Methylation-blocked enzymatic recycling amplification for highly sensitive fluorescence sensing of DNA methyltransferase activity†
Feng
Chen
and
Yongxi
Zhao
*
Key Laboratory of Biomedical Information Engineering of Education Ministry, School of Life Science and Technology, Xi'an Jiaotong University, Xi'an, Shaanxi 710049, P. R. China. E-mail: yxzhao@mail.xjtu.edu.cn; Fax: +86-29-82663454; Tel: +86-29-82663454
First published on 19th October 2012
Abstract
Herein, using DNA adenine methylation (Dam) methyltransferase (MTase) as a model analyte, a novel fluorescence sensing strategy was developed for facile, rapid and highly sensitive detection of the activity and inhibition of the target based on methylation-blocked enzymatic recycling amplification. In this sensing system, nicking endonuclease Nt.AlwI with the methylation-sensitive property was selected to achieve signal amplification. In addition, a DNA heteroduplex probe is specially designed to contain the recognition sequences for both Dam MTase and Nt.AlwI. In the absence of Dam MTase, Nt.AlwI cleaves the DNA heteroduplex at only the top strand. At the reaction temperature, the cleaved heteroduplex is unstable and readily separates. The released bottom strand can hybridize with the molecular beacons (MB) and subsequently trigger Nt.AlwI-mediated recycling cleavage of MBs, providing a dramatically amplified fluorescence signal. However, when the heteroduplex is methylated by Dam MTase, the cleaving operation is blocked, resulting in an inconspicuous fluorescence enhancement. Unlike existing signal amplified assays which use at least two enzymes, only one is involved in this amplified strategy. Under optimized conditions, the sensing system reveals a detection limit of 0.05 U mL−1 in a short assay time (65 min), which is much superior to all presently reported methods except for two electrochemical biosensors (0.04 U mL−1). Furthermore, the application of the assay in human serum and screening of Dam MTase inhibition were demonstrated with satisfactory results. Overall, the proposed sensing system shows great potential for further application in biological research, early clinical diagnosis and designed drug therapy.
Introduction
DNA methyltransferase (MTase) transfers the methyl group from S-adenosyl methionine (SAM) to target adenine or cytosine residues in the recognition sequences during DNA methylation,1 a process which plays a critical regulatory role in many biological processes by altering gene expression.2–4 Aberrant DNA methylation has been found to be a new cancer biomarker in mammals and is associated with levels of bacterial virulence,2,3,5–7 DNA MTase has been treated as a potential target in anticancer therapy and for new antimicrobial drugs.5,8,9 Thus, evaluation of DNA MTase activity and inhibition has drawn an explosion of attention over the past several decades. Classical DNA MTase assays are performed with radioactive labeling, gel electrophoresis and high performance liquid chromatography.10–13 Although they are well-established, these approaches are DNA-consuming, labor-intensive, require radiolabeled substrates or are of insufficient sensitivity. These intrinsic drawbacks significantly limit the broad application of these methods.Recently, several techniques including electrochemical biosensors,14–17 electrogenerated chemiluminescence methods,18 bioluminescence detection,19 and strand displacement amplification (SDA)-based DNAzyme colorimetric assay20 have been developed to overcome these drawbacks. All of the above approaches offer high sensitivity, although they suffer from a lengthy assay time (155–420 min). Some are also limited by complex handling procedures as well as complicated designs due to the inevitable introduction of at least two enzymes.16,20 To further improve analytical performance, Li's group described a rapid gold nanoparticles (AuNPs)-based colorimetric assay for DNA MTase activity with acceptable sensitivity.21 The detection finished within 6 min. However, it takes several days to perform the preparation and functionalization of AuNPs. In addition, the nonspecific aggregation of the functionalized AuNPs in a complex biological sample causes a false positive, limiting its practical application. The features of simplicity and real-time monitoring are advantageous, and a fluorescence-based sensing strategy was developed for the detection of DNA MTase activity within a relatively short assay time.22 Unfortunately, its lack of sensitivity does not meet trace target analysis in real samples. Recently, another fluorescent assay for MTase activity was reported with a low detection limit, but its disadvantage was its long assay time.23 Accordingly, these are still a challenge in the development of facile, rapid and highly sensitive methods for the determination of DNA MTase activity.
