Fibrinogen nanofibers for guiding endothelial cell behavior†
Dencho
Gugutkov
ab,
Johan
Gustavsson
a,
Maria Pau
Ginebra
bc and
George
Altankov
*acd
aInstitute for Bioengineering of Catalonia, Barcelona, Spain
bUniversitat Politècnica de Catalunya, Barcelona, Spain
cCentro de Investigación Biomédica en Red en Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN), Zaragoza, Spain
dInstitució Catalana de Recerca i Estudis Avancats (ICREA), Barcelona, Spain. E-mail: George.Altankov@icrea.cat; Fax: +34 93 403 98 73; Tel: +34 93 403 97 09
First published on 8th July 2013
Abstract
This paper describes the biological consequences of presenting electrospun fibrinogen (FBG) to endothelial cells as a spatially organized nanofibrous matrix. Aligned and randomly oriented FBG nanofibers with an average diameter of less than 200 nm were obtained by electrospinning of native FBG solution. Electrophoretic profiling confirmed that the electrospun FBG resembled the native protein structure, and fluorescent tracing of FITC-labeled FBG showed that electrospun fibers withstood immersion in physiological solutions reasonably well for several days. With respect to cellular interactions, the nanofibrous FBG matrix provided better conditions for initial recognition by human umbilical vein endothelial cells compared to pre-adsorbed FBG on a flat surface. Furthermore, the spatial organization of electrospun FBG fibers presented opportunities for guiding the cellular behavior in a way that is not possible when the protein is presented in another form (e.g. adsorbed or soluble). For example, on aligned FBG fibers, cells rapidly oriented themselves along the fibers, and time-lapse recordings revealed pronounced cellular movements restricted to the fiber direction. In great contrast, on randomly deposited fibers, cells acquired a stellate-like morphology and became locally immobilized by the fibers. We also show that the FBG fiber orientation significantly influenced both the cytoskeleton organization in confluent cell layers and the orientation of the extracellular fibronectin matrix secreted by the cells. In conclusion, this study demonstrates that electrospun FBG nanofibers can be a promising tool for guiding endothelial cell behavior for tissue engineering applications.
Introduction
Cellular interactions with foreign materials initially rely on fundamental processes such as protein adsorption1 and ligand–receptor interactions2 which determine the subsequent cellular interaction and response. In fact, those initial events mimic to a certain extent the natural interaction of cells with the extracellular matrix (ECM),3 a notion that is required not only for understanding biocompatibility, but also for biology and medicine in general, since tissue development in multicellular animals heavily relies on the ability of cells to synthesize and organize ECM – a fact that cannot be underestimated in biomaterials science.Basically, the ECM is a structure consisting of glycosaminoglycans (a gel-like matter) and spatially organized matrix fibrils (e.g. collagen, laminin, fibronectin, etc.) whose length in some cases greatly exceeds that of individual cells.4 The importance of correct regulation of fibril deposition is exemplified by diseases such as osteogenesis imperfecta (caused by mutations in collagen genes), fibrosis (i.e., ectopic accumulation of ECM), as well as by its frequent appearance in degenerative tissue disorders.5,6 The biological relevance of fibrillar nanostructures is also clearly reflected in the efforts made in the fields of biomaterials and tissue engineering to reproduce and apply ECM-like fibers, e.g. by electrospinning7 or by self-assembly.8 Despite the promise of such techniques, there is still a need to understand how cells respond to complex structures that contain both spatial and biological information.
Among the different fibrillar proteins, fibrinogen (FBG)9 is of particular interest for tissue engineering applications due to its many biological functions. FBG is a soluble protein secreted into the plasma in high amounts mainly by hepatocytes, and it plays a key role in the coagulation cascade and as a bridging molecule for platelet aggregation.10 FBG and its derivative fibrin also serve as a provisional ECM in some injured tissues,11 and is involved in wound healing,12 tissue regeneration,13 inflammatory cell response,14 and angiogenesis.15 Thus, the consistency of using fibrin(-ogen) as a tissue sealant and/or scaffold for cellular therapies16 has been explored for example in cardiovascular, bone, cartilage, and skin tissue engineering applications.17–20 Interestingly, extrahepatic cells such as lung, intestinal and cervical epithelial cells also secrete FBG21–23 which then can integrate as a recognizable part of the natural ECM of these cells in the form of non-covalently assembled fibrils.24 Finally, considering the previously established fact in our laboratory that FBG undergoes significant spatial organization and remodeling into fibrillar structures in contact with endothelial cells,25 we anticipated that the organization of FBG molecules in matrix-like configurations could have significant impact on subsequent cell behavior. Therefore the aim of this paper was to carefully explore how endothelial cells, here using primary human umbilical vein endothelial cells (HUVECs) as a model system, respond to spatially organized nanofibers produced from electrospinning of pure FBG solution. For that purpose, we built on the work of Bowlin and coworkers26 who first succeeded in preparing electrospun native FBG nanofibers. Although it has been previously described that cells such as fibroblasts easily colonize electrospun FBG scaffolds,27 the fiber stability in aqueous environments and the cellular recognition and locomotor activity toward spatially organized FBG nanofibers are unknown. In this study we address these questions, which comprise the novelty of the obtained results.
