Nanotopography – potential relevance in the stem cell niche
Lesley-Anne
Turner
 * and
Matthew
J. Dalby
* and
Matthew
J. Dalby
Centre for Cell Engineering, Institute of Molecular, Cell and Systems Biology, College of Medical, Veterinary and Life Sciences, Joseph Black Building, University of Glasgow, Glasgow G12 8QQ, Scotland, UK. E-mail: lesley-anne.turner@glasgow.ac.uk
First published on 11th September 2014
Abstract
Understanding signals in the niche that regulate stem cell behaviours is important for applications such as tissue engineering, and limitations in ex vivo/in vitro recapitulation of some stem cell niches, particularly the bone marrow, have led researchers to attempt to delineate signalling mechanisms present in vivo using a reductionist approach. This is especially important as ‘stemness’ is not solely an intrinsic property of stem cells but a result of the reciprocal interactions between stem cells and their niches. Physical stimuli such as mechanical stiffness and topography are known to significantly impact stem cell behaviours; being translated through adhesions, intracellular tension and mechanotransduction, which can alter gene expression and thus cell fate. In this review general properties of the stem cell niche are initially described, using intestinal and bone marrow niches as examples. The lesser-described physical stimuli of nanotopography and the mechanisms by which stem cells respond and interact with it are described, including biochemical and physical mechanotrasduction, chemical and physical signal integration and adhesion mechanisms in both anchorage-dependent and -independent cells. Specific examples of nanotopographical influence over stem cell differentiation are highlighted and parallels drawn between the stem cell niche and these ‘synthetic’ in vitro observations. Ultimately if the complex stem cell niche is to be mimicked in vitro or stem cells exploited for medical applications the physical microenvironment, including nanotopography, must be optimised.
1. Introduction
The stem cell niche2 can be considered as a basic unit of tissue physiology,3 which both defines the specific anatomical location in which a stem cell resides and confers functionality (regulating stem cell survival, self renewal and differentiation).4 Conversely, the attributes that enable stem cells to be so fundamental for tissue generation and repair also prime them for undesirable effects such as unregulated proliferation resulting in tumour formation. Thus, in the body fine control of stem cells through the niche is critical for tissue homeostasis; the niche provides a structured environment that protects the stem cell from the body (protection from depletion or undesired signalling) and the body from stem cells (providing an environment that precisely controls stem cell self renewal in order to avoid tumourigenesis). In the niche stem cells are maintained in a metabolically quiescent state that protects them from DNA damage until they are required, at which point they produce progenitor/transit amplifying cells that go forward to regenerate damaged tissue.5 This niche control over progenitor production is a result of the integration of complex, dynamic signals from surrounding tissue, enabling the niche to respond to the needs of the organism in a balanced manner. This specialised niche function, of precisely controlling stem cell self-renewal whilst, which enables differentiation when required, is mediated by a specific microenvironment. The niche microenvironment is composed of cell and non-cellular elements that present chemical and physical cues in order to facilitate stem cells central roles in tissue generation and repair (Fig. 1). Understanding these cues will enable us to characterise the stem cell niche, which is essential if we are to exploit stem cells for therapeutic and research use, e.g. in regenerative medicine.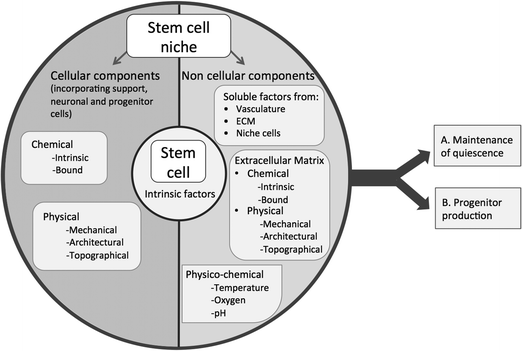 | ||
| Fig. 1 Schematic diagram illustrating potential factors within the stem cell niche that control stem cell quiescence and progenitor production. | ||
2. The stem cell niche
In humans somatic niches, which are required for tissue repair and are thought to be necessary to ensure stem cell longevity and multipotency, are found in many tissues including: muscle (satellite stem cells located beneath muscle fibre basal lamina), brain (hippocampus), skin (bulge of hair follicle), intestines (epithelium) and bone marrow (putatively the sinusoidal surface). The later two are the most well described and investigated in humans, and thus will be used as the main examples in this review. They can be considered as examples of 2D and 3D niches – intestinal stem cells are exposed to a basal lamina on one side and lumen on the other (2D), and bone marrow stem cells encompassed by extracellular matrix (ECM)/cells on all sides (3D). Although there is heterogeneity both between niches in different tissues and also within the same type of niche depending on functional state (e.g. homeostasis, regeneration, development), similarities between different niches in humans as well as other organisms are thought to exist,1,6,7 including:1. Niches contain specialised cells (be that niche cells of different lineages and/or stem cell progeny) and are found in specific anatomical locations (both of which differ depending on stem cell type).
2. The niche functions as a physical anchor for stem cells, which may be through cell–cell or cell–ECM adhesions.
3. The niche appears to function to regulate stem cells and orchestrate their behaviour, be that quiescence, self-renewal or differentiation, in response to signals from the body.
4. The niche is a dynamic structure that integrates cues from potentially many sources, e.g. ECM, chemical factors, metabolic cues, mechanical stimuli, architectural constraints, cell–cell contacts, and the stem cells themselves.
5. Blood vessels are often found near niches, which potentially enable the transport of long range signals and enable recruitment/egress of cells.
The intestinal niche
One of the most well defined stem cell niches is the intestinal niche, where adult intestinal stem cells enable the epithelial lining of the intestine to renew and repair daily throughout life in response to the aggressive luminal environment, where in humans up to 1011 epithelial cells are lost every day.8 This extensive cell renewal arises from relatively small populations of leucine-rich repeat-containing heterotrimeric guanine nucleotide-binding protein-coupled receptor 5 (LGR5+) stem cells. These LGR5+ stem cells reside in protected niches formed by crypts at villi bases (Fig. 2). During tissue renewal transit amplifying cells proliferate upwards towards the top of crypts, where they differentiate into epithelial cells. Thus, the intestinal crypt niche enables LGR5+ stem cells to indefinitely self-renew whilst at the same time regenerating functional epithelia. Although a complex mechanism that is incompletely understood, a number of components in the niche that regulate this bi-directional behaviour have been described. Paneth cells that neighbour LGR5+ stem cells secrete niche specific factors (e.g. epidermal growth factor, WNT3A and notch ligand) and are thought to be essential for the translation of tissue/body factors into signals for stem cells (e.g. calorie restriction induces Paneth cells to reduce mTOR complex 1 signalling, which in turn reduces the size of the stem cell pool). The ECM component laminin is thought to be important in the intestinal niche; along the crypt-villus axis variations in laminin composition in the basement membrane are thought to play a role in establishing and maintaining stem cell distributions.9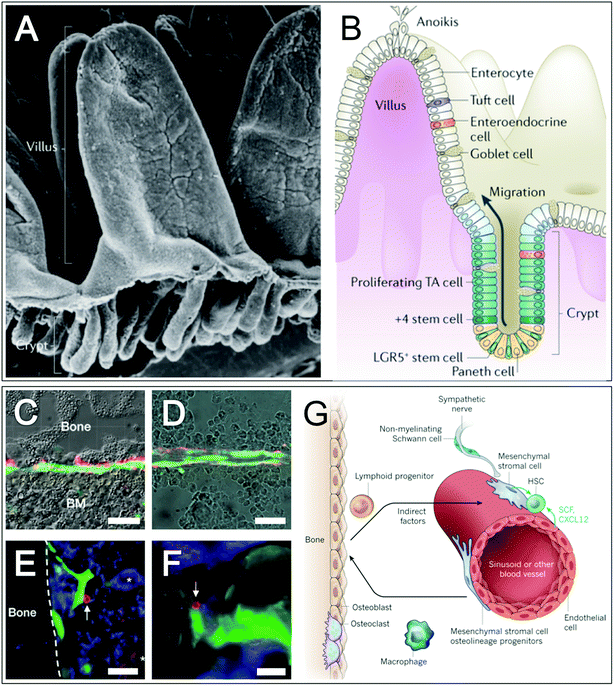 | ||
| Fig. 2 Intestinal and bone marrow stem cell niches. Intestinal villi and crypts are shown in scanning electron microscope (A) and schematic (B) images, highlighting the location of the intestinal stem cell niche at crypt bases, TA = transit amplifying. Optical images of the endosteal region (C) and bone marrow parenchyma (D) in mice with fluorescent images overlaid show mesenchymal stem cells (nestin+ cells) (MSCs) and neuronal cells (catecholaminergic fibres) in green and red respectively. Fluorescent images of the endosteal region (E) and sinusoids (F) of mice, highlighting haematopoietic stem cells (CD150+CD48−Lin−), haematopoietic progenitors cells (CD48+) and MSCs in red with arrows, blue and green respectively. Megakaryotes (CD150+CD48+Lin+) are highlighted with asterisks. Schematic (G) of a proposed bone marrow stem cell niche. Scale bars are 50 μm (C–F). Images A and B,8 adapted by permission from Macmillan Publishers Ltd: Nat. Rev. Mol. Cell Biol., 15(1), 19–33, DOI: 10.1038/nrm3721, copyright (2013). Images C–F,13 adapted by permission from Macmillan Publishers Ltd: Nature, 466(7308), 829–834, DOI: 10.1038/nature09262, copyright (2010). Image G,10 adapted by permission from Macmillan Publishers Ltd: Nature, 16(505), 327–34, DOI: 10.1038/nature12984, copyright (2014). | ||
The bone marrow niche
In contrast to the intestinal stem cell niche, the bone marrow supports two stem cell types – mesenchymal stem cells (MSCs) and haematopoietic stem cells (HSC), which have a co-dependant relationship. This niche is less well characterised and far less linear than the simpler intestinal niche and as such a number of competing models have developed to describe it.10 MSCs supply the local tissues with skeletal cells (fat, bone, cartilage, reticular and HSC supporting).11 It should be noted that MSCs can be derived from other tissues (e.g. fat, umbilical cord and dental tissue) and thus have other niches, and furthermore the term MSC is a contested one. A perivascular location for MSCs is hypothesised (suggesting crossover with pericytes12), however as MSCs are also extractable from non-vascularised niche tissue (e.g. cartilage) this may indicate differing populations of MSCs. HSCs replace more than 500 billion blood cells everyday (including platelets and red, myeloid and lymphoid cells). In the bone marrow, HSC numbers are kept fairly constant except in times of haematopoietic stress when HSCs are able to mobilise and move to extramedullary sites. During development, the HSC niche is found in diverse tissues including the liver where the number of HSCs expands daily. The MSC/HSCs niche is perivascular and often, although not always, located near trabecular bone10,13 (Fig. 2). Endothelial cells (which share a common lineage with haematopoietic cells) are thought to make up the cellular element of the niche and synthesise multiple factors that promote HSC maintenance and localisation (including stem cell factor and C–X–C motif chemokine). There is a great deal of evidence to suggest that other cell types are important indirect regulators of the bone marrow niche, including: sympathetic neurons that confer a circadian rhythm of HSC retention and mobilisation,14 non-myelinating Schwann cells, osteoclasts, monocytes/macrophages and haematopoietic cells.15,16In addition to the sinusoidal niche, the endosteum has previously been proposed as a bone marrow MSC/HSC niche. However, research now suggests that rather than comprising an integral part of the niche, i.e. being niche constituent cells, osteoblastic and osteolineage cells of the endosteum are thought to contribute to the HSC niche as indirect regulatory components.17–19 That said, it should be noted that HSCs are predominantly found in the trabecular region of bone marrow and the endosteal region is highly vascularised, suggesting a degree of overlap.