Recently, to improve sensitivity, several signal amplification strategies have been developed for the detection of enzyme activities,24,25 nuclear acids,26,27 proteins,28,29 small molecules,30,31 and metal ions.32,33 Owing to the high sensitivity, simple design and rapid response time, more significant research effort has been devoted to constructing various sensing platforms based on nicking endonuclease (NEase) assisted amplification methods. Encouraged by these studies, we report a novel biosensor for the analysis of MTase activity on the basis of the unique technology reported in our recent work.34 Though it is highly sensitive, this method suffers from a complex reaction system due to the use of two endonucleases (one restriction endonuclease and one NEase). Herein, using DNA adenine methylation (Dam) MTase as a model analyte,35 we developed a facile and rapid approach for highly sensitive fluorescence detection of Dam MTase activity and inhibition based on methylation-blocked enzymatic recycling amplification. In the sensing strategy, only NEase Nt.AlwI is employed to mediate signal amplification. This proposed strategy combines low-background molecular beacon (MB) and NEase-mediated recycling amplification, which significantly improved the sensitivity of the biosensor. Furthermore, to demonstrate the potential application of the proposed biosensor in complex biological samples, a human serum sample spiked with different concentrations of Dam MTase was measured.
Experimental
Materials and chemicals
Dam MTase, Nt.AlwI and S-adenosylmethionine (SAM) were obtained from New England Biolabs Inc. (Beijing, China). The inhibitors, gentamycin, 5-fluorouracil and benzyl penicillin were purchased from Sangon Biotechnology Co., Ltd. (Shanghai, China). All of the other chemicals were of analytical reagent grade and used without further purification. The ultrapure water with an electrical resistance larger than 18.2 MΩ was obtained through a Milli-Q system (Millipore, Bedford, MA). All oligonucleotides used in this study were synthesized by Sangon Biotechnology Co., Ltd. (Shanghai, China) and sequences were as follows:Oligo A.
5′-GCGGGATCACGCTACAGCACC-3′ Oligo B.
5′-GGTGCTGTAGCGTGATCCCGC-3′ MB.
5′-FAM-TGAGCGGGATCACGCTACAGCACTTCGCTCA-DABCYL-3′
The bolded and italic letters in MB, Oligo A and Oligo B represent the recognition sequence for Nt.AlwI, and the stem of the MB is italicised.
Apparatus
All fluorescence measurements were carried out on a FluoroMax-4 fluorescence spectrometer (Horiba Jobin Yvon, Edison, NJ) at room temperature. Excitation and emission wavelengths were set at 494 nm and 518 nm with the slit widths of 5 nm, respectively. The emission spectra were obtained by scanning the emission from 510 to 590 nm in steps of 1 nm.Optimization of assay conditions
To achieve the best sensing performance of the system, we optimized the molar ratio of Oligo A to B in the DNA heteroduplex (1:1, 1.3:1, 1.6:1 and 2:1) with a defined Oligo B concentration of 17 nM, the DNA methylation time (0, 30, 45 and 60 min) and the nicking time (0, 15, 20, 25 and 30 min), respectively.Amplified assay of Dam MTase activity
To prepare the DNA heteroduplex, a mixture containing 1333 nM Oligo A and 833 nM Oligo B in the annealing buffer (10 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA, pH = 7.8) was heated to 90 °C for 5 min and then 45 °C for another 10 min. It was stored at 4 °C for further use.The NEBuffer 2 (10 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, pH = 7.9) was used for both DNA methylation reaction and nicking reaction. In a typical experiment, 50 μL of this buffer containing 1 μL of the DNA heteroduplex, 160 μM SAM and different target concentrations was first incubated at 37 °C for 45 min. After this, 5 U of Nt.AlwI and 100 nM MB were introduced into the resulting mixture. The nicking reaction was allowed to proceed at 50 °C for 20 min prior to making the fluorescence measurement.
Dam MTase activity inhibition evaluation
To investigate whether inhibitors (gentamycin, 5-fluorouracil and benzyl penicillin) influence Nt.AlwI, a control experiment was carried out. Prior to the addition of 5 U of Nt.AlwI, inhibitors were introduced into 50 μL of the NEBuffer 2 containing 1 μL of the DNA heteroduplex, 100 nM MB and 160 μM SAM. The solution was incubated at 50 °C for 20 min prior to making the fluorescence measurement.For Dam MTase inhibition experiments, the procedure was similar to detection of its activity noted above, except that different concentrations of inhibitors were added together with Dam MTase.