Materials and methods
Electrospinning of fibrinogen nanofibers
Fibrinogen nanofibers were prepared as previously described.26 Briefly, fibrinogen from bovine plasma (Sigma-Aldrich) was dissolved at 100 mg mL−1 in 1-1-1,3-3-3-hexafluoroisopropanol (HFIP) (Sigma-Aldrich) mixed (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with 10× concentrated DMEM (Invitrogen). The obtained FBG solution was centrifuged at 4000 rpm for 10 minutes and the supernatant was loaded in a conventional 10 mL syringe (BD-Scientific). For electrospinning of that solution we used a home-made setup based on a high voltage supply (Glassman High Voltage Inc.), a syringe pump (Aitecs), and a grounded collector. Randomly deposited FBG nanofibers were obtained by vertical electrospinning of the polymer solution onto glass coverslips (ϕ 15 mm, Thermo Scientific) placed on aluminum foil that covered the collector. The applied voltage was 20–25 kV, the distance between the needle tip and the collector was 125 mm, and the pump flow rate was 0.3 mL h−1. Aligned fibers were obtained using the same spinning conditions, but with an original and simple method of collection based on a rotating drum (Fig. S1†). Basically, two plastic discs (ϕ 120 mm) were mounted on a common axis and separated by 100 mm. Thin metal wires were stretched between the disc peripheries to form parallel strings at 16 mm distance from each other. While rotating the common axis (600–800 rpm), the nanofibers aligned between the metal wires, and they could thereafter be easily collected onto glass coverslips.
1) with 10× concentrated DMEM (Invitrogen). The obtained FBG solution was centrifuged at 4000 rpm for 10 minutes and the supernatant was loaded in a conventional 10 mL syringe (BD-Scientific). For electrospinning of that solution we used a home-made setup based on a high voltage supply (Glassman High Voltage Inc.), a syringe pump (Aitecs), and a grounded collector. Randomly deposited FBG nanofibers were obtained by vertical electrospinning of the polymer solution onto glass coverslips (ϕ 15 mm, Thermo Scientific) placed on aluminum foil that covered the collector. The applied voltage was 20–25 kV, the distance between the needle tip and the collector was 125 mm, and the pump flow rate was 0.3 mL h−1. Aligned fibers were obtained using the same spinning conditions, but with an original and simple method of collection based on a rotating drum (Fig. S1†). Basically, two plastic discs (ϕ 120 mm) were mounted on a common axis and separated by 100 mm. Thin metal wires were stretched between the disc peripheries to form parallel strings at 16 mm distance from each other. While rotating the common axis (600–800 rpm), the nanofibers aligned between the metal wires, and they could thereafter be easily collected onto glass coverslips.
Morphological fiber characterization
The electrospun FBG fibers were morphologically characterized by scanning electron microscopy (SEM, Jeol JSM-5410) at the Center for Biomaterials and Tissue Engineering at the Technical University of Valencia (Spain). Before analysis, the samples were coated with a conductive layer of sputtered gold. The SEM micrographs were taken at an accelerating voltage of 15 kV in order to ensure good image resolution. Fast Fourier Transformation (FFT) outputs of the scanning electron micrographs were then used to characterize the alignment of the fibers.28 The analysis of the SEM micrographs was performed with ImageJ (NIH) and the oval profile plug-in (authored by William O'Connnell). Briefly, a circular projection was placed on the FFT frequency distribution outputs of the micrographs and the radial sums of the pixel intensities for each degree between 0 and 180° were collected. The pixel intensity was then plotted as a function of its angle of acquisition. Distribution data were normalized to a baseline value and plotted in arbitrary units, allowing different data sets to be directly compared.Chemical fiber characterization
The effect of the electrospinning process on the protein configuration was investigated using classical SDS-PAGE for pure FBG, electrospun FBG nanofibers, and fibrin gel under reducing conditions. Fibrin gel was obtained by adding thrombin (Sigma-Aldrich; 2.0 NIHU mL−1) and 2.5 mM CaCl2 to a 2 mg mL−1 fibrinogen solution. The reduced protein samples were prepared by adding 10% (v/v) of 5% β-mercaptoethanol in sample buffer to the protein samples, and after boiling the obtained samples at 100 °C for 5 minutes, they were loaded in a 10% polyacrylamide gel, electrophoresed using vertical slab apparatus (BioRad), and thereafter stained by Coomassie Blue.To estimate the chemical stability of the FBG nanofibers under physiological-like conditions, fluorescein isothiocyanate-labeled FBG (FITC-FBG) was added to the FBG polymer solution (0.14 mg mL−1) before electrospinning. In this way, FITC-FBG/FBG nanofibers were obtained as random fibers on glass coverslips. The weight of each sample was measured before and after the electrospinning process to ensure equal mass of nanofibers on each coverslip (1.0 mg). The samples were then incubated in a standard culture environment (37 °C, 5% CO2) in 0.5 mL of either PBS or culture medium (EndoGRO, Invitrogen) for different time periods: 5, 24, and 72 hours, respectively. After incubation, the supernatant was removed and the remaining nanofibers were dried at room temperature. Then all the samples, including fresh control samples not exposed to any aqueous environments, were immersed in 0.2 M NaOH until the fibers were completely dissolved and no visible debris were present in the solution (48 hours at 37 °C). Finally, the sample extracts were resuspended and analyzed fluorometrically (FluoroMax-4, Horiba-Jobin Yvon; excitation 494 nm, emission 525 nm).
