Proteomic identification of organic additives in the mortars of ancient Chinese wooden buildings†
Huiyun
Rao
ab,
Bo
Li
c,
Yimin
Yang
*ab,
Qinglin
Ma
d and
Changsui
Wang
ab
aKey Laboratory of Vertebrate Evolution and Human Origins of Chinese Academy of Sciences, Institute of Vertebrate Paleontology and Paleoanthropology, Chinese Academy of Sciences, Beijing 100044, PR China. E-mail: yiminyang@ucas.ac.cn
bDepartment of Scientific History and Archaeometry, University of Chinese Academy of Sciences, Beijing 100049, PR China
cSchool of Architecture, Tsinghua University, Beijing 100084, PR China
dChinese Academy of Cultural Heritage, Beijing 100029, PR China
First published on 21st October 2014
Abstract
Mortars are the layers paved on the surface of timber, earth or stone before painting and drawing. The analysis of their material composition and manufacture technology is necessary for revealing old technological approaches, selecting a suitable technological process in restoration and protection, and guiding the development of traditional technology of Chinese painting and colored drawings. According to ancient literature, crop flour and blood have been used as binders in the mortars of Chinese wooden buildings. However, little work is published on their scientific identification, and the reported methods could not determine their precise origins, which is important to understanding ancient mortar technology. In this study, Fourier Transform Infrared Spectroscopy (FTIR), Liquid Chromatography Tandem Mass Spectrometry (LC/MS/MS) and starch grain analysis were employed to analyze the three mortars taken from the Old Summer Palace (18th and early 19th centuries), the Eastern Royal Tombs of the Qing Dynasty (middle 17th to early 20th centuries) and the Taiyuan Confucius Temple (late 19th century), respectively. FTIR analysis indicated the presence of proteins, and then different organic additives, namely, wheaten flour, cattle blood and pig blood, were identified respectively in the three mortars by LC/MS/MS analysis. Starch grain analysis also confirmed the proteomic results. Thus, proteomic analysis is highly effective for identifying the nature and origin of organic additives in the mortars of ancient painting.
Introduction
Ancient Chinese buildings are mainly made of wooden structures and famous for their carved beams and painted rafters. Thus, painting and colored drawings are very valuable parts of ancient Chinese architectural heritage. Ancient artists used mortars as ground layers on wood in order to protect the lumber and prepare it for painting.1 The analysis of historical mortars is necessary for revealing old technological approaches, understanding their unusual properties and subsequently selecting suitable technological processes in restoration.The mortars applied in ancient Chinese wooden buildings exist as a rather complex system of inorganic and organic components, including brick ash, lime, fiber, flour, blood and tung oil,2 among which brick ash and lime have been used as filling materials, fiber as taut material, and flour, blood and tung oil as binding materials.3 The inorganic components of mortars are well studied. In several cases, different combinations of X-ray diffraction analysis, X-ray fluorescence analysis, scanning electron microscopy (SEM) with energy dispersive X-ray (EDX) analysis, high-temperature burning-acid dissolution method, and thermal analysis have been used to analyze the inorganic matters qualitatively and quantitatively.3–5 In addition, fibers can be identified by microscopic observation according to their morphological characteristics and structural differences.6
As for the organic additives, pyrolysis-gas chromatography-mass spectrometry (Py-GC/MS) has been successfully employed to detect the presence of tung oil,7 but the identification of flour and blood remains challenging because of their low contents and the susceptibility to decay during burial. Starch–iodine staining tests, starch grain microscopic analysis and spectrophotometric methods have been used for the qualitative and quantitative analysis of flour in ancient mortars.8,9 Bloodstain tests in forensic science have been applied to examine short-period samples, and crude protein content could be determined by the Kjeldahl nitrogen method or organic elementary analysis.9–11 However, these analytical methods cannot determine the precise origins of these binders, which should be important for understanding mortar technology as flour and blood remarkably improve the anti-fluting property and durability of the mortars.12
On the other hand, the binders in various archaeological contexts have been well identified by micro-chemical analysis and staining methods,13,14 spectroscopic techniques such as infrared spectroscopy and Raman spectroscopy,15 starch grain analysis,16 chromatographic methods including gas-phase and liquid-phase chromatography (coupled with mass spectrometry),17–19 immunoassay techniques,20 and recently developed proteomic methods.21–23 In terms of composition, flour and blood binders all contain certain amounts of protein components, which record abundant biological information. Because proteomic approaches can determine the precise origin of the proteins, i.e. protein species as far as they are available in the databases, even if the content is very low in ancient samples,24 then the methods might be used to analyze the nature and origin of organic additives (flour and blood) in the mortars of ancient Chinese wooden buildings.