3. Signalling in the stem cell niche
The impact and control that stem cell niche signalling has over stem cell behaviour is exemplified by experiments carried out on Drosophilia20,21 and in mice22 whereby committed progenitor cells can resort to stem cells (dedifferentiate) if returned to the niche – suggesting self-renewal and multipotency control by the niche rather than cell intrinsic control. It therefore follows that stemness is considered as not solely an intrinsic property of stem cells but a result of the reciprocal interactions between stem cells and their niches.1 The complex stem cell niche microenvironment is composed of cell (be that niche cells of different lineages and/or stem cell progeny) and non-cellular elements that present chemical and physical cues on macro, micro and nano scales. Using a ‘reductionist’ approach we can delineate the complex signalling of the stem cell niche and gain deeper understanding of the factors controlling stem cell renewal and differentiation. Much of the analysis on the impact of specific signals on stem cell behaviours has been carried out in vitro, e.g. with the aim of recapitulating the stem cell niche.Mimicking the niche in vitro – intestinal organoids
With the great potential of stem cells in regenerative medicine, recapitulating the stem cell niche in vitro/ex vivo is a point of interest. The simpler intestinal crypt is a focus of bioengineering and a number of methods have been used for the growth of intestinal organoids in vitro/ex vivo; the most successful of which8 used lamin-rich Matrigel™ (an animal derived ECM product) and a cocktail of growth factors found in the endogenous stem cell niche.23 The crypt-villus organoid units that formed (Fig. 3) were organised into discrete crypts with both stem and Paneth cells at their base and villus-like structures at the apex. As these organoids were grown from single mouse LGR5+ stem cells embedded in a laminin matrigel the importance of both ECM molecules and niche relevant chemicals for intestinal niche formation are highlighted. In addition the observation that laminin was important in this system correlates with its enrichment at the crypt base in vivo.24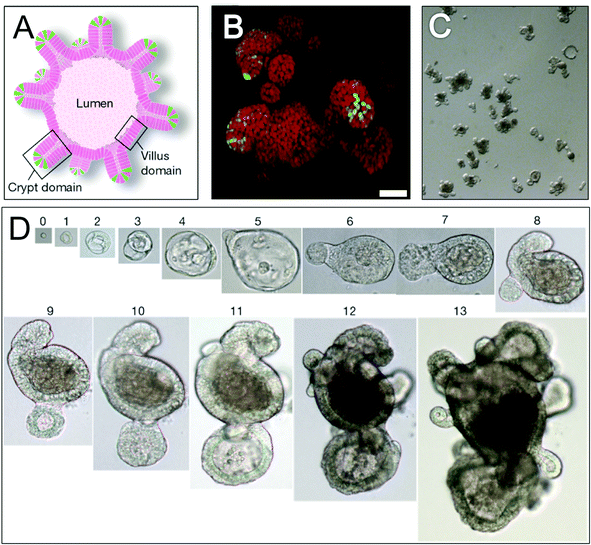 | ||
| Fig. 3 Mimicking the intestinal niche in vitro: organoids. Schematic representation (A) and three-dimensional reconstructed confocal image (B) of an intestinal organoid, highlighting leucine-rich repeat-containing heterotrimeric guanine nucleotide-binding protein-coupled receptor 5 (LGR5+) stem cells (green) at crypt bases and other intestinal cells (red). Optical image (C) shows an organoid suspension derived from a single cell organoid. Optical images (D) showing organoid growth with time from a single LGR5+ stem cell exposed to appropriate conditions (see text). Numbers above images = days of growth. Scale bar is 50 μm (B) and magnification (D) is ×40, ×20, ×10 and ×4 for days 0–4, 5–7, 8–11 and 12–13 respectively. All images,23 adapted by permission from Macmillan Publishers Ltd: Nature, 459(7244), 262–265, DOI: 10.1038/nature07935, copyright (2009). | ||
Mimicking the niche in vitro – bone marrow
Although in vitro recapitulation of the bone marrow HSC niche has been extensively investigated, researchers have had less success than that observed with the intestinal niche and as of yet HSCs are unable to be expanded in vitro. It has been known since the 70s that co-culture with stromal cells, such as MSCs, promotes HSC survival.25 Ex vivo expansion protocols often contain varying concentrations of growth factors (although many other factors are involved in vivo).16 A number of ECM molecules have been identified with structural roles in the HSC niche and some are thought to have specialised, niche-specific functions, including fibronectin, laminin, hyaluronic acid and osteopontin.16 Although clearly beneficial to HSC survival and expansion, neither growth factors, cytokines, chemokines, individual cells or individual ECM molecules have proven sufficient to produce environments conducive to significant HSC maintenance and expansion in vitro. Rather it is likely that a complex combination of factors will be required and research is moving in this combinatorial direction.Together with a cocktail of cytokines, nanofibre scaffolds have been shown to increase the expansion efficiency of HSCs, with enhancement of stemness thought to be related to increased adhesion26 (Fig. 4). In another study, when cultured with cytokines on fibronectin coated microwells that were 10 μm in height and 15–80 μm in diameter, HSCs remained quiescent in smaller diameter wells where the cavity housed individual cells27 (Fig. 4). These observations suggest that cell–substrate contact area is important for HSC renewal and retention of the immature state, which in turn may be related to the number of engaged adhesion structures. The importance of dimensionality/adhesion for maintenance of HSC stemness has been further described by Raic et al., whereby HSC stemness was retained to a greater degree in HSCs co-cultured with MSCs, using a cytokine cocktail, in a three dimensional system compared to a standard two dimensional culture system28 (Fig. 4).
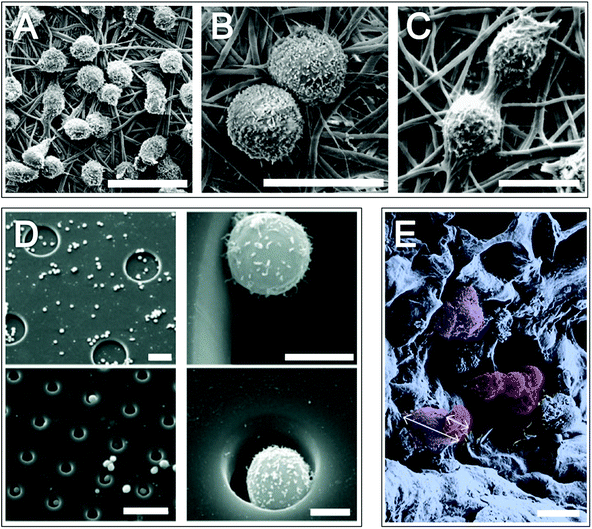 | ||
| Fig. 4 Mimicking the bone marrow niche in vitro: haematopoietic stem cell growth on materials. Scanning electron micrograph images (A–C) of haematopoietic stem cells (HSCs) growing on polyethersulfone 1,4-butanediamine nanofibre meshes, highlighting cell filopodia (B) and division (C). HSC adhesion on fibronectin coated microwells fabricated in poly(ethane-alt-maleic anhydride) on silicone (D). Images on right hand side are magnifications of left hand side images. Pseudo-coloured SEM image of mesenchymal stem cells (purple) and HSCs (red) on a porous hydrogel (E); white arrows highlight different cell dimensions. Scale bars = 20 μm (A), 10 μm (B and C) 50 μm (left images in D) and 5 μm (right images in D) and 20 μm (E). Images A–C,26 reprinted from Chua et al., Functional nanofibre scaffolds with different spacers modulate adhesion and expansion of cryopreserved umbilical cord blood hematopoietic stem/progenitor cells, Exp. Hematol., 2007, 35(5), 771–781, copyright (2007), with permission from Elsevier. Image D,28 reprinted from Raic et al., Biomimetic macroporous PEG hydrogels as 3D scaffolds for the multiplication of human hematopoietic stem and progenitor cells, Biomaterials, 2014, 35(3), 929–940, copyright (2014), with permission from Elsevier. Image E reproduced in part from ref. 27 with permission from The Royal Society of Chemistry. | ||
Effect of physical stimuli on stem cell behaviours
The effects of chemical stimuli (be that solid or free states) on stem cell behaviours are relatively well characterised29–36 and defined growth factors are routinely used in vitro to control stem cell renewal and differentiation.37–39 Accordingly, the remainder of this review will focus on the effects of physical stimuli on stem cells, with emphasis on how these signals may be interpreted by cells and the relevance of nanotopography in the stem cell niche, which are less well defined.40Physical signals in the stem cell niche, and tissue in general, come from both cells and the ECM. The ECM is recognised as an important signalling factor in tissues,41 especially in the form of basement membranes in the niche.3 For example in mice hyaluronic acid42 appears to have an important role in maintaining HSCs43 and neuronal stem cell populations44 and in humans different integrin expression on skin stem cells constrains them to what is presumed to be ECM glycoprotein ligands.45,46 Aside from it's important role in chemical signalling (both through intrinsic chemistry and binding of soluble factors), the physical influence that ECM has over cells can be broken down into architectural, mechanical and topographical. ECM architecture can confine cells, expose them to 2D or 3D environments and regulate their geometry, e.g. cell shape is known to influence stem cell differentiation with well spread flattened cells and rounded cells differentiating down osteogenic and adipogenic lineages respectively.47 Mechanical stimuli, e.g. shear flow, compression or substrate stiffness, influence stem cell differentiation. Finally topography, which is the least studied of the three and in particular nanotopography, will be reviewed in the following section with emphasis on niche specific behaviours such as differentiation and proliferation.