Cells
Human umbilical vein endothelial cells (HUVECs) were purchased from MilliPore (Spain), and cultured at 37 °C and with 5% CO2 in complete EndoGRO medium containing 2% fetal bovine serum (FBS). For experiments, cells were harvested and washed two times in FBS-free medium. Typically, the cell seeding density was 5 × 104 cells per sample in 2.0 mL of culture medium in 24-well TCPS plates (Nunc) containing the samples.Fast cell adhesion assay
To investigate the initial cellular recognition of the nanofibers, a fast cell adhesion assay was performed as previously described.29 Briefly, cells were seeded in serum-free EndoGRO medium on randomly deposited FBG nanofibers and on glass coverslips coated with FBG (50 μg mL−1). At 5 and 20 minutes after plating, cells were washed, fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton-X 100, and stained for the actin cytoskeleton and nuclei with FITC-phalloidin (Invitrogen) and Hoechst 34580 (Invitrogen) dissolved in PBS containing 1% albumin. Stained cells were photographed using an inverted fluorescent microscope (Axio Observer Z1, Zeiss), and cells were counted from 20 randomly chosen microscopic fields on each sample.Overall cell morphology and alignment to fibers
The overall morphology of HUVECs cultured on randomly and aligned electrospun FBG fibers was evaluated after 2 hours of incubation under serum-free conditions. For that purpose, cells were washed, fixed, and stained for actin and nuclei as described in the previous section, and at least three representative images of cells were acquired under each magnification and sample condition. Cell orientation on both aligned and random nanofibers was evaluated by analyzing 40 randomly chosen cells from one representative image using ImageJ (NIH). Briefly, the image of the cell on aligned nanofibers was rotated so that the approximate fiber direction was set perpendicular to the horizontal image baseline which was used as a reference direction (i.e. 0°). Images of cells on random samples were not rotated because of the absence of a reference direction. The longitudinal axis of each cell and its angle against the baseline were then determined. The data were finally normalized and plotted in a distribution histogram between 0 and 180° at 20° intervals. In this way, a perfect cell alignment to the fibers occurred at 90° (i.e. the interval 80–100°).Visualization of focal adhesion complexes
To visualize focal adhesions, fixed and permeabilized cells were saturated with 1% BSA in PBS for 15 minutes and stained with monoclonal anti-vinculin antibody (Sigma) (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800) for 30 minutes followed by another 30 minutes of incubation with secondary goat anti-mouse AlexaFluor® 555-conjugated antibody (Abcam). In addition, FITC-phalloidin was used as described above to stain for the actin cytoskeleton.
800) for 30 minutes followed by another 30 minutes of incubation with secondary goat anti-mouse AlexaFluor® 555-conjugated antibody (Abcam). In addition, FITC-phalloidin was used as described above to stain for the actin cytoskeleton.
Time-lapse recording
To monitor cellular movements on the random and aligned FBG nanofibers, cells growing in the culture flask were fluorescently labeled overnight with CellTracker Green CMFDA (Invitrogen) according to the supplier's instructions. The stained cells were harvested and seeded onto random and aligned nanofiber samples for 2 h in serum-free medium. Live imaging of the cells was then initiated using an inverted fluorescent microscope (Axio Observer Z1) equipped with an on-stage mini-chamber providing appropriate culture conditions (37 °C, humidified atmosphere, 5% CO2). Consecutive images were recorded using AxioVision software (Zeiss) every five minutes during a total period of six hours.Cell motility
To characterize the patterns of cellular movement, the time-lapse movies were analyzed using the ImageJ Manual Tracking plugin (developed by FP Cordelières, Institut Curie, Orsay, France). Path trajectories of 20 randomly chosen cells were analyzed, and from it the main track length was calculated. The main track orientation was measured as for cell alignment (see above) and plotted in the distribution histogram between 0 and 180° at 20° intervals.Long-term cultures
To follow the long-term behaviour of HUVECs on FBG nanofibers, 3 × 104 cells were cultured on random and aligned samples for up to 7 days in complete EndoGRO medium. At the end of the incubation the cells were fixed and stained for actin and the nucleus as described above.Fibronectin matrix formation
HUVECs secretion of the fibronectin (FN) matrix was examined via immunofluorescence. For that purpose, 3 × 104 cells per sample were cultured for three days in complete EndoGRO medium. At the end of the incubation, the cells were washed, fixed, and stained with a polyclonal rabbit anti-FN antibody (Santa Cruz) for 30 minutes, followed by secondary goat anti-rabbit AlexaFluor® 555-conjugated antibody (Abcam).Statistical analysis
Each datum is expressed as mean ± standard deviation if not indicated otherwise. Statistical significance was determined by a two-tailed independent Student's t-test and p < 0.05.Results
Characterization of electrospun FBG fibers
Electrospinning of 100 mg mL−1 FBG in HFIP allowed for the production of homogeneous nanofibers with an average diameter of 192 ± 46 nm (n = 50) as determined from SEM micrographs. By changing the way the fibers were collected during the electrospinning process, it was possible to obtain fibers that were either randomly deposited over the sample or aligned along a common direction (Fig. 1). Specifically, the pixel intensity distribution obtained from FFT analyses of the SEM micrographs revealed that 50% more fibers were oriented in the range of 85 to 95° on aligned samples compared to random samples.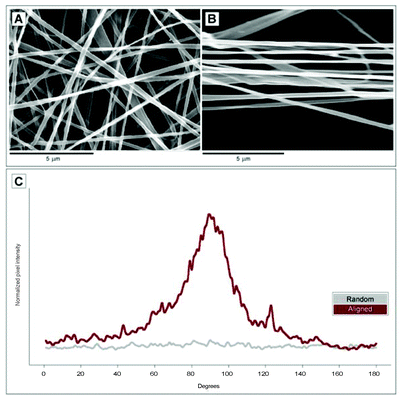 | ||
| Fig. 1 SEM imaging depicted the spatial organization of electrospun FBG fibers in random (A) and aligned (B) configurations. The spatial organization of fibers was characterized by FFT analysis and then plotted as pixel intensity against the angle of acquisition (C). | ||
To study the influence of the electrospinning process on the FBG structure, a SDS-PAGE was performed (Fig. 2a). The electrophoretic profile of electrospun FBG (dissolved in sample buffer) was observed to be identical to the profile of native FBG protein. In contrast, fibrin (i.e. thrombin-treated FBG) displayed an additional band (between B′β′ and γ) due to fibrinopeptide loss from the polymer backbone.