In this study, we proposed a multi-method approach for the identification of organic additives in three ancient mortars. Fourier transform infrared spectroscopy (FTIR) was first implemented to evaluate protein presence in the samples, and the proteomic method was then employed to distinguish the proteins from organic additives.
Experimental section
Sample description
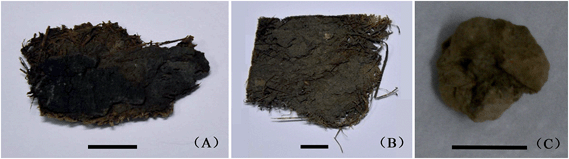
The three mortars were collected by the Chinese Academy of Cultural Heritage. Sample A (Fig. 1A), taken from the peeling color painting of the Old Summer Palace, was a black–brown mortar piece (∼4 cm width) with a fibrous layer. The Old Summer Palace, known in Chinese as Yuanmingyuan, is a large royal palace with both Western and Chinese architectural styles, noted for the “Garden of Gardens”. Located in the northwestern suburbs of Beijing, it was built in the 18th and early 19th centuries as a place where the emperors of the Qing Dynasty resided and processed government affairs. In 1860, it was destroyed and looted during the Second Opium War, and only ruins remain now.Sample B (Fig. 1B) was a brown mortar fragment (∼5 cm width) with a fibrous layer. It was sampled from the Eastern Royal Tombs of the Qing Dynasty, located in Zunhua City (Hebei Province, China), 125 km northeast of Beijing. These tombs were built since the mid-17th century. It presents the most advanced technology of ancient Chinese architecture and has significant value for history, art and science. Longendian, where the archaeological sample was extracted, is the largest ground building of the site, which served as the main place for ritual activities.
Sample C (Fig. 1C) was a yellow mortar of granular appearance (∼1.5 cm width). It was taken from the west wind-room of the Taiyuan Confucius Temple in Shanxi Province, China. After the original buildings were destroyed by flood in 1881, the temple was reconstructed on the Chongshan Temple ruins in the following year. During the period of the Republic of China, it was known as a place for sacrifices to Confucius. In 1919, the Museum of Educational Books in Shanxi Province was established inside the temple, which was the first museum in Taiyuan City and renamed the Shanxi Museum in 1953.
FTIR analysis
The samples were analyzed as KBr micropellets with a Nicolet 6700 (Thermo Scientific) FTIR spectrometer working in a transmission mode. Spectra were acquired over the range of 4000–400 cm−1 using a resolution of 4 cm−1, with 32 scans per spectrum. The software OMNIC 8.0 was applied to deal with the data.Protein extraction
The extraction procedure was modified from published ref. 25 and 26 and successfully carried out on modern mortars, thus it was applied on ancient samples. 100 μl of the extracting solution (tris-HCl, pH 8.0, 10 mM dithiothreitol, 10% sodium dodecylsulfate and 0.0025% bromphenol blue) was added to approximately 20 mg of ancient sample. The mixture was subjected to ultrasonic baths (3 × 15 min) followed by incubation for 1 h at 56 °C, then sonicated again for 15 min and centrifuged for 15 min at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g.
000 g.
Sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE)
The protein extraction was separated and purified by electrophoresis. 45 μl of the supernatant was mixed with 5 μl of glycerol, heated at 95 °C for 5 min, cooled to room temperature and loaded onto the gel (5% stacking gel, 12% separating gel) with 25 μl of the mixture in each well. The electrophoresis apparatus was first connected to an 80 V power source and switched to a 120 V power when the sample arrived at the separating gel. As the sample ran on the separating gel for approximately 3 cm, the power was turned off and the gel removed. A microwave-assisted Coomassie Blue staining protocol was followed. The gel immersed in the staining solution (0.25% Coomassie Blue w/v, 50% ethanol, and 10% acetic acid) was incubated in a microwave oven at medium-low heat for 45 s, followed by slow shaking for 10 min. The staining solution was then dumped. The gel was washed with distilled water several times, immersed in the destaining solution (25% ethanol, 8% acetic acid) and slowly shaken overnight until the blue-stained protein area was visible. Each sample was run on individual gels to avoid horizontal carryover of the proteins.27In-gel digestion
The procedures of in-gel digestion and the followed LC/MS/MS analysis were modified from published ref. 28. The blue-stained protein area of the gel was cut into small particles of 1 mm3. The gel particles were washed three times with distilled water, destained with 50% acetonitrile/25 mM NH4HCO3, dried with 100% acetonitrile and alkylated under darkness with 50 mM iodoacetamide at room temperature for 30 min. After the solution was removed, the gel particles were washed twice with 25 mM NH4HCO3 buffer, dried with 100% acetonitrile and immersed in the trypsin solution (12.5 ng μl−1 trypsin in 25 mM NH4HCO3) to ensure that the gel particles were covered with liquid. The digestion was incubated in a microwave oven at 850 W for 1 min, and the peptides were then extracted with 100% acetonitrile. The extraction was vacuum dried and cryopreserved for further identification by MS.LC/MS/MS
The digested sample was re-dissolved in 0.1% formic acid (buffer A) before MS analysis. A 2 μl sample was injected and analyzed by an RP C18 capillary LC column from Michrom Bioresources (100 μm × 150 mm, 3 μm). The eluted gradient was 5–30% buffer B (0.1% formic acid and 99.9% acetonitrile; flow rate, 0.5 μl min−1) for 30 min. The MS data were acquired on an LTQ Orbitrap Velos mass spectrometer using CID (sample A) or HCD (samples B and C) mode. The parameters were set in the following manner: 20 data-dependent CID MS/MS scans per every full scan for the CID mode and 10 HCD scans for the HCD mode; full scans were acquired in Orbitrap at a resolution of 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000; 35% normalized collision energy for the CID mode and 40% for the HCD mode; internal mass calibration (445.120025 ion as lock mass with a target lock mass abundance of 0%); charge-state screening (excluding precursors with unknown charge state or +1 charge state); and dynamic exclusion (exclusion size list, 500; exclusion duration, 30 s).
000; 35% normalized collision energy for the CID mode and 40% for the HCD mode; internal mass calibration (445.120025 ion as lock mass with a target lock mass abundance of 0%); charge-state screening (excluding precursors with unknown charge state or +1 charge state); and dynamic exclusion (exclusion size list, 500; exclusion duration, 30 s).
Database search
The MS/MS spectra of samples were searched against the SwissProt database using Mascot software version 2.3.02 (Matrix Science, UK). Trypsin was chosen for cleavage specificity with a maximum number of two allowed missed cleavages. Carbamidomethylation (C) was set as a fixed modification, whereas deamidation (NQ) and oxidation (M) were set as variable modifications. The searches were performed using a peptide tolerance of 10 ppm and a product ion tolerance of 0.5 Da. The data were then filtered at a p-value <0.05.Starch grain analysis
Starch grain analysis was implemented to study the starch component of the flour additives in the mortars and to confirm the proteomic results. A small amount of material was scraped into a 5 ml centrifuge tube with a scalpel, and 2 ml deionized water was added. The mixture was subjected to ultrasonic baths (2 × 15 min) and left for several hours. After shaking, a drop of suspension was pipetted onto a slide. Before it had dried, one drop of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 water/glycerin solution was added and a cover slip applied. The slide was examined with polarized and transmitted light at 500× to identify and photograph the starch grains present. 100 grains were measured to obtain data on the length of the starch grains. Since the starch grains less than 5 μm in size always show negligible morphological differences and could not offer considerable information for identification,29 only those exceeding 5 μm in size were counted and calculated.
50 water/glycerin solution was added and a cover slip applied. The slide was examined with polarized and transmitted light at 500× to identify and photograph the starch grains present. 100 grains were measured to obtain data on the length of the starch grains. Since the starch grains less than 5 μm in size always show negligible morphological differences and could not offer considerable information for identification,29 only those exceeding 5 μm in size were counted and calculated.
