Cyclen-based cationic lipids with double hydrophobic tails for efficient gene delivery†
Hai-Jiao
Wang
,
Yan-Hong
Liu
,
Ji
Zhang
*,
Yang
Zhang
,
Yan
Xia
and
Xiao-Qi
Yu
*
Key Laboratory of Green Chemistry and Technology (Ministry of Education), College of Chemistry, Sichuan University, Chengdu 610064, PR China. E-mail: xqyu@scu.edu.cn; jzhang@scu.edu.cn
First published on 11th July 2014
Abstract
Cationic lipids have been regarded as an important type of non-viral gene vector; to develop novel lipids with high transfection efficiency (TE) and biocompatibility is of great importance. A series of cyclen-based cationic lipids bearing double hydrophobic tails were synthesized herein. To study their structure–activity relationship (SAR), several analogs including the amide-contained double-tailed lipids, lipids containing ester bonds with the reverse direction, and lipids with a single tail were also prepared. Several assays were used to study their interactions with plasmid DNA, and results reveal that these lipids could smoothly condense DNA into nanosized particles. CCK-8-based cell viability assays showed relatively lower cytotoxicity of the lipoplexes compared to commercially available Lipofectamine 2000. In vitro transfection assays exhibited that some of the lipids (5a, 5b and 8b) may give excellent TEs, which were up to 10 times higher than Lipofectamine 2000. SAR of these lipids was studied in detail by investigating the effects of several structural aspects including the chain length and saturation degree, chain numbers, the type of linkage bond, and orientation of ester bonds. The results not only demonstrate that these lipids might be promising non-viral gene vectors, but also afford us clues for further optimization of lipidic gene delivery materials.
Introduction
As a new, powerful approach for curing diseases such as severe combined immunodeficiency, cancer, cystic fibrosis, AIDS and cardiovascular ailments, gene therapy has emerged as an alternative method to traditional chemotherapy.1,2 Many gene delivery systems have been developed to transfer the therapeutic gene to diseased cells. Compared to viral vectors, non-viral gene delivery systems may offer a safer and simpler method of delivery of therapeutic genes. As a result, they have gained significant attention over the past two decades. However, non-viral vectors still have some shortcomings including relatively low transfection efficiency (TE) and potential toxicity.3,4 Therefore, developing novelly designed chemical gene vectors with high TE and low toxicity is of great importance.Non-viral gene delivery systems include cationic polymers, cationic surfactants (also known as cationic lipids), gold nanoparticles,5,6etc. Among these materials, cationic lipids are regarded as one type of important and potential vector compound for their simple structure, ease of preparation, good repeatability and biocompatibility.7,8 In solution, cationic lipids may form liposomes, which can bind anionic DNA molecules to form lipoplexes via electrostatic interactions, preventing DNA from degradation by nucleases in the transfection process.8,9 Although many cationic lipids have been developed and some of them were commercially available (such as Lipofectamine 2000™), their potential cytotoxicity, lack of targeted ability, relatively low stability, poor capacity to pass the blood–brain barrier, etc.2,10,11 limit their clinical applications.
Generally, cationic lipids consist of three parts: a cationic headgroup, a hydrophobic domain, and a linker. The cationic headgroup always have one or more cationic moieties, such as quaternary ammonium,12–14 polyamines,15–17 amino acids,18–20 and guanidinium.21,22 The hydrophobic domain can help the lipids to form micelles via the hydrophobic effect. Even a minor structural change in the cationic lipids might significantly influence their gene delivery behavior.23,24 In our ongoing research, 1,4,7,10-tetraazacyclododecanes (cyclen) were found to be excellent candidates for the headgroup of cationic lipids. Cyclen has four amine groups with a wide range of pKa values, and its cyclic backbone is hard to self-fold, which may hinder their electrostatic interaction with DNA.25–27 Our previous studies mainly focused on mono-substituted cyclen lipidic molecules.27–29 In fact, several commercially available lipids serving as gene vectors have two hydrophobic tails in their structures. It is known that double-tailed lipids can form lamellar phases even without a helper lipid and thus lead to increased efficiency in gene delivery. In contrast, single-tailed lipids tend to form micelles due to their surfactant nature and may subsequently increase their cytotoxicity.3,30 In this report, we further investigate the cyclen-based double-tailed molecules, which may also be referred to as gemini-type lipids. Their structure–activity relationship in DNA interaction and gene delivery were discussed in detail. Results show that these lipids may serve as efficient non-viral gene delivery vectors with low toxicity.