Results and discussion
Strategy of amplified Dam MTase assay
The principle of the amplified sensing system is illustrated in Scheme 1. To clear our design, it is necessary to recall the specific properties of Nt.AlwI. As one of the best-characterized NEase, Nt.AlwI recognizes the double-stranded 5′-GGATC-3′ site (incorporating the recognition site 5′-GATC-3′ for Dam MTase) and only cleaves at the 4th nucleotide downstream from the recognition site on the top strand. In addition, Nt.AlwI is sensitive to Dam MTase-catalyzed methylation, namely this cleavage reaction will be blocked after the methylation of A residues at the 5′-GGATC-3′ site by Dam MTase. | ||
| Scheme 1 Schematic illustration of methylation-blocked enzymatic recycling amplification for the detection of Dam MTase activity. The recognition sequences for Nt.AlwI in both the heteroduplex and MB are highlighted in red and the small arrows (blue) represent the cleavage site for Nt.AlwI. | ||
Taking advantage of these novel merits, the DNA heteroduplex containing a recognition site for Nt.AlwI is prepared by the hybridization of Oligo A and B, and MB is elaborately designed to share the same sequence with Oligo A in most portions of the loop and in part of the stem. When the NEase Nt.AlwI as well as the MB is mixed with the DNA heteroduplex, it will bound to the heteroduplex and split only Oligo A into two short fragments. Due to the low melting temperature, the cleaved heteroduplex is unstable and readily separated under the nicking reaction temperature of 50 °C. The released Oligo B will hybridize with the MB and open its hairpin structure to form a stable duplex, which contains the full recognition site for Nt.AlwI. Once the MB is cleaved by Nt.AlwI, it is dissociated from the duplex, restoring the fluorescence signal. Then the released Oligo B can form a new duplex with another MB and trigger the second cycle of cleavage. In this way, the NEase can go through a cyclical MB-cleaving operation by the repetition of hybridization, cleavage, and release, which provides an amplified fluorescence signal. In contrast, in the presence of Dam MTase, the 5′-GATC-3′ site in the DNA heteroduplex can be methylated at the A residues, which prevents Nt.AlwI from scissoring Oligo A. Then, the MB-cleaving operation will be resisted, thereby inhibiting fluorescence activation. Since Nt.AlwI is disabled to cleave the single-stranded DNA (such as MB), no fluorescence is triggered. As a consequence, Nt.AlwI-mediated recycling cleavage of MBs, as indicated by the amplified fluorescence, is dynamically blocked by Dam MTase-catalyzed DNA methylation.
Feasibility and amplification capability of the proposed strategy
To confirm the sensing mechanism of the proposed method, several experiments were carried out. In the mixture containing only DNA heteroduplex and MB, a very weak fluorescence signal was observed (Fig. 1, curve a). When the same mole concentration of Oligo B was mixed with the MB instead of the heteroduplex, it caused an obviously enhanced fluorescence response (Fig. 1, curve b) owing to the open of the MB's hairpin structure. On the other hand, after the addition of Nt.AlwI into the mixture containing the DNA heteroduplex and MB, a strong fluorescence response (about 4-fold higher than that of curve b) was yielded (Fig. 1, curve c), indicating that Nt.AlwI-mediated recycling cleavage of MBs had occurred. However, when Dam MTase was present, it seriously inhibited the increase of fluorescence intensity (Fig. 1, curve d), which is only about 1/9 of that without Dam MTase (curve c), suggesting that Dam MTase-catalyzed DNA methylation prevented Nt.AlwI-mediated cleavage operation. Combining these findings, we conclude that our developed strategy provided substantial signal amplification for Dam MTase activity and inhibition assay.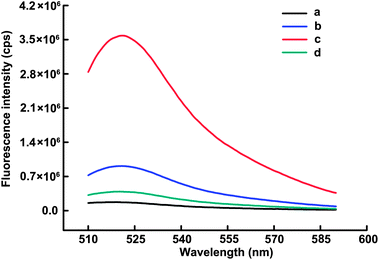 | ||