4. Mechanisms by which cells may interpret nanotopographical signals
Cells interact with topographic stimuli via cell adhesion molecules at their periphery, which allow the cell to physically connect with the adjacent structure and functionally provide a route for bi-directional communication by which the topographical environment can be translated into intracellular messages (mechanotransduction). Mechanotransduction can propagate through the cell as biochemical (indirect) or physical (direct) mechanotransduction (discussed below) and effect behaviours as diverse as growth, apoptosis, morphology, proliferation and differentiation. It is the type and arrangement of adhesions that determines cell responses to topographical stimuli.Adhesions
Different cells have differing types and concentrations of adhesion molecules mediating cell–extracellular and cell–cell attachments, which include: cadherins, integrins, CD44, syndecans and discoidin domain receptors (DDRs) (although syndecans and DDRs ‘true’ status as adhesion receptors remains to be clarified).48 Integrin proteins and CD44 proteoglycans are thought to provide high affinity cell–ECM adhesions in anchorage dependent cells. Integrins are well-characterised; they bind a number of ECM molecules, including laminins and fibrillar collagens.49 The cell surface proteoglycans CD44 and syndecan bind, respectively: glycoproteins, glycosaminoglycans and hyaluronic acid50,51 and a wide range of molecules including adhesive ECM molecules and growth factors.52 Whilst the structure of individual ECM components vary widely, many share common motifs; for example, the arginine-glycine-aspartic acid (RGD) motif found on the hydrophilic loops of a number of ECM molecules including fibronectin, vitronectin and tenascin. It is these motifs, or ligands, which cell–substrate adhesion molecules bind to with specificity, for example integrins bind RGD. Adhesions can be enhanced by synergistic cell binding to additional ECM structures/motifs, for example syndecans and DDRs in particular are thought to synergistically cooperate with integrins.53,54 Adhesions (be that cell–cell or cell–substrate) have been identified as important in stem cell functions such as maintenance, proliferation and differentiation42,55,56 as well as niche functions such as cell anchoring, recruitment/egress and potentially control of division (whereby adhesion formation potentially regulates cell polarisation and thus switching between symmetrical and asymmetrical stem cell division).57 In particular, integrin mediated adhesions are thought to be important in both HSC and skin stem cell niches.58In non anchorage-dependent cells adhesion is less pronounced, which when viewed in vitro manifests as spherical cells rather than flattened as is observed with anchorage dependent cells (Fig. 5). Although less pronounced, cell–substrate adhesions are specific. Research carried out by Franke et al. demonstrated that when HSCs (non anchorage dependent cells) were cultured upon a range of ECM molecules stronger adhesion was observed between fibronectin and integrins, weaker adhesion between heparin and selectins and in some instances, for example on tropocollagen, no adhesion was observed at all.59
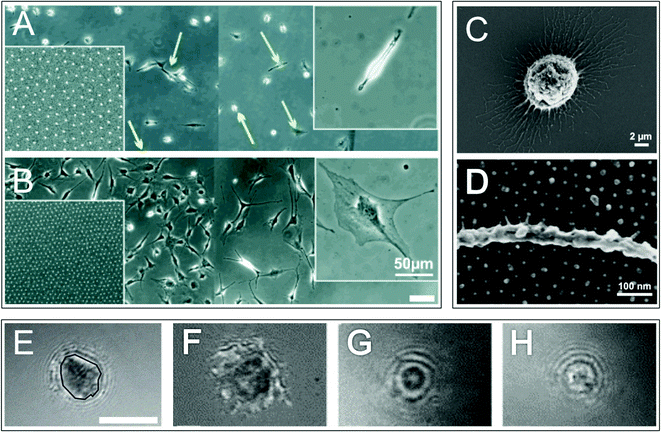 | ||
| Fig. 5 Adhesion in anchorage dependent and independent cells. Optical micrographs of osteoblasts cultured on a non-adhesive surface patterned with adhesive arginine-glycine-aspargic acid (RGD) ligands separated by approximately 85 nm (A) and 28 nm (B) (the right side of the main images are non-adhesive areas). Left insets show nanodot patterns and right insets show magnified images of a typical cell on each surface. Green and yellow arrows (A) highlight migrating and quiescent cells respectively. Optical micrograph images (C and D) of human acute myeloid leukemia cell line KG-1a cells (model cell line for immature haematopoietic stem cells) on a non-adhesive surface patterned with adhesive RGD ligands separated by 36 nm show cell protrusions and adhesion points. Reflectance interference contrast microscopy (RICM) images of haematopoietic progenitor cells grown on: fibronectin (E and F), heparin (G) and tropocollagen (H). An example of a cell–substrate adhesion area is highlighted with a black line (E). Three types of adhesion are illustrated: irregularly shaped areas with dark contrast highlight tight membrane–substrate contact indicating integrin mediated adhesion (F), small circular shaped dark areas show a smaller contact zone and indicate selectin mediated adhesion (G) and small circular shaped bright areas highlight non-adherent cells (H). Scale bars are 100 μm (A and B) and 5 μm (E–H). Images A and B,61 reproduced with permission from Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, copyright 2004. Images C and D,64 reproduced from Muth et al., Regulation of hematopoietic stem cell behavior by the nanostructured presentation of extracellular matrix component, PLoS One, 2013, 8, e54778. Images E–H59 reprinted from Franke et al., Engineered matrix coatings to modulate the adhesion of CD133+ human hematopoietic progenitor cells, Biomaterials, 2007, 28(5), 836–843, copyright 2007, with permission from Elsevier. | ||
In addition to the type of cell adhesion molecule and cell/ECM ligand, cell adhesion is dependent on ligand density and arrangement. Integrin clustering (multimerisation or gathering), which enables more complex adhesive structure formation (please see next section ‘adhesion structures’ for more details), only happens in anchorage dependent cell adhesion to RGD sequences if a maximum inter-ligand distance of 58–72 nm between attachment sites is presented.60,61 In addition, the minimum unit of clustered integrins that forms a functional cell adhesion has been described as a tetrameric configuration.62 In contrast, non-anchorage dependent cells such as HSCs have different threshold dimensions, for example HSCs adhering to RGD required ligand density thresholds in the range of 32–45 nm63,64 (Fig. 5). Interestingly, threshold distances have been found to vary depending on the type of ligand that HSCs are adhering to, with osteopontin requiring less than approx. 75 nm and FNRGD approximately 100 nm.64 Differences in threshold distances between anchorage dependent cells and non-anchorage dependent HSCs are hypothesised to be a result of differences in the actin cytoskeleton and adhesion related proteins in the two cell types. In integrin mediated focal adhesions of anchorage-dependent cells, the principle adhesion kinase is focal adhesion kinase (FAK), whereas in integrin-linked multiprotein complexes of HSCs the primary kinase is Pyk2.65–67 The actin cytoskeletons of the two cell types differ, being fibrous in anchorage dependent cells but ring like in HSCs, which enable them to maintain their round shape even when adhered.64 We note that the different ligand specificity of different RGD formats (linear, place in sequence etc.) may lead to alterations in some of these values and this needs to be checked between reports.68
Adhesion structures
In the integrin adhesome (‘the network of protein interactions that potentially link integrins to the actin cytoskeleton’) with integrin clustering comes structural and signalling protein recruitment and connection with the cell cytoskeleton, which leads to the formation of larger adhesion structures (adhesion maturation). Many adhesion structures have been described, including: nascent adhesions, focal complexes, focal adhesions, podosomes, fibrillar adhesions and three dimensional matrix adhesions69 (Fig. 6). All are dense, complicated structures (for a detailed review see ref. 70) and although heterogeneous, they share common features. One of the best-defined adhesions is the focal adhesion, comprised of integrins, structural proteins, adaptor proteins, signalling molecules, and cytoskeletal components.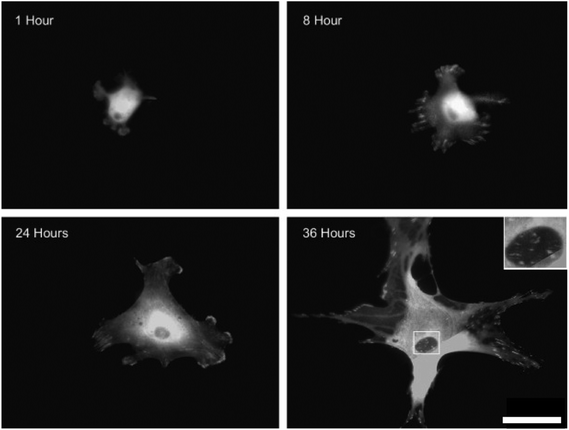 | ||
| Fig. 6 Integrin mediated adhesion structures. Fluorescent microscopy images of vinculin stained human osteoblasts cultured on planar poly(methyl methacrylate) substrates. As the osteoblasts spread, the focal complexes present 8 hours after seeding were slowly replaced by focal adhesions and then super mature adhesions by 36 hours. Boxed area shows sub nuclear adhesion formation on the cellular ventral membrane. Scale bar = 50 μm. Images,69 reprinted from Biggs et al., The use of nanoscale topography to modulate the dynamics of adhesion formation in primary osteoblasts and ERK/MAPK signaling in STRO-1+ enriched skeletal stem cells, Biomaterials, 2009, 30(28), 5094–5103, copyright 2009, with permission from Elsevier. | ||
There are differing theories as to how early adhesions nucleate and elongate (there is likely to be overlap), including: initiation by integrin binding and clustering followed by assembly of the cytoplasmic adhesome complex (including actin binding) and alternatively, assembly of cytoplasmic adhesome components, co-localisation and then integrin binding.71 Integrin adhesion structures are dynamic and can change with time, potentially maturing from small and transient nascent adhesions to progressively larger and more stable focal complexes then focal adhesions and finally fibrillar adhesions that are able to rearrange ECM. As focal contacts mature, in addition to increasing in size they are thought to change in composition, structure and function; going from contacts that transmit strong tractional forces to adhesions that are thought to be more passive anchoring structures that maintain a spread cell morphology.72 Adhesion size appears to be an important factor for cell mechanics; increasing adhesion size has been found to correlate with increased cellular tension.17 Recruitment of molecules and cross linking expands and strengthens the adhesion as well as decreasing the force generated/experienced per molecule,18 thus feeding back into force generated signalling controlling adhesion maturation.73,74 Adhesive structures vary between cell types, for example: migrating cells don't have fibrillar adhesions, neutrophils/macrophages show nascent adhesions and focal complexes and fibroblasts and smooth muscle cells display focal complexes and focal adhesions.71 Cells are able to bridge adhesions between integrin clusters – i.e. if integrin clusters are large enough cell structural proteins can bridge between the integrin clusters to form larger adhesions. This appears to be dependent on ECM composition with cells on fibronectin able to bridge between 500 nm diameter clusters and cells on vitronectin only requiring 200 nm diameter clusters.75,76 It should be noted that there is some evidence that the adhesions formed in vitro may differ in some ways to those formed in vivo, especially when considering cells that exist naturally in a three dimensional environment. Differences are thought to arise in adhesion composition, localization and function; for example three dimensional adhesions generally show enhanced biological activities, less integrin usage, fewer stress fibres and less stringent cytoskeletal organisation compared to their two dimensional counterparts.77
Biochemical (indirect) mechanotransduction
Upon ligand binding intracellular integrin domains undergo conformational change, which, through signalling protein activation (such as FAK) and/or physical transmission through cytoskeletal proteins (such as talin and alpha actinin), results in signal propagation through the cell. Both direct signalling protein activation and cytoskeletal protein interactions can result in activation of downstream biochemical signalling cascades, hence the term biochemical/indirect mechanotransduction. Mechanotransduction via signalling protein activation follows mechanisms similar to signalling cascades generated by soluble chemicals such as growth factors – once a mechanotransducer is activated, multiple pathways can cause inhibition or enhancement of any one particular signalling cascade (for example FAK activation of the ERK/MAPK (extracellular signal related kinase/mitogen activated protein kinase) pathway) and is known to effect various cellular responses such as proliferation and differentiation.78–80Many molecules and structures are immobilised on the insoluble cytoskeleton anchored to adhesion sites; the cytoskeleton is thought to act as both a scaffold for solid-state biochemical reactions and as a reservoir for sequestered soluble factors.81,82 It therefore follows that if integrin signalling is sufficient to influence cytoskeletal molecular conformation thus modifying cytoskeleton shape, tensions, structure or kinetics, any of the structures associated with the cytoskeleton may also be influenced.83 Where these structures are signalling molecules, if sequestered molecules are released or binding sites exposed or hidden, downstream biochemical signalling cascades may be enhanced or inhibited following mechanisms similar to biochemical signalling cascades initiated by soluble chemicals e.g. cytokines and growth factors.84 In this way, mechanical force can be transduced through the cytoskeleton and using solid-state chemistry can be converted into biochemical reactions (Fig. 7).