Experimental section
Materials and methods
All chemicals and reagents were obtained commercially and were used as received. Absolute chloroform (CHCl3) and dichloromethane (CH2Cl2) were distilled after being dried with calcium hydride (CaH2). Anhydrous acetonitrile (CH3CN) was distilled after being dried with P2O5. Trifluoroacetic acid (CF3COOH) was dried and purified under nitrogen by using standard methods. Column chromatography was performed using 200–300 mesh silica gel. All aqueous solutions were prepared from deionized or distilled water. [4,7,10-Tris(tert-butoxycarbonyl)-1,4,7,10-tetraaza-cyclododecan-1-yl]acetic acid31 and 1,7-bis(tert-butyloxycarbonyl)-1,4,7,10-tetraazacyclodocane32 were synthesized according to the literature. The 1H NMR and 13C NMR spectra were measured using a Bruker AM400 NMR spectrometer. Proton chemical shifts of NMR spectra were given in ppm relative to internal reference TMS (1H, 0.00 ppm). HRMS spectral data were recorded on a Bruker Daltonics Bio TOF mass spectrometer. MicroBCA protein assay kit was obtained from Pierce (Rockford, IL, USA). Luciferase assay kit was purchased from Promega (Madison, WI, USA). Endotoxin-free plasmid purification kit was purchased from TIANGEN (Beijing, China). The plasmids used in the study were pGL-3 (Promega, Madison, WI, USA) coding for luciferase DNA and pEGFP-N1 (Clontech, Palo Alto, CA, USA) coding for EGFP DNA. The Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum were purchased from Invitrogen Corp. 293 T human embryonic kidney cell lines, A549 lung cancer cell lines and BEL-7402 cells (human hepatocellular carcinoma cell) were purchased from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences.Synthesis of target lipids
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The chemical yield and analysis data are listed in ESI.†
1). The chemical yield and analysis data are listed in ESI.†
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The chemical yields and analysis data are listed in ESI.†
1). The chemical yields and analysis data are listed in ESI.†
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). After that, the deprotection procedure was used to obtain the desired lipid 6. The chemical yields and analysis data are listed in ESI.†
1). After that, the deprotection procedure was used to obtain the desired lipid 6. The chemical yields and analysis data are listed in ESI.†
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). After that, the deprotection procedure was used to obtain the desired lipid 8. The chemical yields and analysis data are listed in ESI.†
1). After that, the deprotection procedure was used to obtain the desired lipid 8. The chemical yields and analysis data are listed in ESI.†
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) After that, the deprotection procedure was used to obtain the desired lipid 10. The chemical yields and analysis data are listed in ESI.†
1) After that, the deprotection procedure was used to obtain the desired lipid 10. The chemical yields and analysis data are listed in ESI.†
Preparation of cationic liposomes
Individual cationic lipid (0.005 mmol) or its mixture with DOPE in the desired mole ratio was dissolved in anhydrous chloroform (2.5 mL) in autoclaved glass vials. Thin films were made by slowly rotary-evaporating the solvent at room temperature. The last trace of the organic solvent was removed by keeping these films under vacuum for more than 8 h. The dried films and Tris-HCl buffer (10 mM, pH 7.4) were preheated to 70 °C, and then the buffer was added to the films resulting in the final lipid concentration of 2.0 mM. The mixtures were vortexed vigorously until the films were completely resuspended. Sonication of these suspensions with an ultrasonic probe for 5 min in an ice bath afforded the corresponding cationic liposomes that were stored at 4 °C.Amplification and purification of plasmid DNA
pGL-3 and pEGFP-N1 plasmids were used. The former one was used as the luciferase reporter gene, which was transformed in JM109 Escherichia coli, and the latter one was used as the enhanced green fluorescent protein reporter gene, which was transformed in E. coli DH5α. Both plasmids were amplified in E. coli grown in LB medium at 37 °C and 220 rpm overnight. The plasmids were purified by an EndoFree TiangenTM Plasmid Kit. Then, the purified plasmids were dissolved in TE buffer solution and stored at −80 °C. The integrity of plasmids was confirmed by agarose gel electrophoresis. The purity and concentration of plasmids were determined by the ratio of ultraviolet (UV) absorbances at 260 nm to 280 nm.Preparation of lipid/DOPE/DNA complexes (lipoplexes)
To prepare the liposome/pDNA complexes (lipoplexes), various amounts of cationic lipids were mixed with a constant amount of DNA by pipetting thoroughly at various N/P ratios, and the mixture was incubated for 30 min at room temperature. The theoretical N/P ratio represents the charge ratio of protonated N atoms on cationic lipid to nucleotide phosphates (in mole) and was calculated by considering the average nucleotide mass of 350.Gel retardation assay
To determine the formation of liposome/pDNA complex (lipoplexes), lipoplexes of various N/P ratios ranging from 0 to 8 were prepared as described above. Constant amount of 0.125 μg DNA was used here; 15 μL of each lipoplexe solution was electrophoresed on the 1% (WV-1) agarose gel containing gel red and Tris-acetate (TAE) running buffer at 135 V for 30 min. DNA was visualized with a UV lamp using a BioRad Universal Hood II. In the experiments with serum, the lipoplex solution (20 μL contained 10% or 50% serum) was obtained by adding Tris-HCl buffer and serum (2 μL or 10 μL) and incubating for a certain time.Dynamic light scattering (DLS)
Particle size and zeta potential of liposomes or lipoplexes at various N/P ratios were measured by a dynamic light scattering system (Zetasizer Nano ZS, Malvern Instruments Led) at 25 °C. Lipoplexes were prepared by following the instructions used for transfection and measured with 1 μg DNA per mL sample.Transmission electron microscopy (TEM)
The morphologies of the lipoplexes were observed by TEM (JEM-100CXa) with an acceleration voltage of 100 kV. 4 μg of pUC-19 was added to the appropriate volume of the liposomes solution (N/P ratio of the lipoplex relative to pDNA, N/P = 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), then diluted to the total volume of 250 μL. The solution of the lipoplexes was incubated at 37 °C for 0.5 h. A drop of lipoplex suspensions was placed onto the copper grid. After a few minutes, the excess solution was blotted away with a filter paper. Then, a drop of 0.5% (w/v) phosphotungstic acid was placed on the above grid. The grid was dried at room temperature at atmospheric pressure for several minutes before observation.
1), then diluted to the total volume of 250 μL. The solution of the lipoplexes was incubated at 37 °C for 0.5 h. A drop of lipoplex suspensions was placed onto the copper grid. After a few minutes, the excess solution was blotted away with a filter paper. Then, a drop of 0.5% (w/v) phosphotungstic acid was placed on the above grid. The grid was dried at room temperature at atmospheric pressure for several minutes before observation.
Cell culture
HEK293 human embryonic kidney cell lines were incubated in Dulbecco's modified Eagle's medium (DMEM), A549 lung cancer cell lines and BEL-7402 cells (human hepatocellular carcinoma cell) were incubated in 1640 containing 10% fetal bovine serum (FBS) and 1‰ antibiotics (penicillin–streptomycin, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mL−1) at 37 °C in a humidified atmosphere containing 5% CO2.