Results and discussion
FTIR characterization of the mortars
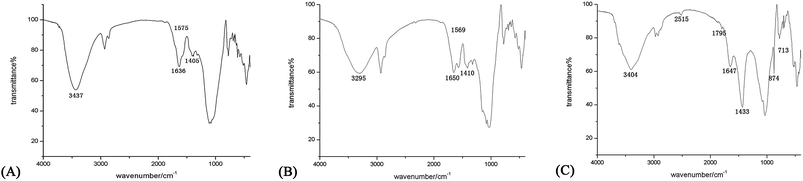
The FTIR results of the three mortars (Fig. 2) all imply the presence of proteins.30,31 As for samples A and B, it is possible to identify the characteristic signals of the amide group (–N(H)–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O–). More specifically, the peaks at 3437 cm−1 and 3295 cm−1 are assigned to the N–H stretching vibration region, 1636 cm−1 and 1650 cm−1 to the C
O–). More specifically, the peaks at 3437 cm−1 and 3295 cm−1 are assigned to the N–H stretching vibration region, 1636 cm−1 and 1650 cm−1 to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration region, 1575 cm−1 and 1569 cm−1 to the N–H bending vibration region, and 1405 cm−1 and 1410 cm−1 to the C–N stretching region. The pattern of the absorption peaks is attributed to the presence of proteins in the samples. However, the spectrum of sample C is slightly different because some inorganic components (calcium carbonate) are present and interfere with the absorption peak pattern of the protein constituents.32 The peaks at 2515 cm−1, 1795 cm−1, 874 cm−1 and 713 cm−1 are all characteristic peaks of calcium carbonate. As to the peaks attributed to proteins, the peaks at 3404 cm−1 and 1647 cm−1 are assigned to the N–H and C
O stretching vibration region, 1575 cm−1 and 1569 cm−1 to the N–H bending vibration region, and 1405 cm−1 and 1410 cm−1 to the C–N stretching region. The pattern of the absorption peaks is attributed to the presence of proteins in the samples. However, the spectrum of sample C is slightly different because some inorganic components (calcium carbonate) are present and interfere with the absorption peak pattern of the protein constituents.32 The peaks at 2515 cm−1, 1795 cm−1, 874 cm−1 and 713 cm−1 are all characteristic peaks of calcium carbonate. As to the peaks attributed to proteins, the peaks at 3404 cm−1 and 1647 cm−1 are assigned to the N–H and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration regions, whereas 1433 cm−1 is considered as a combination of the C–N stretching region of proteins and asymmetric stretching vibration region of carbonate. The absorption peak pattern indicates the presence of proteins and some inorganic components (calcium carbonate) in sample C.
O stretching vibration regions, whereas 1433 cm−1 is considered as a combination of the C–N stretching region of proteins and asymmetric stretching vibration region of carbonate. The absorption peak pattern indicates the presence of proteins and some inorganic components (calcium carbonate) in sample C.
Identification of flour and blood proteins
The gel figures after staining have been given as ESI in Fig. S1–S3.† As the gel figures show, proteins are separated not only in a vertical dimension but also spread horizontally. A blue-stained protein area on the gel was shown as whole, instead of individual protein bands, which should result from protein degradation in the sample. Considering the detection limit of Coomassie Blue staining,33 the protein content reserved in the area is sufficient for subsequent mass spectrometry analysis.The LC/MS/MS results displayed in Table 1 show that one protein from wheaten flour was detected in sample A, whereas two proteins from cattle blood and three from pig blood were identified in samples B and C, respectively. At least two peptides were identified in each protein. The fragmentation of digested peptides in the collision cell mainly occurs at the peptide bond position, giving rise to y and b ions. Fig. 3 shows the MS/MS spectra of two specific peptides EAVLGLWGK and VLQSFSDGLK from the porcine hemoglobin subunit beta identified in sample C (Table 1), in which y and b represent the single-charged mass fragments, y++ and b++ the double-charged fragments, y0 and b0 the dehydrated fragments and y* and b* the deaminated fragments. The y and b ions have good continuity, suggesting the data is reliable.