Physical (direct) mechanotransduction
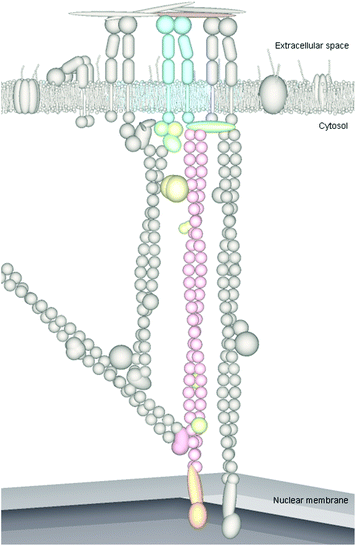
As well as considering cells as biochemical units, they can also be considered as mechanical units with perhaps both features contributing to cell responses to materials. Perhaps the best theory presented to date of how cells could respond as mechanical units can be found in tensegrity theory. The cell cytoskeleton can be described as a tensegrity structure, whereby opposing forces act in unison to maintain the shape of the structure, whilst providing strength and resilience; this means that cells are maintained in a prestressed state and are in equilibrium under a balance of forces.85 That is, the cytoskeletal network is under isometric tension, which removes any slack in the system; ‘lack of slack’ means that any mechanical stresses applied to the system (cytoskeleton) are immediately sensed. The tensegrity architecture model therefore provides a mechanism whereby mechanical stimulation of the cell cytoskeleton via attachment proteins, at a point on a cell's membrane, causes the cell to react as a whole and results in integrated, global changes in cytoskeletal structure.86 Cytoskeletal stresses tend to dissipate less in stiffer structures, thus the stiff prestressed cytoskeleton is able to concentrate and focus stresses and facilitates longer distance force propagation compared to less stressed structures.As an extension to physical mechanotransduction through the cytoskeleton, mechanical pull on integrins can be further propagated through the cytoskeleton to cell nuclei, causing nuclear envelope86 and whole nuclei87 distortion. The cytoskeleton is contiguous with the nucleoskeleton (also called the karyoskeleon and nuclear matrix88) and is connected via LINC (linker of nucleoskeleton and cytoskeleton) complexes (Fig. 7). LINC complexes span the nuclear envelope; on the cytosol side they bind cytoskeletal proteins including F-actin, intermediate filaments and microtubules, and intranuclearly LINC complexes transmit forces to the nucleoskeleton and chromatin (for example by directly binding lamins, which in turn can bind DNA either directly or indirectly through matrix attachment regions (MARS) on telomeres). Cytoskeletal forces are transduced through LINC complexes into the nucleus, analogous to force transmission through focal adhesions to the cytoskeleton. Also similarly to the adhesion complexes described previously, LINC complexes can be transient or stable and are thought to form a range of ‘adhesion’ types. Stiffness differentials between the cytoskeleton and nucleus (the nucleus is the stiffest organelle and is approximately nine times stiffer than the surrounding cytoskeleton86) are thought to facilitate force propagation over the relatively large distances between the cell membrane and nuclear envelope.89 Within the cytoskeleton intermediate filaments are thought to play a more significant role in force transfer to the nucleus compared to actin microfilaments due to mediation of force transfer at both high and low strains, whereas actin mediation is only conferred at low strains.86
The nucleoskeleton is a permanent network of core filaments underlying thicker fibres and is composed of proteins including lamins, titin, actin, nuclear myosins and kinesins. Not only does the nucleoskeleton confer specific shape, mechanical properties and functionality to the nucleus and genome, similarly to the cytoskeleton, many molecules and structures are immobilised on the insoluble structure. This immobilisation is proposed to act as a platform for functional complexes required for nuclear activities, such as transcription and DNA repair.88,90,91 The nucleoscaffold appears to exhibit precise spatial order to the nucleus in terms of chromosome organisation, DNA replication, transcription and processing of RNA.83 Distortion of the cell nucleus or nuclear envelope may alter the molecular conformation of the nucleoskeleton similarly to the manner in which the cytoskeleton is altered in response to adhesion dynamics. Nucleoskeletal proteins have been shown to influence a number of processes, including: transcription, replication and DNA repair, thus alterations in their shape, tensions, structure or kinetics may also influence these associated properties.92 Interestingly, mutations in the genes encoding A-type lamins of the nuclear envelope have been linked to ageing of adult stem cells and their niches.93,94
Cytoskeleton mediated physical mechanotransduction may present a number of advantages over soluble biochemical mechanotransduction conferred via diffusion. The quick response system means that mechanical forces can cause rapid effects, more so than biochemical signalling that takes diffusion time. In addition to quick response times, cytoskeletal mediated signalling channels forces along discrete fibres of the cytoskeleton providing a mechanism by which to concentrate stresses on specific molecules at particular locations, whilst protecting other cellular components from these mechanical forces, meaning that only a subset of molecules may experience force levels strong enough to alter their activities.91
Alterations in gene expression as a result of mechanotransduction and intracellular tension
Alterations in intracellular tension are thought to have important consequences for stem cells, with high, low and intermediate levels of tension promoting osteogenic differentiation, adipogenic differentiation and stem cell renewal respectively.47,95,96 The mechanisms that translate mechanotransduction into alterations in gene expressions are less well understood compared to cytosolic mechanotransduction, although a number of mechanisms have been proposed. One mechanism hypothesises that when nuclei are deformed, chromosome positions within the nuclei are distorted.96–100 Chromosomes occupy discrete territories within the interphase nucleus and because transcription factors and machinery are thought to be in varying concentrations in different regions of the nucleus, altering chromosome positions may affect their accessibility for transcription and therefore alter gene expression.101 Nuclear envelope distortion alone may be a factor in the mechanotransduction of forces;102,103 Itano et al. proposed that calcium ion (Ca2+) release from the perinuclear space after envelope distortion acts on calcium ion regulated transcription factors, which in turn alter gene expression levels.103 Other mechanisms include the translocation or activation of transcription factors. Focal adhesion signalling may result in translocation of focal adhesion proteins to the nucleus where they can act as transcription factors; for example once released, research has shown that zyxin may translocate and alter transcription of genes such as endothelin-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 78 and that FAK can shuttle from focal adhesions to operate within the nucleus, where it targets ubiquitination of the cell-cycle mediator p53 (tumour protein 53) and can act as a transcription co-regulator with the GATA4 zinc-finger transcription factor, which is linked to embryogenesis.104–106 Two transcriptional co-activators involved in altering gene expression regulating cell growth are YAP and TAZ. Mechanical properties have been shown to alter the locations and activity of YAP and TAZ: in stiff cells they are active and located in the nucleus whereas in compliant cells they are inactive and located in the cytosol.107–109 YAP and TAZ are also proposed to mediate a mechanical memory in cells, acting as rheostats that store information about a cells physical environment and influence cell fate.107
78 and that FAK can shuttle from focal adhesions to operate within the nucleus, where it targets ubiquitination of the cell-cycle mediator p53 (tumour protein 53) and can act as a transcription co-regulator with the GATA4 zinc-finger transcription factor, which is linked to embryogenesis.104–106 Two transcriptional co-activators involved in altering gene expression regulating cell growth are YAP and TAZ. Mechanical properties have been shown to alter the locations and activity of YAP and TAZ: in stiff cells they are active and located in the nucleus whereas in compliant cells they are inactive and located in the cytosol.107–109 YAP and TAZ are also proposed to mediate a mechanical memory in cells, acting as rheostats that store information about a cells physical environment and influence cell fate.107
Signal integration
Signals mediated through adhesions can be integrated with other local signals, such as those initiated by soluble chemical factors. For example, integrins co-localise with other signalling structures, including growth factor receptors and stress-sensitive ion channels.110 Many molecules involved in signalling cascades other than those initiated by integrin signalling (such as growth factors and cytokines) are immobilised on the focal adhesion cytoskeleton, bringing these downstream molecules and mechanotransduced pathways into close proximity.81,111–113 In addition to co-localisation and focal adhesion orientation, different signalling molecules and biochemical pathway components can be brought into close association by scaffolding proteins, which act as ‘hubs’ facilitating the recruitment and organisation or target proteins, resulting in tethering of multiple components of signalling pathways to one location. This effect is further extended by scaffold proteins interacting directly with one another or through bridging proteins to form higher order macromolecular complexes.114In focal adhesions, scaffolding proteins include FAK, paxillin and RACK1. Scaffolding proteins are an integral part of adhesome signal organisation and their interactions follow specific trends, which can be described using network motifs (defined as unique patterns of interactions between proteins that appear significantly more often in the real network compared with randomised networks115). The adhesome has been found to be composed of network motifs that consist of binding interactions regulated by on/off switches, the most common of which were three-node motifs consisting of a scaffolding protein, signalling molecule and its down-stream target.70 That these were the most common of the adhesome motifs highlights the importance of the adhesome in integrating mechanical and chemical signalling.
Complimentary to spatial control over signal integration, tensegrity architecture offers a model by which mechanical signals can be globally integrated. Tensegrity architecture describes a way in which forces transmitted through localised adhesion sites induce rearrangements throughout the cell within a tensionally integrated cytoskeleton.113 The sensitivity of the system is dependent on the concentration of prestresses in the cytoskeleton91 and connection to adhesion molecules.86 Taken together these two mechanisms illustrate how signal integration can be achieved on both local and global scales, being mediated by for example focal adhesions (integration ‘hubs’) and the cytoskeleton respectively.
In order for stem cells to effectively function in the stem cell niche, the different signals that they are exposed to must be integrated to give a ‘whole-cell’ response. In addition the complex signals relayed to the niche itself must be integrated, e.g. by niche support cells, to ensure precise temporal and spatial control over stem cell proliferation and differentiation.
Synthetic materials
Using techniques traditionally associated with electronic engineering and materials science, such as photolithography and reactive ion etching,116–118 defined topographic patterning can be achieved in biomaterials enabling researchers to home-in on specific physical cues influencing cell behaviours. This is particularly useful for characterising the effects of topography and mechanotransduction on the stem cell niche.Where biomaterials are incorporated into culture systems, new functionality will also be conferred. Cells do not tend to interact directly with biomaterials, rather, when cell solutions are brought into contact with biomaterials (be they naturally derived or synthetic) a layer of proteins adheres to the surface within milliseconds.119 Protein adsorption is affected by the properties of the material120 (Fig. 8), including surface charge, wettability (hydrophobicity) and topography. It is this adsorbed layer of protein that is thought to influence cell interactions with the underlying material.121
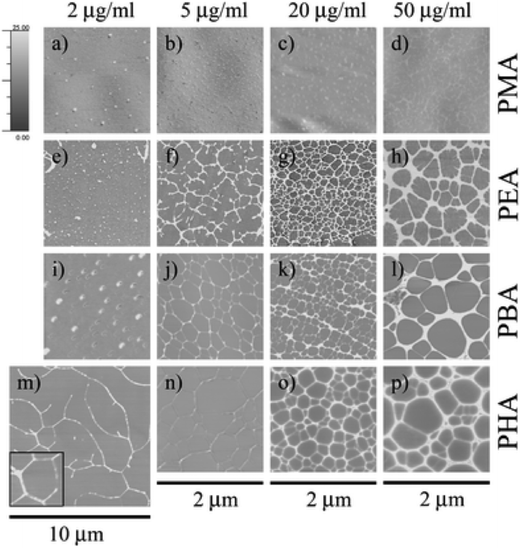 | ||
| Fig. 8 Protein adsorption on different materials. Atomic force microscopy images of fibronectin adsorbed onto poly(methyl acrylate), poly(butyl acrylate), poly(ethyl acrylate) and poly(hexyl acrylate) (PMA, PEA, PBA and PHA respectively) after 10 minutes of incubation in different solution concentrations. Images reproduced in part from ref. 120, with permission of The Royal Society of Chemistry. | ||
5. Stem cell responses to nanotopography
The importance of scale
Many researchers have shown that cells are able to respond to differences in topography with behaviours as diverse as adhesion, morphology, proliferation and differentiation being influenced (e.g. ref. 100 and 122–126). Over 100 years ago, early work described cells as being responsive to topographic features127 and the term ‘contact guidance’ was first published by Weiss in 1945![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 128 (Fig. 9).
128 (Fig. 9).