000 U mL−1) at 37 °C in a humidified atmosphere containing 5% CO2.
Cellular uptake of plasmid DNA
The cellular uptake of the polymer/fluorescein labeled-DNA complexes was analyzed by flow cytometry. The Label IT Cy5 Labeling Kit was used to label pDNA with Cy5 according to the manufacturer's protocol. Briefly, HEK293 cells were seeded in 12-well plates (1.5 × 105 cells per well) and allowed to attach and grow for 24 h. For transfection in the absence of serum, the medium was exchanged with a serum-free medium. As for transfection in the presence of serum, the medium was exchanged with a serum-containing medium. Cells were incubated with Cy5 labeled DNA complexes (2 μg DNA per well, optimal N/P ratio of each sample) in media for 4 h at 37 °C. Subsequently, the cells were washed with 1× PBS and harvested with 0.25% trypsin/EDTA and resuspended in 1× PBS. Mean fluorescence intensity was analyzed using a FACSCalibur flow cytometer (Becton Dickinson and Company). Cy5-labeled plasmid DNA uptake was measured in the FL4 channel using the red diode laser (633 nm). Data from 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were gated using forward and side scatter parameters to exclude cell debris. The flow cytometer was calibrated for each run to obtain a background level of w1% for control samples (i.e., untreated cells).
000 events were gated using forward and side scatter parameters to exclude cell debris. The flow cytometer was calibrated for each run to obtain a background level of w1% for control samples (i.e., untreated cells).
Cytotoxicity assays
Toxicity of lipoplexes toward HEK293, A549 and Bel-7402 cells was determined by using a Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan). About 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well were seeded into 96-well plates. After 24 h, optimized lipid/DOPE formulations were complexed with 0.2 μg of DNA at various N/P ratios for 30 min; 100 μL of lipoplexes were added to the cells in the absence of serum. After 4 h of incubation, lipoplex solutions were removed, and 100 μL of media with 10% FBS was added. After 24 h, 10 μL CCK-8 was added to each well and the plates were incubated at 37 °C for another 1 h. Then, the absorbance of each sample was measured using an ELISA plate reader (model 680, BioRad) at a wavelength of 450 nm. The cell viability (%) was obtained according to the manufacturer's instructions. Lipoplex prepared from Lipofectamine 2000™ was used as the control.
000 cells per well were seeded into 96-well plates. After 24 h, optimized lipid/DOPE formulations were complexed with 0.2 μg of DNA at various N/P ratios for 30 min; 100 μL of lipoplexes were added to the cells in the absence of serum. After 4 h of incubation, lipoplex solutions were removed, and 100 μL of media with 10% FBS was added. After 24 h, 10 μL CCK-8 was added to each well and the plates were incubated at 37 °C for another 1 h. Then, the absorbance of each sample was measured using an ELISA plate reader (model 680, BioRad) at a wavelength of 450 nm. The cell viability (%) was obtained according to the manufacturer's instructions. Lipoplex prepared from Lipofectamine 2000™ was used as the control.
Transfection procedure
Gene transfection of a series of complexes was investigated in HEK293, A549 and Bel-7402 cells. Cells were seeded in 24-well plates (7.5 × 104 cells per well for HEK293, 1.2 × 105 cells per well for A549 and Bel-7402) and grown to reach 70–80% cell confluence at 37 °C for 24 h in 5% CO2. Before transfection, the medium was replaced with a serum-free or a 10% serum-containing culture medium containing liposomes/pDNA (0.8 μg) complexes at various N/P ratios. After 4 h under standard incubator conditions, the medium was replaced with a fresh medium containing serum and incubated for another 20 h.For fluorescence microscopy assays, cells were transfected by complexes containing pEGFP-N1. After 24 h incubation, GFP-expressed cells were observed using an inverted fluorescence microscope (Nikon Eclipse TE 2000E) equipped with a cold Nikon camera. Control transfection was performed in each case using a commercially available transfection reagent Lipofectamine 2000™ based on the standard conditions specified by the manufacturer.
For luciferase assays, cells were transfected by complexes containing pGL-3. For a typical assay in a 24-well plate, 24 h post transfection as described above, cells were washed with cold PBS and lysed with 100 mL 1× Lysis reporter buffer (Promega). The luciferase activity was measured by the microplate reader (Model 550, BioRad, USA). The protein content of the lysed cell was determined by BCA protein assay. Gene transfection efficiency was expressed as the relative fluorescence intensity per mg protein (RLU per mg protein). All the experiments were done in triplicates.
Results and discussion
Synthesis of the cyclen derived lipids
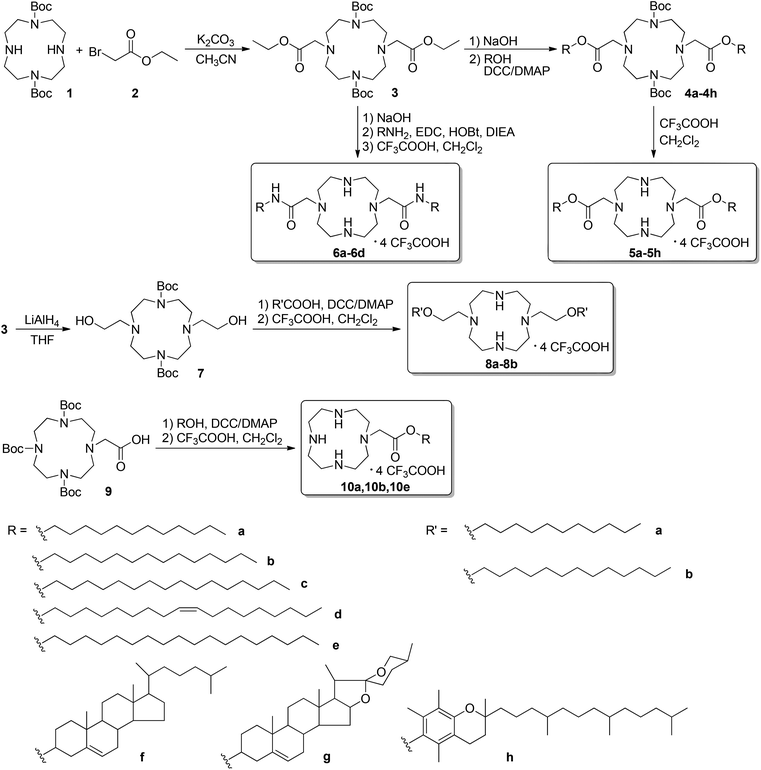