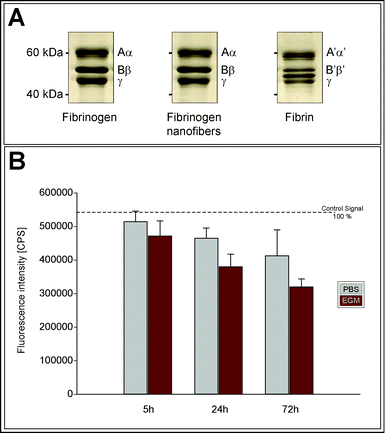 | ||
| Fig. 2 Characterization of the stability of electrospun FBG fibers. (A) SDS-PAGE profiles of pure FBG, electrospun FBG, and fibrin. (B) Fluorescence intensity was measured in counts per second (CPS) of extracted electrospun FBG/FITC-FBG incubated in PBS or EndoGRO medium (EGM) for different time periods. Error bars show standard error. | ||
The stability of the FBG nanofibers in aqueous solutions was estimated by extracting FITC-FBG from electrospun FBG/FITC-FBG nanofibers that had been immersed in PBS or EndoGRO medium for different time periods (Fig. 2b). It was found that the fluorescent signal measured in the extracted samples decreased with time in both solutions, representing a loss of FITC-FBG from the electrospun fibers into solution. After 72 hours of incubation, 24% of fluorescent signal was lost in fibers incubated in PBS. In EndoGRO medium, the fluorescent signal was, in general, weaker at all time points, and by 72 hours the fluorescent signal had decreased to 36% of its original intensity.
Cellular interactions with FBG nanofibers
The cellular recognition and response to the FBG nanofibers were first evaluated using the so-called “fast adhesion” assay, i.e. cell adhesion was measured when any differences in the affinity of integrin receptors to their ligand might be easily distinguished, and which typically occurs within a few minutes after seeding.29 As shown in Fig. 3, after five minutes of incubation almost two times more cells were counted on FBG nanofibers compared to regular FBG-coated samples. However, already after 20 minutes of incubation, cell adhesion on FBG nanofibers and regular FBG-coating was equilibrated.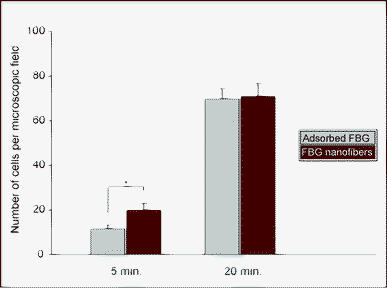 | ||
| Fig. 3 HUVEC attachment to FBG-coated glass coverslips (grey bars) and electrospun FBG nanofibers (red bars) at 5 and 20 minutes after plating. Data show the average cell number per microscopic field. | ||
After two hours of incubation, cells had been given enough time to develop their shape, and we then examined the overall cell morphology. When adhered to regular FBG-coated samples, the cells presented a typical flattened morphology (Fig. 4a) comprising random cell polarization and a well-developed cytoskeleton with actin fibers that inserted into focal adhesions complexes (Fig. 4d). In sharp contrast, adhesion to random nanofibers promoted an irregular cell shape with multiple cytoplasmic projections extending towards differently oriented fibers (Fig. 4b). The cell protrusions showed high accumulation of actin that co-localized with vinculin in the focal adhesions (Fig. 4e) which often inserted long actin fibers with centripetal organization, suggesting that cells insert some traction over the nanofibers. On aligned FBG nanofibers, the cells acquired a morphology strongly associated with the spatial organization of the underlying nanofibers. They typically presented an extended morphology that strongly followed the fibers orientation (Fig. 4c). The elongated cell shape corresponded to the highly extended actin stress fibers that were inserting into relatively small focal adhesion complexes located mostly at the polarized cell edges (Fig. 4f). Analysis of cell orientation with respect to the underlying fibers showed that cells oriented along the aligned fibers (Fig. 4i), while on random fibers and the regular FBG-coated sample the cell orientation was equally distributed in all directions (Fig. 4g and 4h).
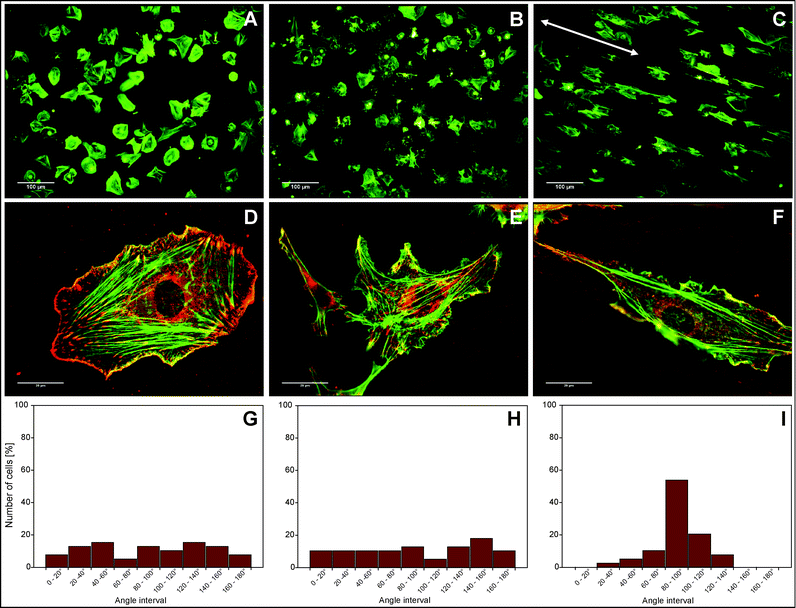 | ||
| Fig. 4 Cellular interactions with FBG nanofibers after 2 hours of incubation in serum-free medium. (A–C) Overall cell morphology of HUVECs on adsorbed FBG (A), randomly deposited nanofibers (B), and aligned nanofibers (C), where the direction of fiber alignment is indicated by the white arrow. (D–F) Immunofluorescent micrographs showing the development of focal adhesion complexes (vinculin in red) and the actin cytoskeleton (phalloidin in green) on adsorbed FBG (D), random nanofibers (E), and aligned nanofibers (F). (G–I) Distribution of cell orientation on adsorbed FBG (G), randomly deposited nanofibers (H), and aligned nanofibers (I), measured as the angle between the longitudinal cell axis and the line perpendicular to the nanofiber direction. | ||
Cell motility
Time-lapse recordings demonstrated that the cells were highly motile on the aligned FBG nanofibers (see ESI videos 1 and 2†), with individual cells undergoing the typical motile cycle of extending a leading cell edge, followed by traction of the tail cell edge. While most of the cells on aligned FBG nanofibers carried out a linear path of translocation, cells on random nanofibers performed rather oscillatory and relatively short movements and in different directions (see ESI videos 3 and 4†). Quantitative analysis of cell motions confirmed that cellular movements on aligned nanofibers clearly coincided with the fiber orientation, while cells on random nanofibers did not display any preferred direction of movement (Fig. 5a). Moreover, cells on aligned nanofibers were confirmed to traverse significantly longer distances (track length) than cells on random nanofibers (Fig. 5b).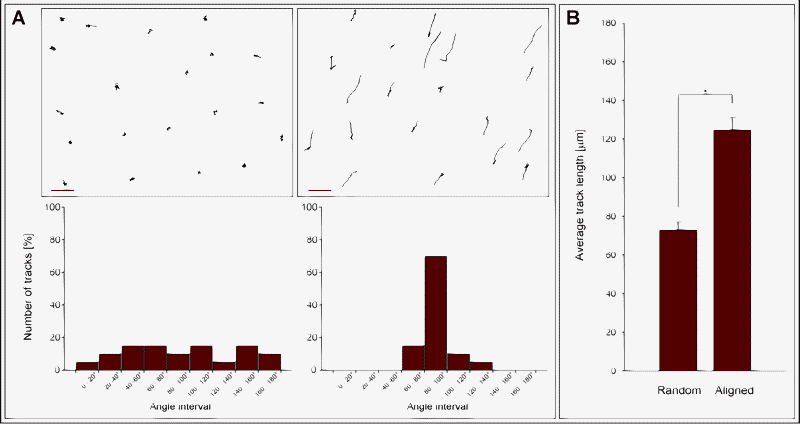 | ||
| Fig. 5 Cell motility on FBG nanofibers. (A) HUVEC cell tracks on random (upper left) and aligned nanofibers (upper right). Scale bars represent 100 μm. The distributions of the cell track orientation on random and aligned fibers are shown below each graph. (B) Average cell track length on random and aligned FBG nanofibers. | ||
Long-term cell culture on FBG nanofibers