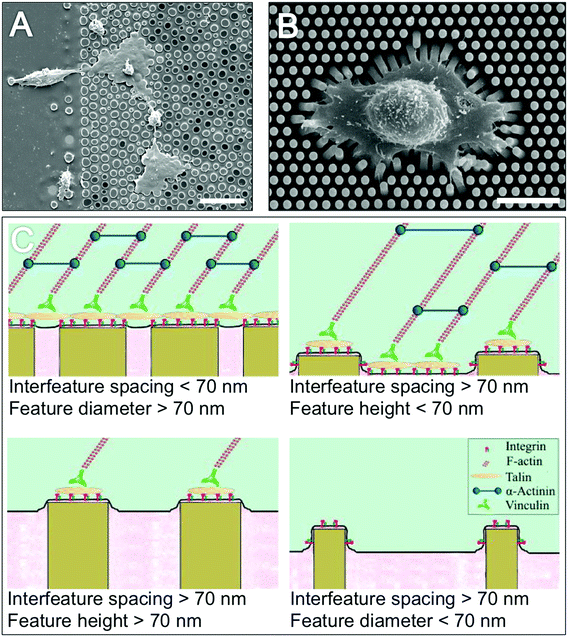 | ||
| Fig. 9 Topographic influences over cell morphology and adhesion. Electron micrograph image (A) of fibroblast contact guidance on poly(styrene) pillars. Electron micrograph image (B) of a fibroblast grown on flexible polydimethylsiloxane pillars illustrate the tractional forces that cells exert on the substrates on which they are grown. Schematic diagrams (C) illustrate the importance of topography feature dimensions for cell adhesion formation, adhesion frequency and cytoskeletal stresses. Scale bar is 20 μm (A) and 15 μm (B). Image A,126 reproduced with kind permission from Springer Science and Business Media. Image B,122 reproduced with permission from Trichet et al., Evidence of a large scale mechanosensing mechanism for cellular adaptation to substrate stiffness, PNAS, 2012, 109(18). Image C,131 reprinted from Biggs et al., Nanotopographical modification: a regulator of cellular function through focal adhesions, Nanomedicine, 2010, 6(5), 619–633, copyright 2010, with permission from Elsevier. | ||
More recently the importance of topography scale has been identified. Adhesion formation and dynamics occurs on the nanoscale; for example integrins span the 10 nm thick plasma membrane and the attached actin cytoskeleton is separated by a 40 nm-high focal adhesion core-region consisting of strata with specific roles. These strata include a signalling layer consisting of integrin cytoplasmic tails, FAK and paxillin (signalling proteins), an intermediate force transducing stratum containing talin and vinculin; and an actin-regulatory strata containing VASP (vasodilator-stimulated phosphoprotein), zyxin and α-actinin.129 Furthermore, in order to connect nanoscale actin filaments to microscale actin contractile stress fibre bundles cells appear to use adhesion related particles which are small enough to fit between gathered integrins.130 Thus the effects of scale are important; due to their sizes being in the same order of magnitude, micron scale features can be considered as ‘housing’ cells whereby cells are confined by substrate features. In contrast, because nanoscale features are an order of magnitude smaller than cells and on the same size scale as their molecular components, nanotopography can be considered as interacting with cells, whereby nanoscale structures act as multiple signalling points (Fig. 9).
Nanotopography is known to influence the way in which adhesions form on substrates and is thought to be the signal initiation point of cell-nanotopography interactions.69,131,132 By controlling adhesion size and type, adhesion composition, function and strength (tractional force as a result of the actin-myosin cytoskeleton) can be influenced. Cytoskeletal tension has been shown as an important transducer of physical stimuli through cells, with e.g. in vitro studies show that MSCs differentiate down differing lineages when cultured on substrates of varying stiffness, and in the developing mouse embryo early embryogenesis is halted if myosins are knocked out.47,73,133 Thus tractional force is thought to play a large part in cell responses to nanotopographical stimuli due to its influence on cytoskeletal tension (which as described previously can effect biochemical and physical mechanotransduction and ultimately niche relevant cell behaviours such as proliferation and gene expression). Tractional force is known to vary between adhesion structure types, for example, focal complexes are under great tractional force (their size is not thought to correlate with the force experienced72,134) whereas focal adhesions are under less tractional force (their size shows linear correlation with tractional force for a given substrate stiffness122). Thus a route between nanotopography influenced adhesions, cytoskeletal tension and niche-relevant stem cell signalling is described.
In vitro Observations
In terms of stem cell responses to nanotopography, MSCs are the most well-described and will be summarised in this section and Table 1. Nanotopography is known to influence cell adhesion; when cultured on nanopits (310 nm deep and 30 μm diameter) increased MSC spreading has been observed compared to cells cultured on both planar and groove patterned (327 nm deep, 50 μm wide) substrates.135 When cultured on nanopillars (6 nm high, 200 nm wide, centre to centre spacings of 290 nm), MSCs showed increased adhesion compared to those grown on planar substrates.136 This trend was also observed when MSCs were cultured on titania nanopillars (28 nm diameter, 15 nm high and 40 nm centre to centre spacing).137 Height was found to be an important factor in MSC adhesion to these nanopillars; for nanopillars with heights ranging from 15 to 100 nm, cell adhesion was increased on smaller pillars and decreased on larger ones.137 One of the most commonly charaterised cell responses to topographies are those of cell alignment and orientation in the direction of, or perpendicular to, grooves. This observation holds true for MSCs; for example cell cytoskeletal and nuclear elongation and alignment has been reported in response to nanogrooves (350 nm deep, 350 nm wide and 700 nm pitch) compared to planar substrates where no elongation or alignment was observed.138 It is noted that grooves wide enough to be step-cues rather than guidance cues can be more osteogenic than planar or narrow grooves.69| Cell type | Topography features | Topography dimensions | Cell response (compared to planar substrates unless stated) | References |
|---|---|---|---|---|
| Mesenchymal stem cells | Nanopits | 310 nm deep, 30 μm diameter. | Increased cell spreading. | 135 |
| 120 nm diameter, 100 nm deep, 300 nm centre to centre spacing, ordered geometry. | Retention of multipotency, linked to focal adhesion formation. | 131,140,142 | ||
| 120 nm diameter, 100 nm deep, 300 nm centre to centre spacing, disordered geometry. | Osteogenic differentiation, linked to focal adhesion formation. | 97,131,149 | ||
| Nanopillars | 6 nm high, 200 nm wide, centre-centre spacings of 290 nm. | Increased adhesion. | 136 | |
| 28 nm diameter, 15–100 nm high, 40 nm centre to centre spacings. | Increased cell adhesion on shorter pillars. | 137 | ||
| Nanogrooves | 350 nm deep, 350 nm wide, 700 nm pitch. | Cytoskeletal and nuclear elongation and alignment. | 138 | |
| 350 nm deep, 350 nm wide, 700 nm pitch. | Increased neuronal marker expression compared to both planar and micro grooved substrates. Differentiation response linked to focal adhesion formation. | 132,138 | ||
| Nanotubes | 30–100 nm diameter. | Larger tubes stimulated osteogenic differentiation compared to smaller diameter tubes. | 150 | |
| Biomimicry – banding | Helical nanoribbons with 63 nm helical periodicity. | Osteogenic differentiation compared to nanoribbons with 100 nm periodicity. Differentiation response dependent on mechanotransduction of mechanical stimuli. | 144 | |
| Embryonic stem cells | Nanopits | 120 nm diameter, 100 nm deep, 300 nm centre to centre spacing, ordered geometry. | Differentiation down a mesodermal lineage towards the osteoblastic lineage. | 146 |
| Nanogrooves | 350 nm wide, 500 nm high, 700 nm pitch (coated with gelatin). | Neuronal differentiation. | 147 |
Yim et al. demonstrated that MSCs respond to nanogrooves (dimensions of 350 nm depth, 350 nm width and 700 nm pitch) by up-regulating expression of neuronal, vascular and muscle cell markers.138 Neuronal marker expression was most significant, suggesting that nanogrooves can induce neuronal lineage differentiation in MSCs.138 It should be noted that this behaviour represents a transdifferentiation and although neural proteins were expressed (MAP2 and beta tubulin), a functional role has yet to be demonstrated. In this work nanogrooves induced significantly greater MSC differentiation compared to micro grooves, highlighting the importance of scale and demonstrating the potential power of nanotopography over micro topography in terms of cell differentiation. This influence of nanotopography was linked to focal adhesion formation, with smaller and more elongated cell focal adhesions on the nanogrooves compared to micro grooves and planar substrates, in addition topography induced gene expression was shown to be dependent on actomyosin contractility and FAK activity.132 The importance of topography scale was also observed in MSCs cultured upon nanotube titanium oxide with diameters ranging from 30–100 nm. Specifically, on tubes with 100 nm diameters cells differentiated down an osteogenic lineage, whereas on 30 nm tubes no noticeable differentiation was observed.139
The degree of disorder in nanopit arrangements have been shown to effect MSC differentiation; ordered nanopits (120 nm diameter, 100 nm depth, 300 nm centre to centre spacing) stimulated MSCs to proliferate within a multipotent state,140 whereas when a degree of disorder was introduced to the pits (a randomly orientated deviation of 50 nm from the centre) MSCs differentiated down an osteogenic lineage141 (Fig. 10). This osteogenic differentiation was comparable to differentiation stimulated using traditional chemically defined media. The responses of MSCs to pit disorder were hypothesised to be related to differences in adhesion formation (with pits being non-adhesive areas) resulting in differences in intracellular tension. MSCs on the highly ordered pattern had smaller adhesions than those on the disordered topographies, which developed super mature, or fibrillar, adhesions that were hypothesised to result in increased intracellular tension and thus differentiation down an osteogenic lineage.131,142
Biomimicry of nanotopography has been investigated in the culture of MSCs on helical synthetic nanoribbons, where helices with a 63 nm periodicity (thus mimicking type 1 collagen, the most common collagen found in bone, which displays a repetitive topographic banding pattern of 67 nm along the length of its fibrils143) induced osteogenic differentiation whereas a periodicity of 100 nm did not.144 Importantly, this differentiation effect was found to be dependent on the mechanotransduction of physical stimuli. Biomimicry has also been shown to influence adipose derived MSC behaviour when cultured on cell-imprinted substrates (the original cells were removed from the substrate before MSC culture, leaving behind a topographic pattern of their surface and residual cellular fragments).145 MSCs adopted the shape and certain gene expression profiles of the original cells, which was hypothesised to be at least partially a result of the micro and nanotopographies of the imprinted substrates.
Embryonic stem cells
The importance of the stem cell niche is exemplified with embryonic stem cells (ESCs); in order to culture ESCs in vitro, ESCs must be grown on specialised substrates, such as feeder layers (for example mouse embryonic fibroblasts) or Matrigel™. The necessity of such substrates makes it difficult to investigate the effects of nanotopography on ESC behavior, although a few studies have been carried out. When cultured on nanopit topographies (120 nm diameter, 100 nm depth, 300 nm centre to centre spacing and a randomly orientated deviation of 50 nm from the centre) ESCs differentiated down a mesodermal lineage towards the osteoblastic phenotype to a greater degree than cells cultured on planar substrates.146 On nanogrooves (350 nm wide, 500 nm high, 700 nm pitch) coated with gelatin ESCs differentiated down a neuronal lineage.147 It should however be noted that this observation is perhaps more complex as there is some evidence that neural differentiation is a default response in ESCs cultured without specific lineage differentiation stimuli.148This overview of stem cell responses to nanotopographical stimuli is summarised in Table 1.