The synthetic route of double-tailed lipids 5a–5h is shown in Scheme 1. For comparison, several series of their analogs were also prepared. These analogs include the amide-contained double-tailed lipids (6a–6d), ester bonds with a reverse direction (8a–8b),24,28 and lipids with single tail (10a, 10b and 10e). The lipids with double hydrophobic tails were prepared from 1,7-diBoc-protected cyclen 1, which was obtained from direct acylation of cyclen with Boc2O. Compound 1 reacted with ethyl bromoacetate 2 to give the central intermediate 3. After hydrolysis of the ester bonds, subsequent condensations between the acid and alcohols or amines containing hydrophobic groups resulted in the diBoc-protected precursors of target lipids 5 and 6, respectively. Final deprotection with trifluoroacetic acid gave the target lipids. The lipids containing ester bonds with reverse direction 8 were also prepared from compound 3 by simple reduction with LiAlH4 and subsequent condensation with fatty acids. The single-tailed lipids 10 were prepared from 1, 4,7-triBoc-protected cyclen via a method similar to that for the preparation of 5. The hydrophobic groups used herein include long carbon chains with different lengths and saturation degrees, steroids, and tocopherol. All novel compounds were characterized by NMR and HRMS.Interactions between the liposomes and plasmid DNA
Cationic liposomes may be formed from either individual cationic lipid or in combination with co-lipids such as 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE). Herein the liposomes were prepared by using lipid and DOPE in the mole ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
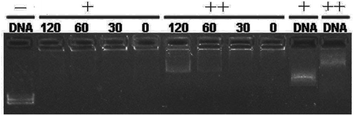
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. DNA binding ability of these liposomes was first studied by gel retardation assay. As shown in Fig. 1, since cyclen has two amine groups with relatively strong basicity, all lipids can completely inhibit the electrophoretic mobility of plasmid DNA at a N/P ratio of 4 except for 5h (gel retardation assay of the other lipids is shown in Fig. S1†), which retard DNA at a N/P ratio of 8, indicating its weaker DNA binding ability.
1. DNA binding ability of these liposomes was first studied by gel retardation assay. As shown in Fig. 1, since cyclen has two amine groups with relatively strong basicity, all lipids can completely inhibit the electrophoretic mobility of plasmid DNA at a N/P ratio of 4 except for 5h (gel retardation assay of the other lipids is shown in Fig. S1†), which retard DNA at a N/P ratio of 8, indicating its weaker DNA binding ability.
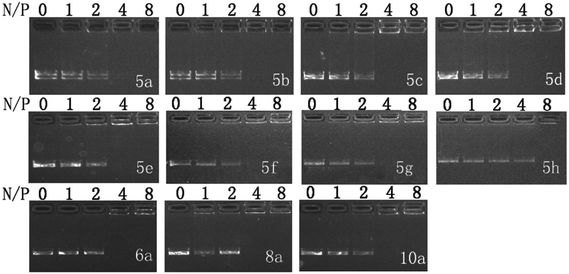 | ||
Fig. 1 Electrophoretic gel retardation assays of pDNA in the presence of liposomes at different N/P ratios. The molar ratio of lipid–DOPE was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Images with other liposomes are shown in Fig. S1.† 1. Images with other liposomes are shown in Fig. S1.† | ||
Gel retardation assay was also carried out to examine the DNA binding ability of the liposomes and the stability of lipoplexes in the presence of serum. Results show that selective liposomes (5a, 5b, 8a and 8b) also effectively retarded DNA at N/P of 4 in the presence of 10% serum (Fig. S2†), indicating that their DNA binding capacities are not influenced by serum. Moreover, we investigated the stability of 5a-derived lipoplex in the presence of serum. Fig. 2 shows that the 5a/DNA complex is stable in 10% serum, and no released or enzymolyzed DNA was found. With the presence of 50% serum, the complex is still stable in the first hour. These results demonstrate that lipoplexes derived from the cyclen-based double tailed lipids were seldom affected by serum, suggesting their good stability.
Proper size and zeta potential are important factors for liposome/DNA complexes (lipoplexes) used as gene vectors. Dynamic light scattering (DLS) was applied to characterize these properties of the blank liposomes and lipoplexes. All liposomes showed their particle sizes in a range of 100–170 nm except 5h, which has relatively poor water solubility. The liposomes have positive surface charges due to the protonated cyclen ring, facilitating their electrostatic interaction with negatively charged DNA (Fig. S3†). The ester bond contained liposomes show good stability. The particle sizes and zeta-potentials after being stored for 2 weeks were found to have little difference compared to the freshly prepared liposomes (Table S1†). Fig. 3A depicts the mean particle sizes of the 5/DNA lipoplexes at different N/P ratios, and the diameters in the range of 120–700 nm were found. In general, less than 400 nm particles were formed from a N/P ratio of 4, at which full retardation was observed in gel electrophoresis. Lipids with shorter chains might give lipoplexes with smaller size. The steroid or tocopherol contained lipids (5f–5h) were prone to form larger particles, indicating that the hydrophobic interaction between these molecules are not strong enough. For the analogs (Fig. S4†), lipids 6 and 8 containing double long chains also gave similar particle behavior. However, lipoplexes formed from 10 with single chain have much larger particle sizes. This phenomenon might be attributed to the shape of the liposomes. In the lipid molecule with single chains, the cross-sectional area of hydrophilic cyclen is much bigger than the hydrophobic domain, leading to the formation of spherical micelles. Meanwhile, the lipids with double chains have the hydrophilic and hydrophobic domains with similar cross-sectional area, thus the liposomes might adopt a bilayer supramolecular structure, which is more liable to compact DNA into small nanoparticles. Zeta potentials of all lipoplexes showed a similar trend along with the increase of N/P ratios (Fig. 3B). The potential reversed to positive and became stable from the N/P of 4, suggesting a full condensation of plasmid DNA. These results are in accordance with those in the gel electrophoresis assay.