Fig. 1 Fluorescence emission spectra of MB (100 nM) under different conditions: (a) MB + H; (b) MB + Oligo B (17 nM); (c) MB + H + Nt.AlwI (5 U); (d) MB + H + Nt.AlwI (5 U) + Dam MTase (80 U mL−1). H represents the DNA heteroduplex with a fixed Oligo B concentration of 17 nM and the 1.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of Oligo A to Oligo B in all these experiments. 1 molar ratio of Oligo A to Oligo B in all these experiments. | ||
Optimization of assay conditions
To achieve the best sensing performance, the effect of the molar ratio of Oligo A to B in the DNA heteroduplex was investigated. The experiment was carried out with a fixed Oligo B concentration of 17 nM, and the result is shown in Fig. 2A. The fluorescence response to target gradually decreased with the increasing concentration of Oligo A, indicating increasing blocking of Nt.AlwI-mediated cleavage due to more formation and subsequent methylation of the DNA heteroduplex. Nevertheless, the fluorescence intensity in the absence of Dam MTase also decreased as the ratio value increased. This unwished for decrease mainly results from the competitive cleavage of Oligo A against MBs. Although the target-induced fluorescence response still followed a decreasing trend with further increase of the ratio value, the maximum fluorescence decrement (F0/F) was observed at the value of 1.6:1. Therefore, the molar ratio of 1.6:1 was used for the following assay. Additionally, to accelerate the reaction while insuring a reasonably high sensitivity, the optimized time for DNA methylation and nicking reaction were set as 45 min (Fig. 2B) and 20 min (Fig. 2C), respectively. This total assay time (65 min) is much shorter than those of most existing reports.14–20,23,36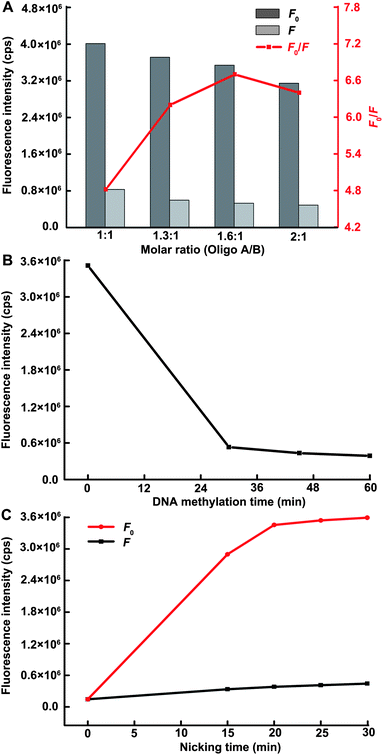 | ||
| Fig. 2 (A) The effect of molar ratio of Oligo A to B in the DNA heteroduplex on the sensing performance of the proposed methods. The DNA methylation time and nicking time are each fixed at 30 min. The descriptors F and F0 represent the fluorescence intensity in the presence and absence of target, respectively. (B) The DNA methylation time-dependent fluorescence response with a fixed nicking time of 30 min. (C) The nicking time-dependent fluorescence response over background fluorescence with a fixed DNA methylation time of 45 min. The Dam MTase concentration in all figures is 80 U mL−1. | ||
Amplified assay of Dam MTase activity
Under optimized conditions, we demonstrated the ability of the described strategy to detect target with high sensitivity. Fig. 3A showed that the fluorescence intensity decreased upon increasing the Dam MTase concentration up to 120 U mL−1, indicating the methylation of the 5′-GATC-3′ site caused the dose-dependent inhibition of Nt.AlwI-mediated cleavage. The change of fluorescence intensity in responding to the different Dam MTase concentrations was depicted in Fig. 3B. A linear correlation (R2 = 0.989) existed between the fluorescence intensity and target concentration over the range of 0.08–26 U mL−1 (Fig. 3B, inset). Based on three times the standard deviation of the background fluorescence signal, the detection limit was estimated to be 0.05 U mL−1, which is lower than those of all existing fluorescence approaches. Furthermore, this sensitivity is 5 and 8 times higher than those of the SDA-based DNAzyme colorimetric method20 and hairpin-shaped DNAzyme-assisted signal amplification,37 respectively. The low detection limit is comparable to two electrochemical biosensors16,36 (both 0.04 U mL−1), one of which coupled with terminal transferase-mediated extension.16 It is worth noting that such a high sensitivity was achieved within a total assay time of 65 min, much shorter than those of the two electrochemical biosensors (173 and 370 min, respectively). The detail of the comparisons is shown in Table 1.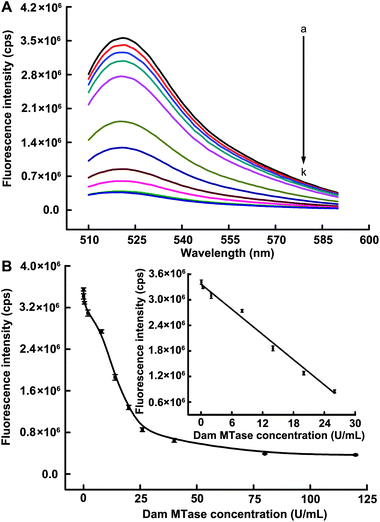 | ||