To follow the long-term fate of HUVECs grown on FBG nanofibers, cells were cultured on both random and aligned samples for up to 7 days. First, to learn whether the orientation of underlining nanofibers affected the ECM deposition, the cell layers were stained for FN after three days of incubation. On random fibers, the cell-secreted FN was rather stochastically distributed (Fig. 6a), while on aligned nanofibers the FN matrix clearly assembled along the main fiber direction (Fig. 6b). When cells were cultured further in time, they formed confluent layers on both aligned and random nanofibers within 5–7 days. By staining the cells for their actin cytoskeleton at the end of the culture period, remarkable differences in the organization of cells within the confluent cell layers depending on the spatial organization of the fibers could be observed. On aligned samples, the orientation of the actin cytoskeleton was dominated by the direction of underlying nanofibers, resulting in a uniform linear pattern coinciding with the cell polarization (Fig. 6d). In contrast, on random samples there was no specific orientation of the actin bundles (Fig. 6c).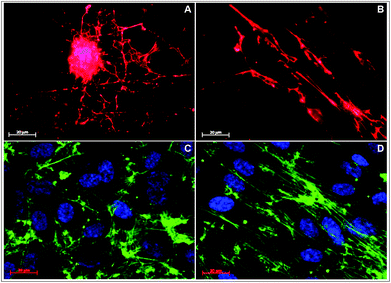 | ||
| Fig. 6 Deposition of fibronectin secreted by HUVECs cultured for 3 days on random (A) and aligned FBG nanofibers (B). Indirect immunofluorescent staining of HUVEC cultured for 7 days on random (C) and aligned FBG nanofibers (D) with phalloidin (green) and Hoechst (blue). | ||
Discussion
Successful cell–biomaterials interaction is critical for all tissue engineering applications, and depends not only on the adsorption of adhesive proteins,1 but also on the spatial organization of the proteins.29 Therefore, spatial engineering of cell–materials interfaces via reconstruction of supramolecular ECM protein structures is, in general, highly sought for,8,30 and precise reproduction of the fibrillar ECM organization is of specific importance as the ECM is by nature a fibrous structure. In that context, electrospinning of polymer solutions is known to be a potent engineering approach for a simple and fast production of biomimetic nanofibers that resemble natural ECM fibrils,31 and it has been repetitively used to create ultrathin fibers from a variety of different matrix proteins,32 including fibrinogen and fibrin.26,33 The latter achievements are of interest for multiple reasons. For example, fibrin(-ogen) is the main component of the provisional ECM during wound healing, and it is also a well-characterized acute phase protein of the innate immune response that attracts many adhesive glycoproteins to serve as a reservoir for growth factors, proteases, and protease inhibitors.34,35 Moreover, FBG acts as regular ECM for several epithelial cells where it, interestingly, assembles into non-covalent fibrils,24 which emphasizes the importance of the spatial organization of the protein itself. That observation raises the question: to what extent cells recognize and respond to spatially organized FBG sized in the range of natural ECM fibrils? At the same time, little is still known about the role of fibrillar FBG structures that are not formed via the traditional conversion of FBG into insoluble fibrin, i.e. by enzymatic cleavage by thrombin and further stabilized by transglutaminase (i.e. factor XIII).9 To address those questions, we herein used tailor-made technology for deposition of FBG nanofibers in either random or aligned configurations. Using established protocols26 we obtained fibers with an average diameter of 192 nm, which compares well with previously reported values between 80 and 700 nm depending on the FBG concentration (80–170 mg mL−1).26 However, two important aspects of electrospun FBG need to be clarified: (1) the influence of the electrospinning process on the protein structure and (2) the long-term stability of the electrospun fibers in physiological aqueous environments. In this context, one has to remember that it is still not clear how FBG becomes insoluble during electrospinning. One possibility is that FBG is normally cleaved to fibrin if traces of thrombin are present in commercial FBG. Here, our electrophoretic data clearly showed that FBG did not convert to fibrin during electrospinning as it did not display the additional band present in fibrin caused by the fibrinopeptide loss.9 This observation confirms that FBG molecules within the electrospun nanofibers assemble and become insoluble by other mechanisms. One can consider the possibility that electrospinning provides the conditions where FBG molecules polymerize by van der Waals forces of attraction that overcome the electrostatic forces of repulsion, giving rise to a peculiar amyloid-like product (aggregate) similar to those described under some pathological conditions (e.g. Alzheimer's disease).36 It should also be kept in mind that extrahepatic FBG assembles into an insoluble matrix fibrils in presence of cells (fibroblasts, lung and mammary epithelial cells), in a process where cryptic beta residues are exposed to form FBG fibrils.35 However, the relevance of such an assembly mechanism in absence of cells is not clear.With respect to the second issue mentioned above, i.e. the long-term stability of electrospun FBG fibers under aqueous conditions, it has been detailed by Baker et al. that the mechanical properties of electrospun FBG fibers decrease significantly when the fibers are immersed in wet environments (e.g., the stiffness decreases more than 1000 times).37 Therefore, to understand how aqueous conditions affected the electrospun FBG fibers, we used FITC-labeled FBG for fluorescent tracing of the protein under physiological solutions. That approach revealed that approximately 5% of the protein fibers dissolved during 24 hours of incubation. In our view, that degradation rate is not an obstacle as initial cellular interactions go very fast, typically within minutes (Fig. 3), and also the endothelial cells will secrete many other matrix proteins (e.g. fibronectin, von Willebrand factor, etc.) during the first 24 hours from seeding, which will follow the initial fiber orientation and thus contribute to the cellular response. Indeed, our study demonstrates coextensive organization of FN matrix fibrils at the third day of incubation (Fig. 6). Nevertheless, upon prolonged incubation times, about 30% of the electrospun FBG fibers were dissolved, and such degradation has to be carefully considered for potential biomedical applications.