| Sample | Identified proteins | Species | Score | Sequence coverage (%) | No. peptides (unique) | Accession number |
|---|---|---|---|---|---|---|
| Sample A | Alpha-amylase/trypsin inhibitor CM3 | Triticum aestivum | 74 | 13% | 2(2) | P17314 |
| Sample B | Serum albumin | Bos taurus | 868 | 23% | 16(4) | P02769 |
| Hemoglobin fetal subunit beta | Bos taurus | 148 | 24% | 3(1) | P02081 | |
| Sample C | Hemoglobin subunit beta | Sus scrofa | 3489 | 83% | 17(11) | P02067 |
| Hemoglobin subunit alpha | Sus scrofa | 4934 | 75% | 12(4) | P01965 | |
| Serum albumin | Sus scrofa | 605 | 13% | 10(4) | P08835 |
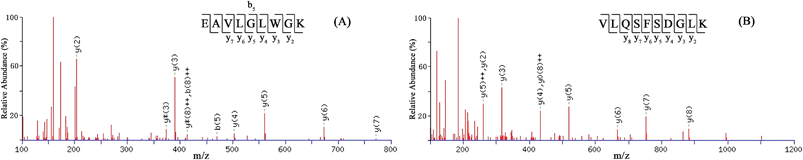 | ||
| Fig. 3 MS/MS spectra of two specific peptides from porcine hemoglobin subunit beta in sample C. (A) EAVLGLWGK. (B) VLQSFSDLK (with a deamidation at Q). | ||
Therefore, it is inferred that different organic additives have been used as binders in the three mortars. In sample A, wheaten flour has been added, but the mortar could have been produced without blood material. In addition, samples B and C have used the additives of cattle and pig blood, respectively. As the results demonstrate, the proteomic identification of organic additives has the advantage that it not only can identify specific proteins in samples but can also accurately verify the origin of the proteins through the specific peptides.
Starch grain analysis vs. proteomic approaches
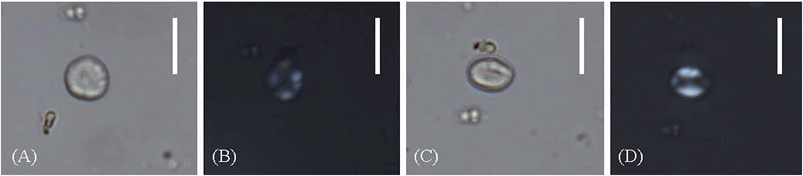
As indicated by the proteomic results, wheaten flour has been added as an organic binder in sample A. Actually, starch is the main component of flour, in addition to proteins. Thus, starch grain analysis was employed in sample A to compare with and confirm the proteomic results. The starch grains found in sample A are circular, subrounded or oval in shape, with centric hilum; some have apparent lamellae. Most of the extinction crosses are bilaterally symmetrical. When rotated to a side view, they become lenticular with a longitudinal fissure (Fig. 4). The maximum lengths of individual grains range from about 10 to 40 μm (Table 2). As the characteristics and size of the present starch grains fit quite well with those of Triticeae,34,35 the starch grains may derive from one or some of the plants from Triticeae, which is consistent with the proteomic results. However, because the characteristics and the size of starch grains are highly similar among several plants from Triticeae, as shown in Table 2, the precise origin of the flour additives (particularly a wheat origin) cannot be determined by starch grain analysis but can be done by proteomic methods.Why are different organic binders added in mortars?
In ancient Egypt and Rome, various organic materials, including proteins, lipids, saccharides and resins, have been added in mortars to improve the properties.36,37 In China, the sticky rice–lime mortar has also been applied in the tombs dated as early as the Northern and Southern dynasties (AD 386–589)38 and served as a representative of traditional Chinese mortars. The ancient scientific book “Tian Gong Kai Wu”, which is noted for “the encyclopedia on Chinese craft in the 17th century”, has recorded the sticky rice–lime mortar used in historical masonry constructions.39 Thus, it could be deduced that ancient craftsmen, domestic and overseas, have already recognized the good mechanical properties and durability of organic/inorganic hybrid material and applied it in historical buildings.In terms of the mortars used in ancient Chinese wooden buildings, the historical book “Ying Zao Fa Shi” written during the Song Dynasty (AD 960–1279), which is the first official book minutely expounding constructional engineering in China, is the earliest literature referring to mortar technology, in addition to documenting wheaten additives. Up to now, the earliest discovered mortar was excavated near Chita City, dating to the Yuan Dynasty (AD 1271–1368).40 An oral source states that blood additives have been used in the mortars from the wooden structure of the Amarbayasgalant Monastry (AD 1727–1735) in Mongolia.41
In this study, three mortars were analyzed and various organic additives were identified as binders. Wheaten flour has been added in the mortar used in the color painting of the Old Summer Palace (sample A) to improve its anti-fluting property,12 whereas blood additives have been used in the other two samples to increase their durability.12 In China, pig blood is the most widely used blood material in the mortars because of its easy accessibility and good viscosity,3 as detected in the mortar from the Taiyuan Confucius Temple (sample C). However, in Western countries, cattle blood is preferred.42,43 On this account, cattle blood identified in the mortar from the Eastern Royal Tombs of the Qing Dynasty (sample B) may indicate a certain degree of cultural interchange. On the other hand, as blood is sometimes used for religious and ritual purposes44 and Longendian, where the mortar was extracted, is the main place for ritual activities in the Eastern Royal Tombs of the Qing Dynasty, the addition of cattle blood may have some sort of religious significance, which needs further investigation.