Signal gradients
Signal gradients are known to be important in morphogenesis during development and may also play a role in the stem cell niche, for example soluble/ECM-tethered chemical gradients may enable fate decisions (e.g. self renewal or differentiation) to be controlled by the distance of the cell from the niche.7 Signal gradients may also be conferred by topographical signalling; for example, studies have shown that in some cell types cultured on grooved substrates topographic cues are able to propagate (in terms of cell migration) up to nine cells away from the original signal.151,1526. Correlations between nanotopographical control of stem cell behaviour in vitro and observations of niche components in vivo
We have described how nanotopography can influence and direct stem cell behaviours including gene expression and illustrated how nanotopographical signals acting via adhesion formation and cytoskeletal tension can be integrated with other chemical signals both locally and globally to give a whole-cell response. In vitro observations have enabled us to delineate and understand how specific nanotopographical cues influence stem cell behaviours and in some cases parallels between in vitro and in vivo observations can be drawn. Nanotopography is present in vivo on both ECM and cells. Endothelial cells form fenestrated sinusoidal capillaries in bone marrow with nanopores between 50 and 300 nm in diameter153 (Fig. 11). Collagen type X, which is found at sites of large fractures and endochondral ossification, displays a nanopattern where nanofeatures are separated by approximately 100 nm in a disordered lattice arrangement154 (Fig. 11). These two topographic structures bear resemblance and dimensions similar to the ordered and disordered 120 nm diameter pit structures described previously that in vitro were found to stimulate MSC phenotype retention and osteogenic differentiation respectively,140,141 highlighting a potential role for nanopits in the bone marrow MSC niche. The cell behaviours observed on these topographies in vitro (retention of multipotency and osteogenic differentiation on ordered and disordered pits respectively) also bear similarities to the cells found in these locations in vivo (perivascular MSCs and osteoblasts on sinusoidal fenestrations and type X collagen respectively).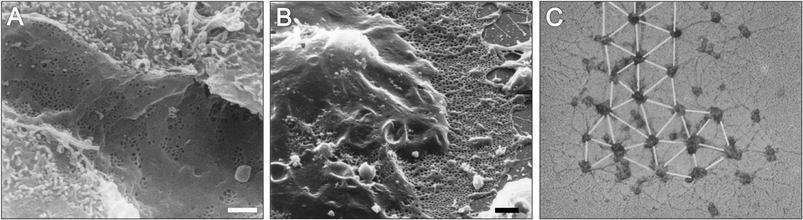 | ||
| Fig. 11 In vivo nanotopographies. Electron microscopy images showing (A) fenestrae in the rat sinusoidal epithelium (B) a liver sinusoidal endothelial cell after treatment with latrunculin highlighting large fenestrated areas and (C) type 10 chicken collagen. Scale bars = 1 μm (A), 2 μm (B) and 200 nm (C). Images A and B,153 reproduced from Braet and Wisse, Comp. Heptol., 2002, 1(1), licensee BioMed Central Ltd., copyright 2002, available at http://www.comparative-hepatology.com/content/1/1/1. Image C,154 copyright Kwan et al., Originally published in J. Cell Biol., 1991, DOI: 10.1083/jcb.114.3.597. | ||
7. Conclusions and perspectives
Traditional tissue engineering whereby cells are cultured on a scaffold in vitro then implanted have shown some success.155 However, future directions for tissue engineering are likely to involve engineering scaffolds with signals that promote in vivo population and potentially even attract and direct the behaviour of endogenous stem cells.4,156Ultimately if the complex stem cell niche is to mimicked in vitro or in vivo for tissue engineering applications the physical environment, including topography, must be optimised. Scale is important in this context because the natural stem cell niche presents topographic cues on macro, micro and nano scales, with each scale have differing types of interactions with cells. Micron scale features can be considered as ‘housing’ cells, where cells are confined by substrate features; in contrast nanotopography can be considered as interacting with cells, whereby nanoscale structures act as multiple signalling points. The power of nanotopography to direct stem cell behaviours is observed in vitro and parallels between these observations and the stem cell niche confound the importance of understanding the role of nanotopography in the stem cell niche.
For nanotopography, future directions will likely include:
1. Development of new tissue culture plastics for stem cell growth. Because topography is a purely physical principle it can be easily injection moulded into the base of traditional cell culture plates, wells and flasks, i.e. the next generation of cell culture materials can appear, at the macro scale, the same as the old generation. This will aid with MSC growth and targeted differentiation using standard culture protocols.
2. High throughput screening 1. For testing of drugs for effects on stem cells, having stem cells in the correct phenotype will be very important. Topographically patterned e.g. 96 well plates will help control the stem cell population while drug trials are undertaken. This should reduce variability and artefact.
3. High throughput screening 2. For understanding the range of stem cell responses achievable, nanotopographical arrays, as has been performed with microtopographical topochips,157 will help increase our understanding of the range of control we can gain over MSCs with nanotopography.
Acknowledgements
Image 10: thanks to Nikolaj Gaadegard for supplying the scanning electron microscopy images. MJD and LAT are funded by BBSRC, EPSRC and MRC.References
- I. Roeder, M. Loeffler and I. Glauche, Towards a quantitative understanding of stem cell-niche interaction: experiments, models, and technologies, Blood Cells Mol. Dis., 2011, 46, 308–317 CrossRef CAS PubMed.
- R. Schofield, The relationship between the spleen colony-forming cell and the haemopoietic stem cell, Blood Cells, 1978, 4, 7–25 CAS.
- D. T. Scadden, The stem-cell niche as an entity of action, Nature, 2006, 441, 1075–1079 CrossRef CAS PubMed.
- D. E. Discher, D. J. Mooney and P. W. Zandstra, Growth Factors, Matrices, and Forces Combine and Control Stem Cells, Science, 2009, 324, 1673–1677 CrossRef CAS PubMed.
- S. Tajbakhsh, Stem cell: what's in a name? 2009 Search PubMed.
- L. Li and T. Xie, Stem cell niche: structure and function, Annu. Rev. Cell Dev. Biol., 2005, 21, 605–631 CrossRef CAS PubMed.
- M. P. Lutolf and H. M. Blau, Artificial stem cell niches, Adv. Mater., 2009, 21, 3255–3268 CrossRef CAS PubMed.
- N. Barker, Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration, Nat. Rev. Mol. Cell. Biol., 2014, 15, 19–33 CrossRef CAS PubMed.
- M. Kedinger, O. Lefebvre, I. Duluc, J. N. Freund and P. Simon-Assmann, Cellular and molecular partners involved in gut morphogenesis and differentiation, Philos. Trans. R. Soc. London, Ser. B, 1998, 353, 847–856 CrossRef CAS PubMed.
- S. J. Morrison and D. T. Scadden, The bone marrow niche for haematopoietic stem cells, Nature, 2014, 505, 327–334 CrossRef CAS PubMed.
- P. Bianco, et al., The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine, Nat. Med., 2013, 19, 35–42 CrossRef CAS PubMed.
- M. Crisan, et al., A perivascular origin for mesenchymal stem cells in multiple human organs, Cell Stem Cell, 2008, 3, 301–313 CrossRef CAS PubMed.
- S. Mendez-Ferrer, et al., Mesenchymal and haematopoietic stem cells form a unique bone marrow niche, Nature, 2010, 466, 829–834 CrossRef CAS PubMed.
- S. Mendez-Ferrer, D. Lucas, M. Battista and P. S. Frenette, Haematopoietic stem cell release is regulated by circadian oscillations, Nature, 2008, 452, 442–447 CrossRef CAS PubMed.
- L. M. Calvi and D. C. Link, Cellular complexity of the bone marrow hematopoietic stem cell niche, Calcif. Tissue Int., 2014, 94, 112–124 CrossRef CAS PubMed.
- M. Hines, L. Nielsen and J. Cooper-White, The hematopoietic stem cell niche: what are we trying to replicate?, J. Chem. Technol. Biotechnol., 2008, 83, 421–443 CrossRef CAS PubMed.
- R. S. Taichman and S. G. Emerson, Human osteoblasts support hematopoiesis through the production of granulocyte colony-stimulating factor, J. Exp. Med., 1994, 179, 1677–1682 CrossRef CAS.
- D. Park, et al., Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration, Cell Stem Cell, 2012, 10, 259–272 CrossRef CAS PubMed.
- L. M. Calvi, et al., Osteoblastic cells regulate the haematopoietic stem cell niche, Nature, 2003, 425, 841–846 CrossRef CAS PubMed.
- T. Kai and A. Spradling, Differentiating germ cells can revert into functional stem cells in Drosophila melanogaster ovaries, Nature, 2004, 428, 564–569 CrossRef CAS PubMed.
- C. Brawley and E. Matunis, Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo, Science, 2004, 304, 1331–1334 CrossRef CAS PubMed.
- T. Nakagawa, Y.-i. Nabeshima and S. Yoshida, Functional Identification of the Actual and Potential Stem Cell Compartments in Mouse Spermatogenesis, Dev. Cell, 2007, 12, 195–206 CrossRef CAS PubMed.
- T. Sato, et al., Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche, Nature, 2009, 459, 262–265 CrossRef CAS PubMed.
- T. Sasaki, R. Giltay, U. Talts, R. Timpl and J. F. Talts, Expression and distribution of laminin alpha1 and alpha2 chains in embryonic and adult mouse tissues: an immunochemical approach, Exp. Cell Res., 2002, 275, 185–199 CrossRef CAS PubMed.
- T. M. Dexter, T. D. Allen and L. G. Lajtha, Conditions controlling the proliferation of haemopoietic stem cells in vitro, J. Cell Physiol., 1977, 91, 335–344 CrossRef CAS PubMed.
- K. N. Chua, et al., Functional nanofiber scaffolds with different spacers modulate adhesion and expansion of cryopreserved umbilical cord blood hematopoietic stem/progenitor cells, Exp. Hematol., 2007, 35, 771–781 CrossRef CAS PubMed.
- I. Kurth, K. Franke, T. Pompe, M. Bornhauser and C. Werner, Hematopoietic stem and progenitor cells in adhesive microcavities, Integr. Biol., 2009, 1, 427–434 RSC.
- A. Raic, L. Rodling, H. Kalbacher and C. Lee-Thedieck, Biomimetic macroporous PEG hydrogels as 3D scaffolds for the multiplication of human hematopoietic stem and progenitor cells, Biomaterials, 2014, 35, 929–940 CrossRef CAS PubMed.
- S. Irion, M. C. Nostro, S. J. Kattman and G. M. Keller, Directed differentiation of pluripotent stem cells: from developmental biology to therapeutic applications, Cold Spring Harbor Symp. Quant. Biol., 2008, 73, 101–110 CrossRef CAS PubMed.
- Y. Xu, Y. Shi and S. Ding, A chemical approach to stem-cell biology and regenerative medicine, Nature, 2008, 453, 338–344 CrossRef CAS PubMed.
- S. Ding and P. G. Schultz, A role for chemistry in stem cell biology, Nat. Biotechnol., 2004, 22, 833–840 CrossRef CAS PubMed.
- W. Li, K. Jiang, W. Wei, Y. Shi and S. Ding, Chemical approaches to studying stem cell biology, Cell Res., 2013, 23, 81–91 CrossRef CAS PubMed.
- T. Xu, M. Zhang, T. Laurent, M. Xie and S. Ding, Concise Review: Chemical Approaches for Modulating Lineage-Specific Stem Cells and Progenitors, Stem Cells Transl. Med., 2013, 2, 355–361 CrossRef CAS PubMed.
- A. E. Boitano, et al., Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells, Science, 2010, 329, 1345–1348 CrossRef CAS PubMed.
- S. Chen, Q. Zhang, X. Wu, P. G. Schultz and S. Ding, Dedifferentiation of lineage-committed cells by a small molecule, J. Am. Chem. Soc., 2004, 126, 410–411 CrossRef CAS PubMed.
- K. A. Hartwell, et al., Niche-based screening identifies small-molecule inhibitors of leukemia stem cells, Nat. Chem. Biol., 2013, 9, 840–848 CrossRef CAS PubMed.
- G. de Haan, et al., In vitro generation of long-term repopulating hematopoietic stem cells by fibroblast growth factor-1, Dev. Cell, 2003, 4, 241–251 CrossRef CAS.
- G. Sauvageau, N. N. Iscove and R. K. Humphries, In vitro and in vivo expansion of hematopoietic stem cells, Oncogene, 2004, 23, 7223–7232 CrossRef CAS PubMed.
- J. Audet, C. L. Miller, C. J. Eaves and J. M. Piret, Common and distinct features of cytokine effects on hematopoietic stem and progenitor cells revealed by dose-response surface analysis, Biotechnol. Bioeng., 2002, 80, 393–404 CrossRef CAS PubMed.
- F. Guilak, et al., Control of stem cell fate by physical interactions with the extracellular matrix, Cell Stem Cell, 2009, 5, 17–26 CrossRef CAS PubMed.
- F. Gattazzo, A. Urciuolo and P. Bonaldo, Extracellular matrix: A dynamic microenvironment for stem cell niche, Biochim. Biophys. Acta, 2014, 1840, 2506–2519 CrossRef CAS PubMed.