Following the initial characterization of the electrospun fibers in acellular environments, we then turned our attention to cellular interactions with the electrospun FBG fibers. It is known that FBG expresses a number of cell-binding domains identified on Aα, Bβ and γ-chains. Specifically, the RGD integrin-binding sequence in the Aα-chain and the heparin binding domain within the Bβ-chain particularly mediate cell–matrix interactions.35,38 According to the electrophoretic profile of FBG fibers, the Aα, Bβ and γ-chains were maintained intact after electrospinning, and thus the electrospun FBG fibers were expected to efficiently promote cell adhesion. Indeed, our data showed that cells adhered equally well on FBG fibers as on glass coverslips coated with FBG. Interestingly, we also showed that the adhesion process went significantly faster on fibrillar FBG compared to FBG adsorbed on a flat surface (Fig. 3). Considering that the fibers were deposited in a random layer dense enough to not permit penetration of cells (i.e. cell adhesion was constrained to a limited surface area), and that the ligand-concentrations on both fibrillar and flat substrata were saturated with respect to optimal cell adhesion (about 30 μg mL−1 is sufficient to give maximal cell adhesion according to our own experience), the observed fast cell adhesion on fibrillar FBG indicates an important role of the spatial ligand organization to rapidly engage in integrin activation. Similar observations were made by Cukierman et al.29 proposing that cell–matrix interactions in three-dimensions occur as a combination of normal focal adhesions and fibrillar adhesions that together mediate cell adhesion much more efficiently than any of those mechanisms do alone. It seems that such advanced matrix-binding structures are pre-synthesized during culture, and when cells are back in a matrix-like environment they rapidly regenerate them.29 Such adhesion mechanisms could explain the improved fast cellular interactions with nanofibrous FBG, although one also has to consider the pure geometrical response to the (nano)fibrous environment.43 Recent studies indicate that cells sense and respond to different nanofiber geometries. For example, Li et al. showed that chondrocytes secrete an increased amount of ECM when seeded on nanofibers compared with the microfiber matrix.44 It is still not clear how cells recognize and respond to certain fibrillar patterns, which should not be equalized with “classical” topographic environments as they differ in many aspects. For example, microscopic analysis and time-lapse imaging revealed that an isotropic topography (having identical values of a property in all directions) did not alter cell morphology but highly induced cell motility.45 Our studies, however, with random nanofibers clearly show that multidirectional fiber orientation affects cell morphology, accompanied by cell immobilization. On the other hand, an anisotropic environment can have variable impact on cell behaviour (e.g. depending on the ridge–groove ratio), while the highly aligned FBG nanofibers induced elongated cell morphology, and importantly provoked directional cell movement. It is generally difficult to match the topographic cell response with the bioactivity of nanofibers. Nevertheless, our results confirm the relevance of studying and learning more about the reciprocal and adaptive interactions between cells and the surrounding matrix in the interface between two and three dimensional environments.39,43
Another important issue that we want to stress on, and which connects with the above observation, is the morphological response of adhering endothelial cells. We found clear differences in the overall cell morphology depending on the spatial organization of fibers. On aligned fibers, cells presented an extended morphology that strongly followed the fiber orientation, while on random nanofibers, cells spread in multiple directions. The presence of highly extended actin stress fibers inserting into focal adhesions suggests that endothelial cells on aligned nanofibers exert traction over the fibers. Also on randomly oriented fibers, cells were shown to form protrusions containing focal adhesions, suggesting strong cellular interaction with the substratum. Interestingly, the overall shape of endothelial cells on random fibers resembles, to a large extent, the stellate-like morphology characteristic for cells residing in three dimensional environments,39 suggesting that cells read the geometry of the underlining nanofibrous substratum (i.e., 2.5D). One should also consider that nanofibers do not provide enough surface area for larger clustering of integrins and thus resemble more the natural 3D ECM environment.40 On both aligned and random fibers we observed small focal adhesion complexes (explained by the small surface area of the fiber itself, rather than insufficient contact), which resemble the natural matrix contacts. Collectively, this study provides morphological evidence for well-established focal adhesion complexes and even for the development of matrix contacts that could explain the facilitated initial adhesion of endothelial cells on FBG nanofibers.