| Fig. 3 (A) Fluorescence emission spectra upon addition of different concentrations of Dam MTase: (a–k) 0, 0.08, 0.4, 2, 8, 14, 20, 26, 40, 80 and 120 U mL−1, respectively. (B) Corresponding fluorescence peak intensities versus Dam MTase concentrations. The inset shows the linearity of the fluorescence response for the sensing system against the low target concentrations. The error bar was calculated from three independent experiments. | ||
| Method | Detection limit (U mL−1) | Time (min) | Ref. |
|---|---|---|---|
| Methylation-blocked enzymatic recycling amplification-based fluorescence biosensor | 0.05 | 65 | This work |
| Hairpin-capture DNA probe-based electrochemical sensor | 0.07 | 420 | 14 |
| Gold nanoparticle-based electrochemical assay | 0.12 | 240 | 15 |
| Terminal transferase-mediated extension-based electrochemical assay | 0.04 | 173 | 16 |
| Methylation-resistant cleavage-based bioluminescence assay | 0.08 | 340 | 19 |
| SDA-based DNAzyme colorimetric assay | 0.25 | 155 | 20 |
| Gold nanoparticle-based colorimetric assay | 0.3 | 6 | 21 |
| Hairpin fluorescence DNA probe | 0.8 | 30 | 22 |
| Carbon nanoparticle-based fluorescent biosensor | 0.1 | 120 | 23 |
| Nicking enzyme-assisted signal amplification-based fluorescence biosensor | 0.06 | 60 | 34 |
| Single wall carbon nanotubes-based electrochemical assay | 0.04 | 370 | 36 |
| Hairpin-shaped DNAzyme-based amplified fluorescence assay | 0.4 | 20 | 37 |
Detection of Dam MTase activity in human serum
A significant challenge for enzyme activity analysis is the ability to realize the detection of target in complex real samples. To test the practicality of the proposed approach, human serum (diluted 20 times with NEBuffer 2) samples spiked with Dam MTase were tested. As shown in Fig. 4, comparable responses were found for Dam MTase in both NEBuffer 2 and diluted human serum. It was observed that the fluorescence intensity decreased with the increase of target concentration. To our surprise, the fluorescence intensities of serum samples were corresponding lower than those of NEBuffer 2 samples. This can probably be described to interferences of serum for cleavage mediated by Nt.AlwI. Moreover, a desirable reproducibility was achieved based on our strategy. The relative standard deviations of fluorescence readouts were 2.4% and 2.7% of 8 U mL−1 and 16 U mL−1 Dam MTase spiked in serum, respectively. These results indicated that the proposed strategy holds a great promise in practical applications for DNA MTase detection with great accuracy and reliability.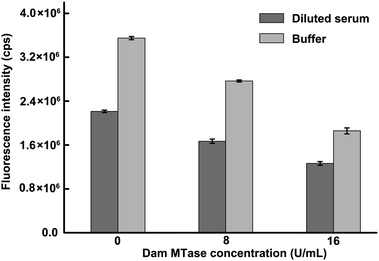 | ||
| Fig. 4 Results obtained from the testing of human serum samples spiked with Dam MTase and Dam MTase in NEBuffer 2. Human serum was diluted in 1:20 ratio with NEBuffer 2. | ||
Dam MTase activity inhibition evaluation
We also demonstrated the capacity of the developed method for screening the inhibition of Dam MTase using gentamycin, 5-fluorouracil and benzyl penicillin as model inhibitors. Before evaluating the inhibitory effects of the drugs on the activity of target MTase, we first investigated whether these drugs had influence on Nt.AlwI activity, and the result is shown in Fig. S1.† It can been seen that there were no remarkable change in the fluorescence intensity in the absence and presence of inhibitors, suggesting these three drugs have no influence on the activity of Nt.AlwI when the concentrations were no more than 38, 1000 and 50 μM, respectively. This result ensured accurate assay for target inhibition. Afterwards, the inhibition assays for Dam MTase (24 U mL−1) were carried out under the concentrations of the several inhibitors mentioned above. As depicted in Fig. 5A, the fluorescence intensity increased with the addition of all inhibitors, indicating a significant inhibition of the methylation on the 5′-GATC-3′ site catalyzed by Dam MTase.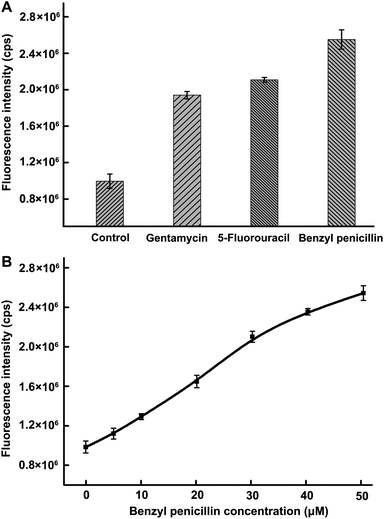 | ||