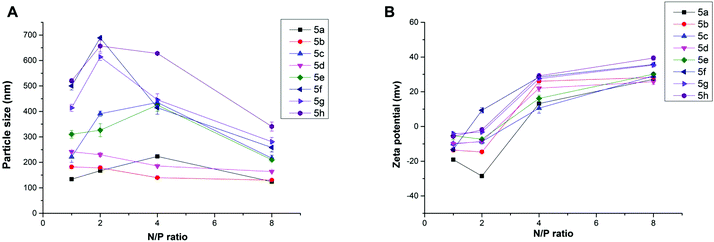 | ||
Fig. 3 Mean particle size (A) and zeta-potential (B) of the lipoplexes formed from lipid 5 under various N/P ratios (DLS at room temperature). The molar ratio of lipid–DOPE was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. 1. | ||
The morphology of lipoplexes formed from 5a, 6b, 8a and 10a was further characterized by transmission electron microscopy (TEM) at a N/P ratio of 8 in deionized water. As shown in Fig. 4, all of the four lipids can effectively condense DNA into homogeneous spherical nanoparticles with a diameter of 30–90 nm. The results once again reflected that these lipids had DNA condensation capability. Like DLS results, these images also show that the lipoplexes formed from 10a gave larger size than others. The particle sizes obtained by TEM were smaller than those measured by DLS, which might be attributed to the fact that the particle sizes measured by DLS were obtained in the hydrated state in deionized water, while those obtained by TEM had been dried after being dropped onto carbon-coated copper meshes.33
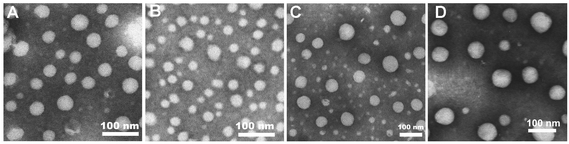 | ||
Fig. 4 TEM images of the lipoplexes formed from 5a (A), 6b (B), 8a (C) and 10a (D) at a N/P ratio of 8 in deionized water. The molar ratio of lipid–DOPE was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. 1. | ||
Cytotoxicity
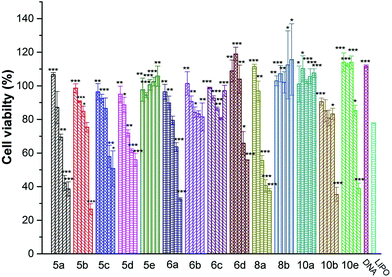
Low cytotoxicity is a prerequisite for an applicable non-viral gene vector. To investigate the cytotoxicity of the cationic lipoplexes, CCK-8-based cell viability assays were executed in several cell lines with Lipofectamine 2000 as a control, and the amount of pDNA condensed with different lipids and Lipofectamine 2000 in the studies was the same. The N/P ratio (1, 2, 4, 8 and 16) and concentration of DNA used here were in accordance with those used in the transfection experiments. As shown in Fig. 5, in HEK293 cells, the cytotoxicity of most lipids increased along with the rise of N/P ratios. Under lower N/P ratios (1–4), the percentages of viable cells were higher than those involving Lipofectamine 2000 in most cases. In other cell lines such as A549 and 7402 (Fig. S5†), although some difference existed in the individual behavior of each lipoplex, their cytotoxicity under N/P ratios used in transfection is low.In vitro gene transfection
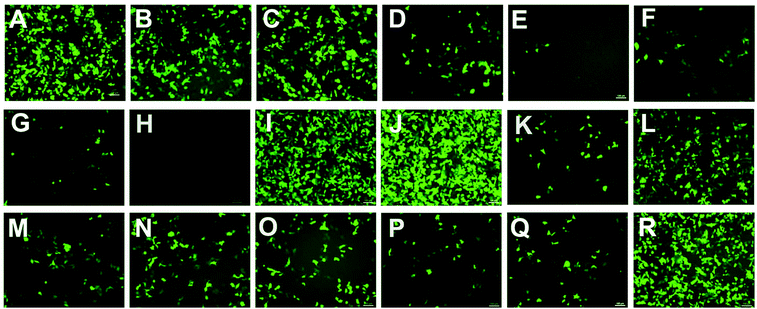
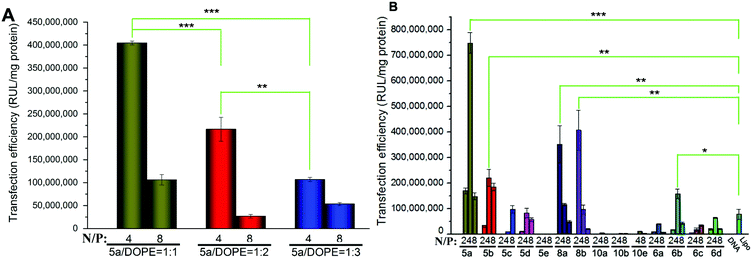
To investigate the gene transfection efficiencies of the synthesized lipids, the fluorescence microscopy assay was first carried out using pEGFP-N1 plasmid DNA as the reporter gene in HEK293 cells, and Lipofectamine 2000 was used as the control. Lipoplexes were prepared at four N/P ratios (1, 2, 4 and 8) for each lipid and the result images (at N/P of 4) are shown in Fig. 6. Evidently, among the lipids, 5a, 8a and 8b showed higher TEs, which were comparable and even higher than Lipofectamine 2000. For the 5 series (Fig. 6A–H), increase of chain length resulted in lower TE, and the C12 saturated chain was found to be optimal for such type of double-tailed lipids. Although the introduction of the unsaturated bond slightly improved the TE (5d, Fig. 6D, relative to 5e), the result is still far from that involving 5a. As expected, the lipids with steroid or tocopherol hydrophobic tails (5f–5h) exhibited poor TE, which might have resulted from their weak intermolecular hydrophobic interactions, and it was difficult to form stable liposomes. Therefore, such lipids were excluded in subsequent studies. The amide-contained analogs 6a–6d (Fig. 6K–N) gave much lower TE than 5a–5d, indicating that the biodegradable ester bonds play an important role in the transfection process. As another type of ester-contained lipids, 8 also gave good TE, especially for 8b (Fig. 6I and 6J). It has been reported that the orientation of the ester bond may largely affect the TE of cationic lipids,24,28 and this would be continuously focussed in following studies. Finally, as an important type of lipid compared here, the single-tailed lipids 10 also showed lower TE than 5 (Fig. 6O–Q), indicating that double tails are necessary for efficient transfection. As described previously, the single-tailed lipids might form spherical micelles, which show more difficulty in condensing DNA compared to the bilayer structure.Quantitative luciferase assays were performed to further investigate the TE of the title lipids. It was known that besides the N/P ratio, the mole ratio of lipid/DOPE in the liposome also affect TE.34 Therefore we started with the optimization of such mole ratios. As shown in Fig. 7A, three ratios were tested by using 5a as a template lipid in HEK293 cells, and it was found that at both N/P ratios (4 and 8), the lipid/DOPE mole ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was the best choice for