Beyond the events of initial cellular adhesion and subsequent spreading, time-lapse studies revealed the patterns of cell migration on the FBG nanofibers. Understanding the factors that control cell migration is, in general, important when designing implants for optimal cellular infiltration and integration with native tissue. In accordance with other studies,41 we show that endothelial cells not only oriented themselves well to aligned nanofibers, but also that they moved significantly along them. Conversely, on randomly deposited fibers, cells were locally immobilized and their accumulated traversed distance was reduced, presumably by their strong anchorage from multiple projections extended to differently oriented fibres. Nevertheless, while guiding the movement of cells in a specific direction is important for recruitment of endothelial cells, e.g. for colonization of scaffolds or angiogenesis, local immobilization of cells may also be of interest for tissue engineering applications, e.g. for controlled cell behaviour on blood contacting devices where up-regulation of the adhesive phenotype might be very important.42 Our data indicate that abrogated motility of HUVECs on random fibers could support their local immobilization to the device. In our present work we are now exploring how the balance of adhesive and motile phenotypes influences the cellular differentiation, trying to address the problem of endothelial cells dedifferentiation.
We finally explored whether FBG nanofibers were able to affect the long-term behavior of endothelial cells. For that purpose we cultured HUVECs on random and aligned nanofibers for one week, and interestingly, the orientations of the underlying nanofibers were clearly reflected in the organization of the confluent cellular layers. Not only was the cellular actin cytoskeleton well-aligned with the fiber direction, but the newly generated fibronectin fibrils also followed the organization of the cytoskeleton and the nanofiber direction. Those observations suggest that the described aligned FBG nanofibers are a promising template for oriented ECM deposition.
Conclusions
This study provides insights into how endothelial cells respond to spatially organized ECM-like stimuli such as pure FBG nanofibers. Being relatively stable for several days under physiological conditions, FBG nanofibers were well recognized by HUVECs and showed a clear ability to modulate cell behaviour via the orientation of the fibers. Collectively, this study shows that FBG nanofibers have potential for tissue engineering applications, but also that they represent a simple model system that can allow for further explorations of the natural interaction between cells and their extracellular environment.Acknowledgements
This work was supported by CIBER-BBN via the European Commission through the FP7 Industry-Academia Partnerships and Pathways (IAPP) project 324386 (FIBROGELNET) and the EuroNanoMed project STRUCTGEL.References
- C. J. Wilson, R. E. Clegg, D. I. Leavesly and M. J. Pearcy, Tissue Eng., 2005, 11, 1 CrossRef CAS.
- A. L. Berrier and K. M. Yamada, J. Cell. Physiol., 2007, 213, 565 CrossRef CAS.
- W. P. Daley, S. B. Peters and M. Larsen, J. Cell Sci., 2008, 121, 255 CrossRef CAS.
- C. Frantz, K. M. Steward and V. M. Weaver, J. Cell Sci., 2010, 123, 4195 CrossRef CAS.
- H. Järveläinen, A. Sainio, M. Koulu, T. N. Wight and R. Penttinen, Pharmacol. Rev., 2009, 61, 198 CrossRef.
- P. Lu, K. Takai, V. M. Weaver and Z. Werb, Cold Spring Harbor Perspect. Biol., 2011, 3, a005058 CrossRef.
- S. Agarwal, J. H. Wendorff and A. Greiner, Polymer, 2008, 49, 5603 CrossRef CAS.
- G. A. Silva, C. Czeisler, K. L. Niece, E. Beniash, D. A. Harrington, J. A. Kessler and S. I. Stupp, Science, 2004, 303, 1352 CrossRef CAS.
- M. W. Mosesson, J. Thromb. Haemostasis, 2005, 3, 1894 CrossRef CAS.
- T. Lisman, C. Weetering and P. G. de Groot, Front. Biosci., 2005, 10, 2504 CrossRef CAS.
- J. W. Weisel, Adv. Protein Chem., 2005, 70, 247 CrossRef CAS.
- R. Raghow, FASEB J., 1994, 8, 823 CAS.
- K. M. Yamada and R. A. F. Clark, in The Molecular and Cellular Biology of Wound Repair, ed. R. A. F. Clark, Plenum, New York, 2nd edn, 1996, pp. 51–93 Search PubMed.