| Fig. 5 (A) Influence of gentamycin (38 μM), 5-fluorouracil (1000 μM) and benzyl penicillin (50 μM) on the activity of Dam MTase. (B) Inhibition of different concentrations of benzyl penicillin on Dam MTase activity. The benzyl penicillin concentrations were 0, 5, 10, 20, 30, 40 and 50 μM, respectively. | ||
We also investigated the dose-dependent inhibition on Dam MTase activity with different concentration of benzyl penicillin (Fig. 5B). With increased inhibitor concentration, the inhibition is enhanced. The concentration causing a 50% fluorescence increase is 27 μM. These results demonstrated that the developed method has potential application in screening MTase-targeted drugs, which has significant value to antibiotics and anticancer therapeutics.
Conclusions
We have developed a facile, rapid and highly sensitive strategy for Dam MTase activity and inhibition assay based for the first time on methylation-blocked enzymatic recycling amplification. In this assay, only one enzyme is brought, yet can afford a significantly amplified signal. Within the short assay time (65 min), a detection limit of 0.05 U mL−1 was obtained with a wide linear range of 3 orders of magnitude. This sensitivity is higher than those of all presently developed methods except for two electrochemical biosensors. In addition, the suitable inhibitor of target MTase can be readily screened and quantified via this new approach. Moreover, the developed method was successfully applied to real sample (human serum) analysis with satisfactory results. It is worth noting that the developed strategy can be used as a universal tool for the detection of other DNA MTases in both prokaryotes and eukaryotes with a minor modification of the DNA heteroduplex and the change of corresponding NEase. Considering the high sensitivity and excellent performance for the detection of serum sample, as well as straightforward design and rapid operation, the proposed strategy holds great promise in the quantitative analysis of DNA MTase activity even in complex real samples, and affords significant information for biological, biomedical research and early clinical diagnosis.Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (21005059) and the Fundamental Research Funds for the Central Universities.Notes and references
- X. Cheng and R. J. Roberts, Nucleic Acids Res., 2001, 29, 3784 CrossRef CAS.
- K. D. Robertson, Nat. Rev. Genet., 2005, 6, 597 CrossRef CAS.
- D. M. Heithoff, R. L. Sinsheimer, D. A. Low and M. J. Mahan, Science, 1999, 284, 967 CrossRef CAS.
- F. Garcia-Del Portillo, M. G. Pucciarelli and J. Casadesus, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 11578 CrossRef CAS.
- M. Szyf, Biochemistry, 2005, 70, 533 CAS.
- M. Toyota, N. Ahuja, M. Ohe-Toyota, J. G. Herman, S. B. Baylin and J. P. J. Issa, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 8681 CrossRef CAS.
- R. Jones, Nat. Rev. Genet., 2002, 3, 900 CrossRef CAS.
- B. Brueckner and F. Lyko, Trends Pharmacol. Sci., 2004, 25, 551 CrossRef CAS.
- M. Szyf, Drug Resist. Updates, 2003, 6, 341 CrossRef CAS.
- E. Boye, M. G. Marinus and A. Løbner-Olesen, J. Bacteriol., 1992, 174, 1682 CAS.
- A. Bergerat, W. Guschlbauer and G. V. Fazakerley, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 6394 CrossRef CAS.
- R. L. P. Adams, A. Rinaldi and C. Seivwright, J. Biochem. Biophys. Methods, 1991, 22, 19 CrossRef CAS.
- W. Messer and M. Noyer-Weidner, Cell, 1988, 54, 735 CrossRef CAS.