high TE. Thus this mole ratio was applied in the following experiments. Subsequently, the TE of all title lipids were investigated under similar transfection conditions, and the results are shown in Fig. 7B. Although the optimal N/P ratios for each lipid were not exactly the same, the relative TE between these lipids accorded well with the results obtained in the eGFP assays (Fig. 6). In the 5 series, 5a gave the best TE, which was 9.5 times higher than Lipofectamine 2000. Besides, the TE decreased with the elongation of chain length. Several other groups also reported that for saturated long chains, increase of chain length would not favor gene transfection, and C12 or C14 chain was optimal for the delivery.30,35–37 Unsaturated chain contained 5d gave much higher TE than its saturated anolog 5e, further suggesting that their ability to increase membrane fluidity would benefit the transfection.30,38–40 Lipids 10 with single tail again exhibited poor TE, while lipids with reverse ester bonds (8a and 8b) showed much higher TE, which was comparable to those obtained in the transfection involving 5a and 5b. In addition, like the eGFP results, the TEs of amide-contained lipids 6 were lower than the lipids with ester bonds (5 and 8). However, 6b also gave twice as higher TE as Lipofectamine 2000.
1 was the best choice for high TE. Thus this mole ratio was applied in the following experiments. Subsequently, the TE of all title lipids were investigated under similar transfection conditions, and the results are shown in Fig. 7B. Although the optimal N/P ratios for each lipid were not exactly the same, the relative TE between these lipids accorded well with the results obtained in the eGFP assays (Fig. 6). In the 5 series, 5a gave the best TE, which was 9.5 times higher than Lipofectamine 2000. Besides, the TE decreased with the elongation of chain length. Several other groups also reported that for saturated long chains, increase of chain length would not favor gene transfection, and C12 or C14 chain was optimal for the delivery.30,35–37 Unsaturated chain contained 5d gave much higher TE than its saturated anolog 5e, further suggesting that their ability to increase membrane fluidity would benefit the transfection.30,38–40 Lipids 10 with single tail again exhibited poor TE, while lipids with reverse ester bonds (8a and 8b) showed much higher TE, which was comparable to those obtained in the transfection involving 5a and 5b. In addition, like the eGFP results, the TEs of amide-contained lipids 6 were lower than the lipids with ester bonds (5 and 8). However, 6b also gave twice as higher TE as Lipofectamine 2000.
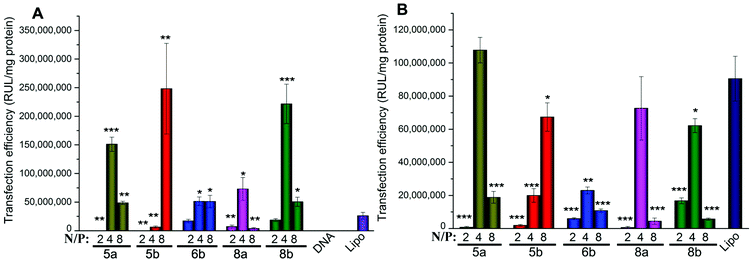
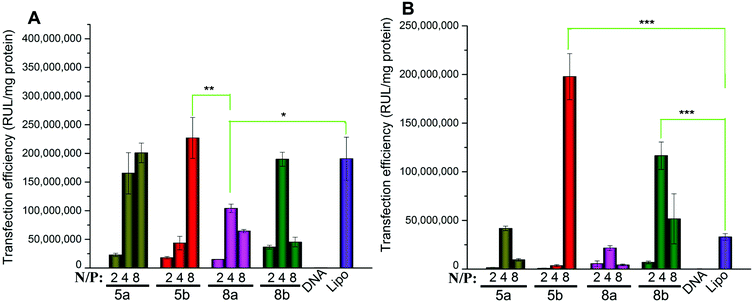
According to the results obtained, in HEK293 cells, the five most efficient lipids (5a, 5b, 6b, 8a and 8b) were applied to the transfection toward other cell lines to further investigate their gene delivery efficacy. As shown in Fig. 8A, in A549 cells, all of the five lipids gave higher TE than Lipofectamine 2000, and the ester-contained lipids with the C14 chain (5b and 8b) showed the best results. The results further proved their excellent gene transfer ability. On the other hand, in Bel-7402 cells, the C12 chain seemed to be the better hydrophobic group, and the optimized TE was relatively lower than those in A549 cells, but still higher than Lipofectamine 2000 (Fig. 8B). The reduced TEs might be due to their higher cytotoxicity in such cell lines.
Biocompatibility is another important factor for a qualified non-viral gene vector. To examine the potential of the lipids for in vivo application, transfections were also carried out in the presence of serum. The eGFP fluorescence microscopy assays involving lipids 5a, 8a and 10a were carried out firstly at a N/P ratio of 4 in the presence of serum. The result images show that the density of cells transfected by 5a and 8a was still higher than that by Lipofectamine 2000 (Fig. S6†), indicating their higher TE. Furthermore, quantitative luciferase assays were also carried out in HEK293 and A549 cell lines with serum. In HEK293 cells, compared with Lipofectamine 2000, although the TE of 5a and 5b were still higher, the relative TEs of the tested lipids all reduced (Fig. 9A). In general, the decreased TE may be ascribed to the electrostatic interaction between negatively charged serum and the lipoplexes with positive surface potential, which might lead to particle aggregation. In A549 cells, the higher relative TEs were kept for the lipids, especially for 5b, which gave 7.7 times higher TE than Lipofectamine 2000 (Fig. 9B). Transfection results in two cell lines both suggest that lipids with C14 chain have higher tolerance against serum. These results show that such types of cyclen-based lipids are promising candidates for non-viral gene delivery vectors.