- S. K. Nilsson, et al., Hyaluronan is synthesized by primitive hemopoietic cells, participates in their lodgment at the endosteum following transplantation, and is involved in the regulation of their proliferation and differentiation in vitro, Blood, 2003, 101, 856–862 CrossRef CAS PubMed.
- M. Ohta, T. Sakai, Y. Saga, S. Aizawa and M. Saito, Suppression of hematopoietic activity in tenascin-C-deficient mice, Blood, 1998, 91, 4074–4083 CAS.
- E. Garcion, A. Halilagic, A. Faissner and C. Ffrench-Constant, Generation of an environmental niche for neural stem cell development by the extracellular matrix molecule tenascin C, Development, 2004, 131, 3423–3432 CrossRef CAS PubMed.
- P. H. Jones and F. M. Watt, Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression, Cell, 1993, 73, 713–724 CrossRef CAS.
- U. B. Jensen, S. Lowell and F. M. Watt, The spatial relationship between stem cells and their progeny in the basal layer of human epidermis: a new view based on whole-mount labelling and lineage analysis, Development, 1999, 126, 2409–2418 CAS.
- R. McBeath, D. Pirone, C. Nelson, K. Bhadriraju and C. Chen, Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment, Dev. Cell, 2004, 6, 483–495 CrossRef CAS.
- S. Schmidt and P. Friedl, Interstitial cell migration: integrin-dependent and alternative adhesion mechanisms, Cell Tissue Res., 2010, 339, 83–92 CrossRef CAS PubMed.
- Y. Takada, X. Ye and S. Simon, The integrins, Genome Biol., 2007, 8, 215 CrossRef PubMed.
- H. Ponta, L. Sherman and P. A. Herrlich, CD44: from adhesion molecules to signalling regulators, Nat. Rev. Mol. Cell. Biol., 2003, 4, 33–45 CrossRef CAS PubMed.
- S. Banerji, et al., Structures of the Cd44-hyaluronan complex provide insight into a fundamental carbohydrate-protein interaction, Nat. Struct. Mol. Biol., 2007, 14, 234–239 CAS.
- M. Bernfield and R. D. Sanderson, Syndecan, a developmentally regulated cell surface proteoglycan that binds extracellular matrix and growth factors, Philos. Trans. R. Soc. London, Ser. B, 1990, 327, 171–186 CrossRef CAS.
- V. Vogel, Mechanotransduction involving multimodular proteins: converting force into biochemical signals, Annu. Rev. Biophys. Biomol. Struct., 2006, 35, 459–488 CrossRef CAS PubMed.
- M. R. Morgan, M. J. Humphries and M. D. Bass, Synergistic control of cell adhesion by integrins and syndecans, Nat. Rev. Mol. Cell. Biol., 2007, 8, 957–969 CrossRef CAS PubMed.
- A. J. Zhu, I. Haase and F. M. Watt, Signaling via β1 integrins and mitogen-activated protein kinase determines human epidermal stem cell fate in vitro, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 6728–6733 CrossRef CAS.
- F. Prosper and C. M. Verfaillie, Regulation of hematopoiesis through adhesion receptors, J. Leukocyte Biol., 2001, 69, 307–316 CAS.
- V. Marthiens, I. Kazanis, L. Moss, K. Long and C. Ffrench-Constant, Adhesion molecules in the stem cell niche–more than just staying in shape?, J. Cell Sci., 2010, 123, 1613–1622 CrossRef CAS PubMed.
- S. Chen, M. Lewallen and T. Xie, Adhesion in the stem cell niche: biological roles and regulation, Development, 2013, 140, 255–265 CrossRef CAS PubMed.
- K. Franke, T. Pompe, M. Bornhauser and C. Werner, Engineered matrix coatings to modulate the adhesion of CD133+ human hematopoietic progenitor cells, Biomaterials, 2007, 28, 836–843 CrossRef CAS PubMed.
- E. A. Cavalcanti-Adam, et al., Lateral spacing of integrin ligands influences cell spreading and focal adhesion assembly, Eur. J. Cell Biol., 2006, 85, 219–224 CrossRef CAS PubMed.
- M. Arnold, et al., Activation of integrin function by nanopatterned adhesive interfaces, ChemPhysChem, 2004, 5, 383–388 CrossRef CAS PubMed.
- M. Schvartzman, et al., Nanolithographic Control of the Spatial Organization of Cellular Adhesion Receptors at the Single-Molecule Level, Nano Lett., 2011, 11, 1306–1312 CrossRef CAS PubMed.
- E. Altrock, C. A. Muth, G. Klein, J. P. Spatz and C. Lee-Thedieck, The significance of integrin ligand nanopatterning on lipid raft clustering in hematopoietic stem cells, Biomaterials, 2012, 33, 3107–3118 CrossRef CAS PubMed.
- C. A. Muth, C. Steinl, G. Klein and C. Lee-Thedieck, Regulation of hematopoietic stem cell behavior by the nanostructured presentation of extracellular matrix components, PLoS One, 2013, 8, e54778 CAS.
- S. Melikova, S. J. Dylla and C. M. Verfaillie, Phosphatidylinositol-3-kinase activation mediates proline-rich tyrosine kinase 2 phosphorylation and recruitment to beta1-integrins in human CD34+ cells, Exp. Hematol., 2004, 32, 1051–1056 CrossRef CAS PubMed.
- I. Dikic, I. Dikic and J. Schlessinger, Identification of a new Pyk2 isoform implicated in chemokine and antigen receptor signaling, J. Biol. Chem., 1998, 273, 14301–14308 CrossRef CAS PubMed.
- H. Avraham, S. Y. Park, K. Schinkmann and S. Avraham, RAFTK/Pyk2-mediated cellular signalling, Cell Signal., 2000, 12, 123–133 CrossRef CAS.
- G. Maheshwari, G. Brown, D. A. Lauffenburger, A. Wells and L. G. Griffith, Cell adhesion and motility depend on nanoscale RGD clustering, J. Cell Sci., 2000, 113(Pt 10), 1677–1686 CAS.
- M. J. Biggs, et al., The use of nanoscale topography to modulate the dynamics of adhesion formation in primary osteoblasts and ERK/MAPK signalling in STRO-1+ enriched skeletal stem cells, Biomaterials, 2009, 30, 5094–5103 CrossRef CAS PubMed.
- R. Zaidel-Bar, S. Itzkovitz, A. Ma'ayan, R. Iyengar and B. Geiger, Functional atlas of the integrin adhesome, Nat. Cell Biol., 2007, 9, 858–867 CrossRef CAS PubMed.
- J. T. Parsons, A. R. Horwitz and M. A. Schwartz, Cell adhesion: integrating cytoskeletal dynamics and cellular tension, Nat. Rev. Mol. Cell. Biol., 2010, 11, 633–643 CrossRef CAS PubMed.
- K. A. Beningo, M. Dembo, I. Kaverina, J. V. Small and Y. L. Wang, Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts, J. Cell Biol., 2001, 153, 881–888 CrossRef CAS.
- A. J. Engler, S. Sen, H. L. Sweeney and D. E. Discher, Matrix elasticity directs stem cell lineage specification, Cell, 2006, 126, 677–689 CrossRef CAS PubMed.
- I. Schoen, B. L. Pruitt and V. Vogel, The Yin-Yang of Rigidity Sensing: How Forces and Mechanical Properties Regulate the Cellular Response to Materials, Annu. Rev. Mater. Res., 2013, 43, 589–618 CrossRef CAS.
- J. Malmstrom, et al., Large area protein patterning reveals nanoscale control of focal adhesion development, Nano Lett., 2010, 10, 686–694 CrossRef PubMed.
- J. Malmstrom, et al., Focal complex maturation and bridging on 200 nm vitronectin but not fibronectin patches reveal different mechanisms of focal adhesion formation, Nano Lett., 2011, 11, 2264–2271 CrossRef CAS PubMed.
- E. Cukierman, R. Pankov, D. R. Stevens and K. M. Yamada, Taking cell-matrix adhesions to the third dimension, Science, 2001, 294, 1708–1712 CrossRef CAS PubMed.
- M. Cattaruzza, C. Lattrich and M. Hecker, Focal adhesion protein zyxin is a mechanosensitive modulator of gene expression in vascular smooth muscle cells, Hypertension, 2004, 43, 726–730 CrossRef CAS PubMed.
- S. Miyamoto, et al., Integrin function: molecular hierarchies of cytoskeletal and signaling molecules, J. Cell Biol., 1995, 131, 791–805 CrossRef CAS.
- X. Zhu and R. K. Assoian, Integrin-dependent activation of MAP kinase: a link to shape-dependent cell proliferation, Mol. Biol. Cell, 1995, 6, 273–282 CrossRef CAS.
- D. Bar-Sagi, D. Rotin, A. Batzer, V. Mandiyan and J. Schlessinger, SH3 domains direct cellular localization of signaling molecules, Cell, 1993, 74, 83–91 CrossRef CAS.
- C. A. Koch, D. Anderson, M. F. Moran, C. Ellis and T. Pawson, SH2 and SH3 domains: elements that control interactions of cytoplasmic signaling proteins, Science, 1991, 252, 668–674 CAS.
- D. E. Ingber, Tensegrity I. Cell structure and hierarchical systems biology, J. Cell Sci., 2003, 116, 1157–1173 CrossRef CAS.
- B. S. Negrutskii and M. P. Deutscher, A sequestered pool of aminoacyl-tRNA in mammalian cells, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 3601–3604 CrossRef CAS.
- D. E. Ingber, The riddle of morphogenesis: a question of solution chemistry or molecular cell engineering?, Cell, 1993, 75, 1249–1252 CrossRef CAS.
- A. J. Maniotis, C. S. Chen and D. E. Ingber, Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 849–854 CrossRef CAS.
- S. Hu, J. Chen, J. P. Butler and N. Wang, Prestress mediates force propagation into the nucleus, Biochem. Biophys. Res. Commun., 2005, 329, 423–428 CrossRef CAS PubMed.
- K. N. Dahl and A. Kalinowski, Nucleoskeleton mechanics at a glance, J. Cell Sci., 2011, 124, 675–678 CrossRef CAS PubMed.
- N. Wang, J. D. Tytell and D. E. Ingber, Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus, Nat. Rev. Mol. Cell. Biol., 2009, 10, 75–82 CrossRef CAS PubMed.
- D. N. Simon and K. L. Wilson, The nucleoskeleton as a genome-associated dynamic ‘network of networks’, Nat. Rev. Mol. Cell. Biol., 2011, 12, 695–708 CrossRef CAS PubMed.
- D. E. Ingber, Cellular mechanotransduction: putting all the pieces together again, FASEB J., 2006, 20, 811–827 CrossRef CAS PubMed.
- K. J. Pienta and D. S. Coffey, Cellular harmonic information transfer through a tissue tensegrity-matrix system, Med. Hypotheses, 1991, 34, 88–95 CrossRef CAS.
- L. B. Boyette and R. S. Tuan, Adult Stem Cells and Diseases of Aging, J. Clin. Med., 2014, 3, 88–134 CrossRef CAS PubMed.
- V. Pekovic and C. J. Hutchison, Adult stem cell maintenance and tissue regeneration in the ageing context: the role for A-type lamins as intrinsic modulators of ageing in adult stem cells and their niches, J. Anat., 2008, 213, 5–25 CrossRef CAS PubMed.
- K. A. Kilian, B. Bugarija, B. T. Lahn and M. Mrksich, Geometric cues for directing the differentiation of mesenchymal stem cells, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4872–4877 CrossRef CAS PubMed.
- P. M. Tsimbouri, et al., A genomics approach in determining nanotopographical effects on MSC phenotype, Biomaterials, 2013, 34, 2177–2184 CrossRef CAS PubMed.
- M. J. Dalby, et al., Nanotopographical stimulation of mechanotransduction and changes in interphase centromere positioning, J. Cell. Biochem., 2007, 100, 326–338 CrossRef CAS PubMed.