Proteomics: an informative technique for the identification of organic additives in mortars
To identify the flour additives, various analytical methods and techniques could be used, including starch–iodine tests,3,45 ordinary microscopic observation,46 starch grain analysis,8,16 analysis of bran fragments,35 and infrared (IR) spectroscopy47,48 as well as the newly developed proteomic approaches.49 Compared with other methods, proteomic approaches have the advantages of precisely identifying the source of flour and also determining its processing technologies through the identification of other proteins from coexisting components. In this study, the proteomic technique was successfully employed to identify the flour additives in the mortars for the first time.Moreover, the methods for characterizing blood in archaeological contexts could be classified into two categories. One is aimed to identify the hemoglobin or haem moiety from blood. This category contains multiple methods, such as the colorimetric test,50 spectroscopic techniques,51 chromatographic methods,52 mass spectrometric analysis,44,53 and so on. These methods can only demonstrate the presence of blood. The other category is the immunoassay technique.54–56 Although immunoassay can determine the origin of used blood, it is limited to specific targeted proteins. However, proteomic approaches can overcome this disadvantage and identify various origins of blood through a single run. In this study, the proteomic technique was introduced to test the blood additives in the mortars, and both cattle and pig blood were successfully identified.
In summary, proteomic approaches are of high sensitivity and can obtain abundant biological information contained in protein residues. As illustrated in this paper, proteomics is unique not only to the proteins themselves but also can offer genus- or species-specific sequence information. Using this technique, the precise origin of the protein additives, in particular flour and blood in the mortars, could be identified simultaneously through a single run. Thus, proteomics is an informative and convenient technique for the identification of organic additives in the mortars.
However, the previous discussion focuses on the qualitative identification of organic additives. Previous studies have shown that the quantification of the blood and flour additives could be determined by the Kjeldahl nitrogen method, organic elementary analysis, and the spectrophotometric method.9–11 Furthermore, if two or more additives have been identified in one sample through proteomics, the relative abundance of protein groups could also be estimated by proteomics,49 which is important for imitating ancient mortars. Thus, proteomic approaches could realize the qualitative and quantitative analysis of organic additives in the mortars, which is of significant importance for unveiling old mortar technology and subsequently selecting suitable technological processes for restoration.
Conclusion
More recently, proteomic approaches have been introduced to archaeology and have been successfully applied in the identification of archaeological pottery remains,49,57,58 binders in artworks,22,59–61 protein additives in building materials,62 and so on. Meanwhile, various types of protein residues have been identified, including animal proteins (meat, egg, milk and collagen from bones or skin), plant proteins (flour and seeds), and so on. The proteomic/genomic databases of protein sequences are developed and updated constantly. Even if a species is not documented and fully sequenced in the databases, the protein identification could be realized via sequence homology to phylogenetically related species.14,57 Furthermore, the precise origin would be determined by species-specific markers assigned based on mass spectrometric characterization of modern samples.58,63 Proteomic approaches are informative techniques for the identification of organic residues in various archaeological contexts.In this paper, proteomic approaches have been successfully applied to approximately 20 mg of ancient sample and have resulted in the identification of different protein additives (flour and blood) in the mortars. This technique has decided not only whether flour/blood had been added but also for the first time identified the precise origin of the flour or blood additives in the mortars. It holds promising potential for the routine identification of organic additives in the mortars from ancient buildings.
Acknowledgements
The authors would like to thank Prof. Sun Wei for the expert technical support and valuable advice. This study was supported by the grants from the National Compass Plans (20111806) from the State Bureau of Cultural Relics in China, the National Science Foundation in China (41172164) and the CAS Strategic Priority Research Program (XDA05130303).References
- IRCMC, Restoration technology of traditional Chinese architecture, Press of Chinese Architectural Industry, Beijing, 1983, in Chinese Search PubMed.
- J. Bian, Painting and colored drawing of Chinese ancient architecture, Chinese Architectural Material Industry Publishing House, Beijing, 2007, in Chinese Search PubMed.
- W. Zhou, Master thesis, Northwest University, 2009, in Chinese with English abstract.
- W. Zhou and L. Wang, Sciences of Conservation and Archaeology, 2010, 22, 1–9 Search PubMed , in Chinese with English abstract.