Cellular uptake
As one of the main barriers in the gene delivery process, the internalization of the nucleic acid complexes largely influence the transfection efficiency. Flow cytometry was carried out to analyze the cellular uptake of DNA. After incubation of the lipoplexes (at the optimal N/P ratio) with HEK293 cells for 4 h in the absence of serum, the percentage of cells positive for Cy5-labeled pDNA was calculated and is shown in Fig. 10. It could be seen that there was only little difference between 5a, 6b and Lipofectamine 2000. However, the TEs of 5a and 6b were quite higher, indicating that the lipoplexes formed from these lipids have better intracellular delivery ability. To our surprise, lipids 10a with poor TE also led to comparable cell internalization, suggesting that lipoplex with relatively large size could also be internalized by the cells, but with worse intracellular fate. Besides, we also did the cellular uptake experiments involving 5b and 8b and found that they have similar cellular uptake as 5a (data not shown). The in-depth transfection mechanism studies of such types of lipids are in progress.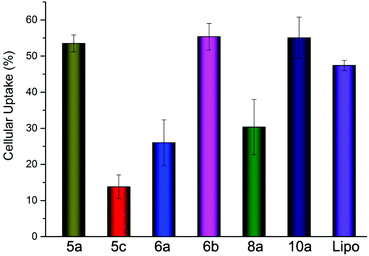 | ||
| Fig. 10 Cellular uptake of lipoplexes at the optimal N/P ratio in HEK293 cells quantified by flow cytometry analysis. | ||
Conclusion
A series of cyclen-based cationic lipids bearing double hydrophobic tails were synthesized. To study their structure–activity relationship as non-viral gene delivery vectors, several types of their analogs were also designed and prepared. Among these lipids, some excellent gene transfection surfactants were obtained, and the optimized transfection efficiency was several times higher than the commercially available Lipofectamine 2000. Results reveal that for such type of cyclen-based lipids, double-tailed structure is necessary for efficient transfection. The length of the hydrophobic chain also largely affects the delivery, and C12 and C14 were found to be the most suitable chain length. Biodegradable ester bond bearing lipids gave better TE than their analogs with amide bond. The orientation of the ester bond has some influence on the transfection ability, but not notable. Additionally, the unsaturated bond on the hydrophobic chain might benefit the transfection. In summary, compared with Lipofectamine 2000, these cationic lipids exhibited much better TE and low cytotoxicity, suggesting their potential to be promising non-viral gene vectors.Acknowledgements
This work was financially supported by the National Program on Key Basic Research Project of China (973 Program, 2012CB720603), the National Science Foundation of China (No. 21232005), and the Specialized Research Fund for the Doctoral Program of Higher Education (20120181130006) in China. J.Z. thanks the Program for New Century Excellent Talents in University (NCET-11-0354). We also thank Analytical & Testing Center of Sichuan University for the TEM analysis.References
- X. Guo and L. Huang, Acc. Chem. Res., 2011, 45, 971–979 CrossRef PubMed.
- M. A. Mintzer and E. E. Simanek, Chem. Rev., 2008, 109, 259–302 CrossRef PubMed.
- C. H. Jones, C.-K. Chen, A. Ravikrishnan, S. Rane and B. A. Pfeifer, Mol. Pharm., 2013, 10, 4082–4098 CrossRef CAS PubMed.
- X.-X. Zhang, T. J. McIntosh and M. W. Grinstaff, Biochimie, 2012, 94, 42–58 CrossRef CAS PubMed.
- E. R. Figueroa, A. Y. Lin, J. Yan, L. Luo, A. E. Foster and R. A. Drezek, Biomaterials, 2014, 35, 1725–1734 CrossRef CAS PubMed.
- L. Han, J. Zhao, X. Zhang, W. Cao, X. Hu, G. Zou, X. Duan and X.-J. Liang, ACS Nano, 2012, 6, 7340–7351 CrossRef CAS PubMed.
- M. Tsukamoto, T. Ochiya, S. Yoshida, T. Sugimura and M. Terada, Nat. Genet., 1995, 9, 243–248 CrossRef CAS PubMed.
- Y. Aoshima, R. Hokama, K. Sou, S. R. Sarker, K. Iida, H. Nakamura, T. Inoue and S. Takeoka, ACS Chem. Neurosci., 2013, 4, 1514–1519 CrossRef CAS PubMed.
- D. Zhi, S. Zhang, S. Cui, Y. Zhao, Y. Wang and D. Zhao, Bioconjugate Chem., 2013, 24, 487–519 CrossRef CAS PubMed.
- P. Kumar, H. Wu, J. L. McBride, K.-E. Jung, M. Hee Kim, B. L. Davidson, S. Kyung Lee, P. Shankar and N. Manjunath, Nature, 2007, 448, 39–43 CrossRef CAS PubMed.
- X. Liang, X. Li, J. Chang, Y. Duan and Z. Li, Langmuir, 2013, 29, 8683–8693 CrossRef CAS PubMed.
- B. K. Majeti, R. S. Singh, S. K. Yadav, S. R. Bathula, S. Ramakrishna, P. V. Diwan, S. S. Madhavendra and A. Chaudhuri, Chem. Biol., 2004, 11, 427–437 CrossRef CAS PubMed.
- B. Kedika and S. V. Patri, Bioconjugate Chem., 2011, 22, 2581–2592 CrossRef CAS PubMed.