- M. J. Flick, X. Du, D. P. Witte, M. Irouskova, D. A. Soloviev, S. J. Busuttil, E. F. Plow and J. L. Degen, J. Clin. Invest., 2004, 113, 1596 CAS.
- V. W. van Hinsbergh, A. Collen and P. Koolwijk, Ann. N. Y. Acad. Sci., 2001, 936, 426 CrossRef CAS.
- M. Dietrich, J. Heselhaus, J. Wozniak, S. Weinandy, P. Mela, B. Tschoeke, T. Schmitz-Rode and S. Joeckenhoevel, Tissue Eng., Part C, 2013, 3, 216 CrossRef.
- Q. Ye, G. Zünd, P. Benedikt, S. Jockenhoevel, S. P. Hoerstrup, S. Sakyama, J. A. Hubbell and M. Turina, Eur. J. Cardio-Thorac., 2000, 17, 587 CrossRef CAS.
- H. E. Davis, S. L. Miller, E. M. Case and J. K. Leach, Acta Biomater., 2011, 7, 691 CrossRef CAS.
- C. J. Hunter, J. K. Mouw and M. E. Levenston, Osteoarthr. Cartilage, 2004, 12, 117 CrossRef.
- M. Hojo, S. Inokuchi, M. Kidokoro, N. Fukuyama, E. Tanaka, C. Tsuji, M. Miyasaka, R. Tanino and H. Nakazawa, Plast. Reconstr. Surg., 2003, 111, 1638 CrossRef.
- S. O. Lawrence and P. J. Simpson-Haidaris, Thromb. Haemost., 2004, 92, 234 CAS.
- E. P. Molmenti, T. Ziambaras and D. H. Perlmutter, J. Biol. Chem., 1993, 268, 14116 CAS.
- S. Y. Lee, K. P. Lee and J. W. Lim, Thromb. Haemost., 1996, 75, 466 CAS.
- M. Pereira, B. J. Rybarczyk, T. M. Odrljin, D. C. Hocking, J. Sottile and P. J. Simpson-Haidaris, J. Cell Sci., 2002, 115, 609 CAS.
- R. Tzoneva, T. Groth, G. Altankov and D. Paul, J. Mater. Sci.: Mater. Med., 2002, 13, 1235 CrossRef CAS.
- G. E. Wnek, M. E. Carr, D. G. Simpson and G. L. Bowlin, Nano Lett., 2003, 3, 213 CrossRef CAS.
- M. C. Mcmanus, E. D. Boland, D. G. Simpson, C. P. Barnes and G. L. Bowlin, J. Biomed. Mater. Res. Part A, 2007, 81, 299 CrossRef.
- C. Ayers, G. L. Bowlin, S. C. Henderson, L. Taylor, J. Shultz, J. Alexander, T. Telemeco and D. G. Simpson, Biomaterials, 2006, 27, 5524 CrossRef.
- E. Cukierman, R. Pankov, D. R. Stevens and K. Yamada, Science, 2001, 294, 1708 CrossRef CAS.
- K. Salchert, U. Streller, T. Pompe, N. Herold, M. Grimmer and C. Werner, Biomacromolecules, 2004, 5, 1350 CrossRef.
- Q. P. Pham, U. Sharma and A. G. Mikos, Tissue Eng., 2006, 12, 1197 CrossRef CAS.
- S. A. Sell, P. S. Wolfe, K. Garg, J. M. McCool, I. A. Rodriguez and G. L. Bowlin, Polymer, 2010, 2, 522 CAS.
- S. R. Perumcherry, K. P. Chennazhi, S. V. Nair, D. Menon and R. Afeesh, Tissue Eng., Part C, 2011, 17, 1121 CrossRef CAS.
- N. Laurens, P. Koolwijk and P. M. de Maat, Thromb. Haemostasis, 2006, 4, 932 CrossRef CAS.
- B. J. Rybarczyk, S. O. Lawrence and P. J. Simpson-Haidaris, Blood, 2003, 102, 4035 CrossRef CAS.
- M. Fändrich, Cell. Mol. Life Sci., 2007, 64, 2066 CrossRef.
- S. Baker, J. Sigley, C. C. Helms, J. Stitzel, J. Berry, K. Bonin and M. Guthold, Mater. Sci. Eng., C, 2012, 32, 215 CrossRef CAS.
- T. M. Odrljin, C. W. Francis, L. A. Sporn, L. A. Bunce, V. J. Marder and P. J. Simpson-Haidaris, Arterioscler., Thromb., Vasc. Biol., 1996, 16, 1544 CrossRef CAS.
- F. Grinnell, Trends Cell Biol., 2003, 13, 264 CrossRef CAS.
- B. Geiger, J. P. Spatz and A. D. Bershadsky, Nat. Rev. Mol. Cell Biol., 2009, 10, 21 CrossRef CAS.
- H. G. Sundararaghavan, R. L. Saunders, D. A. Hammer and J. A. Burdick, Biotechnol. Bioeng., 2013, 110, 1249 CrossRef CAS.
- A. de Mel, G. Jell, M. M. Stevens and A. M. Seifalian, Biomacromolecules, 2008, 9, 2969 CrossRef CAS.
- L. G. Griffith and M. Swartz, Nat. Rev. Mol. Cell Biol., 2006, 7(3), 211 CrossRef CAS.
- W.-J. Li, Y. J. Jiang and R. S. Tuan, Tissue Eng., 2006, 12, 1775 CrossRef CAS.
- E. Lamers, R. Van Horssen, J. Te Riet, F. C. Van Delft, R. Luttge, X. F. Walboomers and J. Jansen, Eur. Cell. Mater., 2010, 20, 329 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Videos 1–4; one figure. See DOI: 10.1039/c3bm60124b |
| This journal is © The Royal Society of Chemistry 2013 |