- W. Zhou, L. Wang, X. Fan, Y. Qi and T. Ma, J. Inner Mongolia Teach. Univ., 2010, 41, 522–526 CAS , in Chinese with English abstract.
- L. Wang, L. Yang, W. Zhou, Q. He, J. Yan, X. Fan, T. Ma and Y. Qi, Relics and Museolgy, 2009, 451–454 Search PubMed , in Chinese with English abstract.
- R. Mazzeo, D. Cam, G. Chiavari, D. Fabbri, H. Ling and S. Prati, J. Cult. Herit., 2004, 5, 273–283 CrossRef PubMed.
- L. Jin and Z. Shao, Identification and Appreciation to Cultural Relics, 2010, 30–35 Search PubMed , in Chinese.
- L. Wang, R. Guo, W. Zhou, X. Fan, L. Zhao and T. Ma, Journal of Lanzhou University (Natural Sciences), 2012, 48, 136–140 CAS , in Chinese with English abstract.
- Y. Hu, D. Hu and B. Kang, Beijing Cult. Relics Mus., 2007, 81–85 Search PubMed , in Chinese with English abstract.
- L. Wang, L. Yang, W. Zhou and X. Fan, Sciences of Conservation and Archaeology, 2011, 23, 59–63 Search PubMed , in Chinese with English abstract.
- D. Hu, Y. Li, J. Li, H. Li, X. Hu and T. Ma, Relics and Museolgy, 2009, 435–450 Search PubMed , in Chinese with English abstract.
- U. Fan, L. He, C. Thieme and E. Emmerling, Dunhuang Research, 1996, 140–153 Search PubMed , in Chinese with English abstract.
- S. Dallongeville, M. Richter, S. Schäfer, M. Kühlenthal, N. Garnier, C. Rolando and C. Tokarski, Analyst, 2013, 138, 5357–5364 RSC.
- H. G. M. Edwards, B. Stern, L. Burgio and M. Kite, Spectrochim. Acta, Part A, 2009, 73, 561–565 CrossRef PubMed.
- T. Li, PhD, Graduate University of Chinese Academy of Sciences, 2010, in Chinese with English abstract.
- W. Fremout, J. Sanyova, S. Saverwyns, P. Vandenabeele and L. Moens, Anal. Bioanal. Chem., 2009, 393, 1991–1999 CrossRef CAS PubMed.
- M. Regert, J. Sep. Sci., 2004, 27, 244–254 CrossRef CAS PubMed.
- S. Wei, V. Pintus, V. Pitthard, M. Schreiner and G. Song, J. Archaeol. Sci., 2011, 38, 2667–2674 Search PubMed.
- D. A. Scott, S. Warmlander, J. Mazurek and S. Quirke, J. Archaeol. Sci., 2009, 36, 923–932 CrossRef PubMed.
- A. Chambery, A. Di Maro, C. Sanges, V. Severino, M. Tarantino, A. Lamberti, A. Parente and P. Arcari, Anal. Bioanal. Chem., 2009, 395, 2281–2291 CrossRef CAS PubMed.
- S. Dallongeville, M. Koperska, N. Garnier, G. Reille-Taillefert, C. Rolando and C. Tokarski, Anal. Chem., 2011, 83, 9431–9437 CrossRef CAS PubMed.
- W. Fremout, M. Dhaenens, S. Saverwyns, J. Sanyova, P. Vandenabeele, D. Deforce and L. Moens, Anal. Chim. Acta, 2010, 658, 156–162 CrossRef CAS PubMed.
- C. Hong, H. Jiang, E. Lü, Y. Wu, L. Guo, Y. Xie, C. Wang and Y. Yang, PLoS One, 2012, 7, e37053 CAS.
- A. Shevchenko, H. Tomas, J. Havlis, J. V. Olsen and M. Mann, Nat. Protoc., 2006, 1, 2856–2860 CrossRef CAS PubMed.
- Y. Yang, A. Shevchenko, A. Knaust, I. Abuduresule, W. Li, X. Hu, C. Wang and A. Shevchenko, J. Archaeol. Sci., 2014, 45, 178–186 CrossRef CAS PubMed.
- A. Knaust, A. Shevchenko and A. Shevchenko, Anal. Biochem., 2012, 421, 779–781 CrossRef CAS PubMed.
- W. Sun, S. Gao, L. Wang, Y. Chen, S. Wu, X. Wang, D. Zheng and Y. Gao, Mol. Cell. Proteomics, 2006, 5, 769–776 CAS.