- J. Su, X. He, Y. Wang, K. Wang, Z. Chen and G. Yan, Biosens. Bioelectron., 2012, 36, 123 CrossRef CAS.
- X. He, J. Su, Y. Wang, K. Wang, X. Ni and Z. Chen, Biosens. Bioelectron., 2011, 28, 298 CrossRef CAS.
- H. Wu, S. Liu, J. Jiang, G. Shen and R. Q. Yu, Chem. Commun., 2012, 48, 6280 RSC.
- W. Li, P. Wu, H. Zhang and C. X. Cai, Anal. Chem., 2012, 84, 7583 CrossRef CAS.
- Y. Li, C. C. Huang, J. B. Zheng and H. L. Qi, Biosens. Bioelectron., 2012, 38, 407 CrossRef CAS.
- C. Jiang, C. Yan, C. Huang, J. Jiang and R. Yu, Anal. Biochem., 2012, 423, 224 CrossRef CAS.
- W. Li, Z. Liu, H. Lin, Z. Nie, J. Chen, X. Xu and S. Yao, Anal. Chem., 2010, 82, 1935 CrossRef CAS.
- T. Liu, J. Zhao, D. Zhang and G. Li, Anal. Chem., 2010, 82, 229 CrossRef CAS.
- J. Li, H. Yan, K. Wang, W. Tan and X. Zhou, Anal. Chem., 2007, 79, 1050 CrossRef CAS.
- X. Y. Ouyang, J. H. Liu, J. S. Li and R. H. Yang, Chem. Commun., 2012, 48, 88 RSC.
- F. F. Andersen, M. Stougaard, H. L. Jorgensen, S. Bendsen, S. Juul, K. Hald, A. H. Andersen, J. Koch and B. R. Knudsen, ACS Nano, 2009, 3, 4043 CrossRef CAS.
- Y. X. Zhao, L. Qi, F. Chen, Y. Zhao and C. H. Fan, Biosens. Bioelectron., 2012 DOI:10.1016/j.bios.2012.10.009.
- R. M. Kong, X. B. Zhang, L. L. Zhang, Y. Huang, D. Q. Lu, W. Tan, G. L. Shen and R. Q. Yu, Anal. Chem., 2011, 83, 14 CrossRef CAS.
- Y. T. Zhou, Q. Huang, J. M. Gao, J. X. Lu, X. Z. Shen and C. H. Fan, Nucleic Acids Res., 2010, 38, e156 CrossRef.
- L. Xue, X. Zhou and D. Xing, Anal. Chem., 2012, 84, 3507 CrossRef CAS.
- A. X. Zheng, J. R. Wang, J. Li, X. R. Song, G. N. Chen and H. H. Yang, Chem. Commun., 2012, 48, 374 RSC.
- Y. X. Zhao, L. Qi, F. Chen, Y. Dong, Y. Kong, Y. Wu and C. H. Fan, Chem. Commun., 2012, 48, 3354 RSC.
- L. M. Lu, X. Zhang, R. M. Kong, B. Yang and W. H. Tan, J. Am. Chem. Soc., 2011, 133, 11686 CrossRef CAS.
- Z. Zhu, Y. Su, J. Li, D. Li, J. Zhang, S. Song, Y. Zhao, G. X. Li and C. H. Fan, Anal. Chem., 2009, 81, 7660 CrossRef CAS.
- L. Qi, Y. X. Zhao, H. Yuan, K. Bai, F. Chen, Y. H. Dong and Y. Y. Wu, Analyst, 2012, 137, 2799 RSC.
- Y. X. Zhao, F. Chen, Y. Y. Wu, Y. H. Dong and C. H. Fan, Biosens. Bioelectron., 2012 DOI:10.1016/j.bios.2012.10.022.
- F. Barras and M. G. Marinus, Trends Genet., 1989, 5, 139 CrossRef CAS.
- Y. H. Wang, X. X. He, K. M. Wang, J. Su, Z. F. Chen, G. P. Yan and Y. D. Du, Biosens. Bioelectron., 2012 DOI:10.1016/j.bios.2012.08.034.
- T. Tian, H. xiao, Y. L. Long, X. Zhang, S. R. Wang, X. Zhou, S. M. Liu and X. Zhou, Chem. Commun., 2012, 48, 10031 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c2an36329a |
| This journal is © The Royal Society of Chemistry 2013 |