- M. J. Dalby, et al., Nanomechanotransduction and interphase nuclear organization influence on genomic control, J. Cell. Biochem., 2007, 102, 1234–1244 CrossRef CAS PubMed.
- L. E. McNamara, et al., The role of microtopography in cellular mechanotransduction, Biomaterials, 2012, 33, 2835–2847 CrossRef CAS PubMed.
- M. J. Dalby, M. O. Riehle, S. J. Yarwood, C. D. Wilkinson and A. S. Curtis, Nucleus alignment and cell signaling in fibroblasts: response to a micro-grooved topography, Exp. Cell Res., 2003, 284, 274–282 CrossRef CAS.
- K. V. Iyer, et al., Modeling and Experimental Methods to Probe the Link between Global Transcription and Spatial Organization of Chromosomes, PLoS One, 2012, 7, e46628 CAS.
- C. H. Thomas, J. H. Collier, C. S. Sfeir and K. E. Healy, Engineering gene expression and protein synthesis by modulation of nuclear shape, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 1972–1977 CrossRef CAS PubMed.
- N. Itano, S. Okamoto, D. Zhang, S. A. Lipton and E. Ruoslahti, Cell spreading controls endoplasmic and nuclear calcium: a physical gene regulation pathway from the cell surface to the nucleus, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 5181–5186 CrossRef CAS PubMed.
- S. T. Lim, Nuclear FAK: a new mode of gene regulation from cellular adhesions, Mol. Cells, 2013, 36, 1–6 CrossRef CAS PubMed.
- S. T. Lim, et al., Nuclear FAK promotes cell proliferation and survival through FERM-enhanced p53 degradation, Mol. Cell, 2008, 29, 9–22 CrossRef CAS PubMed.
- S. T. Lim, et al., Pyk2 inhibition of p53 as an adaptive and intrinsic mechanism facilitating cell proliferation and survival, J. Biol. Chem., 2010, 285, 1743–1753 CrossRef CAS PubMed.
- C. Yang, M. W. Tibbitt, L. Basta and K. S. Anseth, Mechanical memory and dosing influence stem cell fate, Nat. Mater., 2014, 13, 645–652 CrossRef CAS PubMed.
- G. Halder, S. Dupont and S. Piccolo, Transduction of mechanical and cytoskeletal cues by YAP and TAZ, Nat. Rev. Mol. Cell. Biol., 2012, 13, 591–600 CrossRef CAS PubMed.
- S. Dupont, et al., Role of YAP/TAZ in mechanotransduction, Nature, 2011, 474, 179–183 CrossRef CAS PubMed.
- M. Shakibaei and A. Mobasheri, Beta1-integrins co-localize with Na, K-ATPase, epithelial sodium channels (ENaC) and voltage activated calcium channels (VACC) in mechanoreceptor complexes of mouse limb-bud chondrocytes, Histol. Histopathol., 2003, 18, 343–351 CAS.
- P. C. Brooks, et al., Insulin-like growth factor receptor cooperates with integrin alpha v beta 5 to promote tumor cell dissemination in vivo, J. Clin. Invest., 1997, 99, 1390–1398 CrossRef CAS PubMed.
- G. E. Plopper, H. P. McNamee, L. E. Dike, K. Bojanowski and D. E. Ingber, Convergence of integrin and growth factor receptor signaling pathways within the focal adhesion complex, Mol. Biol. Cell, 1995, 6, 1349–1365 CrossRef CAS.
- N. Wang, J. P. Butler and D. E. Ingber, Mechanotransduction across the cell surface and through the cytoskeleton, Science, 1993, 260, 1124–1127 CAS.
- C. Q. Pan, M. Sudol, M. Sheetz and B. C. Low, Modularity and functional plasticity of scaffold proteins as p(l)acemakers in cell signaling, Cell Signal., 2012, 24, 2143–2165 CrossRef CAS PubMed.
- R. Milo, et al., Network motifs: simple building blocks of complex networks, Science, 2002, 298, 824–827 CrossRef CAS PubMed.
- A. del Campo and E. Arzt, Fabrication approaches for generating complex micro- and nanopatterns on polymeric surfaces, Chem. Rev., 2008, 108, 911–945 CrossRef CAS PubMed.
- B. D. Gates, et al., New approaches to nanofabrication: molding, printing, and other techniques, Chem. Rev., 2005, 105, 1171–1196 CrossRef CAS PubMed.
- N. Gadegaard, et al., Arrays of nano-dots for cellular engineering, Microelectron. Eng., 2003, 67–68, 162–168 CrossRef CAS.
- A. Atala, R. Lanza, J. A. Thompson and R. M. Nerem, Principles of regenerative medicine, Elsevier, 2008 Search PubMed.
- N. B. Guerra, et al., Subtle variations in polymer chemistry modulate substrate stiffness and fibronectin activity, Soft Matter, 2010, 6, 4748–4755 RSC.
- C. J. Wilson, R. E. Clegg, D. I. Leavesley and M. J. Pearcy, Mediation of biomaterial-cell interactions by adsorbed proteins: a review, Tissue Eng., 2005, 11, 1–18 CrossRef CAS PubMed.
- L. Trichet, et al., Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 6933–6938 CrossRef CAS PubMed.
- E. K. Yim, et al., Nanopattern-induced changes in morphology and motility of smooth muscle cells, Biomaterials, 2005, 26, 5405–5413 CrossRef CAS PubMed.
- M. J. Dalby, D. McCloy, M. Robertson, C. D. Wilkinson and R. O. Oreffo, Osteoprogenitor response to defined topographies with nanoscale depths, Biomaterials, 2006, 27, 1306–1315 CrossRef CAS PubMed.
- J. Kim, et al., Synergistic effects of nanotopography and co-culture with endothelial cells on osteogenesis of mesenchymal stem cells, Biomaterials, 2013, 34, 7257–7268 CrossRef CAS PubMed.
- L. A. Turner, S. Downes, E. Hill and I. Kinloch, Investigating the suitability of electrohydrodynamic lithography for the fabrication of cell substrates, J. Mater. Sci., 2014, 49, 4045–4057 CrossRef CAS.
- R. G. Harrison, On the stereotropism of embryonic cells, Science, 1911, 34, 279–281 CAS.
- P. Weiss, Experiments on cell and axon orientation in vitro; the role of colloidal exudates in tissue organization, J. Exp. Zool., 1945, 100, 353–386 CrossRef CAS PubMed.
- P. Kanchanawong, et al., Nanoscale architecture of integrin-based cell adhesions, Nature, 2010, 468, 580–584 CrossRef CAS PubMed.
- I. Patla, et al., Dissecting the molecular architecture of integrin adhesion sites by cryo-electron tomography, Nat. Cell Biol., 2010, 12, 909–915 CrossRef CAS PubMed.
- M. J. Biggs, R. G. Richards and M. J. Dalby, Nanotopographical modification: a regulator of cellular function through focal adhesions, Nanomedicine, 2010, 6, 619–633 CrossRef CAS PubMed.
- B. K. K. Teo, et al., Nanotopography Modulates Mechanotransduction of Stem Cells and Induces Differentiation through Focal Adhesion Kinase, ACS Nano, 2013, 7, 4785–4798 CrossRef CAS PubMed.
- M. A. Conti, S. Even-Ram, C. Liu, K. M. Yamada and R. S. Adelstein, Defects in cell adhesion and the visceral endoderm following ablation of nonmuscle myosin heavy chain II-A in mice, J. Biol. Chem., 2004, 279, 41263–41266 CrossRef CAS PubMed.
- J. L. Tan, et al., Cells lying on a bed of microneedles: An approach to isolate mechanical force, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 1484–1489 CrossRef CAS PubMed.
- M. J. Dalby, et al., Osteoprogenitor response to semi-ordered and random nanotopographies, Biomaterials, 2006, 27, 2980–2987 CrossRef CAS PubMed.
- H. L. Khor, et al., Response of cells on surface-induced nanopatterns: fibroblasts and mesenchymal progenitor cells, Biomacromolecules, 2007, 8, 1530–1540 CrossRef CAS PubMed.
- T. Sjostrom, et al., Titanium nanofeaturing for enhanced bioactivity of implanted orthopedic and dental devices, Nanomedicine, 2013, 8, 89–104 CrossRef PubMed.
- E. K. Yim, S. W. Pang and K. W. Leong, Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage, Exp. Cell Res., 2007, 313, 1820–1829 CrossRef CAS PubMed.
- S. Oh, et al., Stem cell fate dictated solely by altered nanotube dimension, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 2130–2135 CrossRef CAS PubMed.
- R. J. McMurray, et al., Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency, Nat. Mater., 2011, 10, 637–644 CrossRef CAS PubMed.
- M. J. Dalby, et al., The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder, Nat. Mater., 2007, 6, 997–1003 CrossRef CAS PubMed.
- P. Tsimbouri, et al., Nanotopographical effects on mesenchymal stem cell morphology and phenotype, J. Cell. Biochem., 2014, 115, 380–390 CrossRef CAS PubMed.
- A. Gautieri, S. Vesentini, A. Redaelli and M. J. Buehler, Hierarchical structure and nanomechanics of collagen microfibrils from the atomistic scale up, Nano Lett., 2011, 11, 757–766 CrossRef CAS PubMed.
- R. K. Das, O. F. Zouani, C. Labrugere, R. Oda and M. C. Durrieu, Influence of nanohelical shape and periodicity on stem cell fate, ACS Nano, 2013, 7, 3351–3361 CrossRef CAS PubMed.
- M. Mahmoudi, et al., Cell-Imprinted Substrates Direct the Fate of Stem Cells, ACS Nano, 2013, 7, 8379–8384 CrossRef CAS PubMed.
- E. Kingham, K. White, N. Gadegaard, M. J. Dalby and R. O. Oreffo, Nanotopographical cues augment mesenchymal differentiation of human embryonic stem cells, Small, 2013, 9, 2140–2151 CrossRef CAS PubMed.
- M. R. Lee, et al., Direct differentiation of human embryonic stem cells into selective neurons on nanoscale ridge/groove pattern arrays, Biomaterials, 2010, 31, 4360–4366 CrossRef CAS PubMed.
- I. Munoz-Sanjuan and A. H. Brivanlou, Neural induction, the default model and embryonic stem cells, Nat. Rev. Neurosci., 2002, 3, 271–280 CrossRef CAS PubMed.
- M. J. Dalby, et al., The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder, Nat. Mater., 2007, 6, 997–1003 CrossRef CAS PubMed.
- S. Oh, et al., Stem cell fate dictated solely by altered nanotube dimension, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 2130–2135 CrossRef CAS PubMed.
- C. Londono, et al., Nonautonomous contact guidance signaling during collective cell migration, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 1807–1812 CrossRef CAS PubMed.
- P. B. Lucker, et al., A microgroove patterned multiwell cell culture plate for high-throughput studies of cell alignment, Biotechnol. Bioeng., 2014, 9999, 1–12 Search PubMed.
- F. Braet and E. Wisse, Structural and functional aspects of liver sinusoidal endothelial cell fenestrae: a review, Comp. Hepatol., 2002, 1, 1 CrossRef PubMed.
- A. P. Kwan, C. E. Cummings, J. A. Chapman and M. E. Grant, Macromolecular organization of chicken type X collagen in vitro, J. Cell Biol., 1991, 114, 597–604 CrossRef CAS.
- P. Jungebluth, et al., Tracheobronchial transplantation with a stem-cell-seeded bioartificial nanocomposite: a proof-of-concept study, Lancet, 2011, 378, 1997–2004 CrossRef CAS.
- M. P. Lutolf, P. M. Gilbert and H. M. Blau, Designing materials to direct stem-cell fate, Nature, 2009, 462, 433–441 CrossRef CAS PubMed.
- H. V. Unadkat, et al., An algorithm-based topographical biomaterials library to instruct cell fate, Proc. Natl. Acad. Sci. U. S. A., 2011, 16565–16570 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |