem. Soc., 2004, 126, 5090–5092 CrossRef CAS.
Footnote |
| † Electronic Supplementary Information (ESI) available: purification and analysis PAGE, CD and Tm spectra, high resolution electrospray mass spectra. See DOI: 10.1039/c2ra21645k/ |
| This journal is © The Royal Society of Chemistry 2012 |