- D. Zhi, S. Zhang, F. Qureshi, Y. Zhao, S. Cui, B. Wang, H. Chen, Y. Wang and D. Zhao, Bioorg. Med. Chem. Lett., 2012, 22, 3837–3841 CrossRef CAS PubMed.
- M. Khan, C. Y. Ang, N. Wiradharma, L.-K. Yong, S. Liu, L. Liu, S. Gao and Y.-Y. Yang, Biomaterials, 2012, 33, 4673–4680 CrossRef CAS PubMed.
- M. Mével, N. Kamaly, S. Carmona, M. H. Oliver, M. R. Jorgensen, C. Crowther, F. H. Salazar, P. L. Marion, M. Fujino, Y. Natori, M. Thanou, P. Arbuthnot, J.-J. Yaouanc, P. A. Jaffrès and A. D. Miller, J. Controlled Release, 2010, 143, 222–232 CrossRef PubMed.
- M. A. Maslov, T. O. Kabilova, I. A. Petukhov, N. G. Morozova, G. A. Serebrennikova, V. V. Vlassov and M. A. Zenkova, J. Controlled Release, 2012, 160, 182–193 CrossRef CAS PubMed.
- S. R. Sarker, S. Arai, M. Murate, H. Takahashi, M. Takata, T. Kobayashi and S. Takeoka, Int. J. Pharm., 2012, 422, 364–373 CrossRef CAS PubMed.
- R. Sheng, T. Luo, H. Li, J. Sun, Z. Wang and A. Cao, Colloids Surf., B, 2014, 116, 32–40 CrossRef CAS PubMed.
- L. Li, H. Song, K. Luo, B. He, Y. Nie, Y. Yang, Y. Wu and Z. Gu, Int. J. Pharm., 2011, 408, 183–190 CrossRef CAS PubMed.
- A. Aissaoui, B. Martin, E. Kan, N. Oudrhiri, M. Hauchecorne, J.-P. Vigneron, J.-M. Lehn and P. Lehn, J. Med. Chem., 2004, 47, 5210–5223 CrossRef CAS PubMed.
- J. Sen and A. Chaudhuri, J. Med. Chem., 2005, 48, 812–820 CrossRef CAS PubMed.
- M. Srujan, V. Chandrashekhar, R. C. Reddy, R. Prabhakar, B. Sreedhar and A. Chaudhuri, Biomaterials, 2011, 32, 5231–5240 CrossRef CAS PubMed.
- M. Rajesh, J. Sen, M. Srujan, K. Mukherjee, B. Sreedhar and A. Chaudhuri, J. Am. Chem. Soc., 2007, 129, 11408–11420 CrossRef CAS PubMed.
- R. M. Izatt, K. Pawlak, J. S. Bradshaw and R. L. Bruening, Chem. Rev., 1991, 91, 1721–2085 CrossRef CAS.
- T. Fujiwara, S. Hasegawa, N. Hirashima, M. Nakanishi and T. Ohwada, Biochim. Biophys. Acta, 2000, 1468, 396–402 CrossRef CAS.
- Q.-D. Huang, W.-J. Ou, H. Chen, Z.-H. Feng, J.-Y. Wang, J. Zhang, W. Zhu and X.-Q. Yu, Eur. J. Pharm. Biopharm., 2011, 78, 326–335 CrossRef CAS PubMed.
- Q.-D. Huang, G.-X. Zhong, Y. Zhang, J. Ren, Y. Fu, J. Zhang, W. Zhu and X.-Q. Yu, PLoS One, 2011, 6, e23134 CAS.
- J.-L. Liu, Q.-P. Ma, Q.-D. Huang, W.-H. Yang, J. Zhang, J.-Y. Wang, W. Zhu and X.-Q. Yu, Eur. J. Med. Chem., 2011, 46, 4133–4141 CrossRef CAS PubMed.
- D. Zhi, S. Zhang, B. Wang, Y. Zhao, B. Yang and S. Yu, Bioconjugate Chem., 2010, 21, 563–577 CrossRef CAS PubMed.
- J. W. Jeon, S. J. Son, C. E. Yoo, I. S. Hong, J. B. Song and J. Suh, Org. Lett., 2002, 4, 4155–4158 CrossRef CAS PubMed.
- L. M. De León-Rodríguez, Z. Kovacs, A. C. Esqueda-Oliva and A. D. Miranda-Olvera, Tetrahedron Lett., 2006, 47, 6937–6940 CrossRef PubMed.
- M. Ma, F. Li, Z.-f. Yuan and R.-X. Zhuo, Acta Biomater., 2010, 6, 2658–2665 CrossRef CAS PubMed.
- A. Bajaj, P. Kondaiah and S. Bhattacharya, J. Med. Chem., 2008, 51, 2533–2540 CrossRef CAS PubMed.
- Z. Hyvönen, A. Plotniece, I. Reine, B. Chekavichus, G. Duburs and A. Urtti, Biochim. Biophys. Acta, 2000, 1509, 451–466 CrossRef.
- V. Gopal, T. K. Prasad, N. M. Rao, M. Takafuji, M. M. Rahman and H. Ihara, Bioconjugate Chem., 2006, 17, 1530–1536 CrossRef CAS PubMed.
- S. Perrone, M. Usai, P. Lazzari, S. J. Tucker, H. M. Wallace and M. Zanda, Bioconjugate Chem., 2013, 24, 176–187 CrossRef CAS PubMed.
- S. Fletcher, A. Ahmad, E. Perouzel, A. Heron, A. D. Miller and M. R. Jorgensen, J. Med. Chem., 2005, 49, 349–357 CrossRef PubMed.
- S. Loisel, V. Floch, C. Le Gall and C. Férec, J. Liposome Res., 2001, 11, 127–138 CrossRef CAS PubMed.
- Q.-F. Zhang, W.-H. Yang, W.-J. Yi, J. Zhang, J. Ren, T.-Y. Luo, W. Zhu and X.-Q. Yu, Bioorg. Med. Chem. Lett., 2011, 21, 7045–7049 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Included as separate formatted document. See DOI: 10.1039/c4bm00174e |
| This journal is © The Royal Society of Chemistry 2014 |