- M. Therin, R. Torrence and R. Fullagar, Aust. Archaeol., 1997, 52–53 Search PubMed.
- R. Zhou and Y. Shen, J. East China Univ. Sci. Technol., 1997, 23, 422–425 CAS , in Chinese with English abstract.
- M. R. Derrick, D. Stulik and J. M. Landry, Infrared spectroscopy in conservation science, Getty Publications, 1999 Search PubMed.
- M. A. Legodi, D. de Waal, J. H. Potgieter and S. S. Potgieter, Miner. Eng., 2001, 14, 1107–1111 CrossRef CAS.
- J. Wang and M. Fan, The Manual of Protein Technology, The Science Press, Beijing, 2000, in Chinese Search PubMed.
- Y. Zhang, Y. Weng, L. Yao, J. Zhang, Y. Zhou, F. Fang and W. Cui, Quat. Sci., 2011, 31, 891–899 Search PubMed , in Chinese with English abstract.
- T. Chen, Y. Wu, Y. Zhang, B. Wang, Y. Hu, C. Wang and H. Jiang, PLoS One, 2012, 7, e45137 CAS.
- E. Cechova, PhD, Università di Bologna, 2009.
- M. P. Colombini and F. Modugno, Organic mass spectrometry in art and archaeology, Wiley Online Library, 2009 Search PubMed.
- ACHP, A Brick Tomb with Colorized Portrait in Deng country, Henan provice, Cultural Relic Press, Beijing, 1958, in Chinese Search PubMed.
- Y. Song, Tian Gong Kai Wu, Commercial Press, Shanghai, 1958, in Chinese Search PubMed.
- R. Ma, The colored painting of China ancient constructs, National Cultural Relic Publishing House, Beijing, 1996, in Chinese Search PubMed.
- R. Lujan, ICCROM mission report, 1988.
- J. Bostock and H. T. Riley, The natural history of Pliny, HG Bohn, London, 1855 Search PubMed.
- N. Davey, A history of building materials, Phoenix House, London, 1961 Search PubMed.
- V. Mazel, P. Richardin, D. Debois, D. Touboul, M. Cotte, A. Brunelle, P. Walter and O. Laprévote, Anal. Chem., 2007, 79, 9253–9260 CrossRef CAS PubMed.
- F. Yang, B. Zhang and Q. Ma, Acc. Chem. Res., 2010, 43, 936–944 CrossRef CAS PubMed.
- J. Pan, History of science and technology in China. Volume papermaking and printing, Science Press, Beijing, 1998, in Chinese Search PubMed.
- B. Harald, The Book and Paper Group Annual, 1991, p. 10 Search PubMed.
- Y. Zeng, B. Zhang and X. Liang, Thermochim. Acta, 2008, 473, 1–6 CrossRef CAS PubMed.
- A. Shevchenko, Y. Yang, A. Knaust, H. Thomas, H. Jiang, E. Lu, C. Wang and A. Shevchenko, J. Proteomics, 2014, 105, 363–371 CrossRef CAS PubMed.
- B. Williamson, J. Archaeol. Sci., 2000, 27, 755–762 CrossRef.
- M. H. Schweitzer, M. Marshall, K. Carron, D. S. Bohle, S. C. Busse, E. V. Arnold, D. Barnard, J. Horner and J. R. Starkey, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 6291–6296 CrossRef CAS.
- D. Gurfinkel and U. Franklin, J. Archaeol. Sci., 1988, 15, 83–97 CrossRef.
- D. Fraser, C. S. DeRoo, R. B. Cody and R. A. Armitage, Analyst, 2013, 138, 4470–4474 RSC.
- A. Högberg, K. Puseman and C. Yost, J. Archaeol. Sci., 2009, 36, 1725–1737 CrossRef PubMed.
- M. F. Seeman, N. E. Nilsson, G. L. Summers, L. L. Morris, P. J. Barans, E. Dowd and M. E. Newman, J. Archaeol. Sci., 2008, 35, 2742–2750 CrossRef PubMed.
- D. A. Scott, M. Newman, M. Schilling, M. Derrick and H. Khanjian, Archaeometry, 1996, 38, 103–112 CrossRef PubMed.
- C. Solazzo, W. W. Fitzhugh, C. Rolando and C. Tokarski, Anal. Chem., 2008, 80, 4590–4597 CrossRef CAS PubMed.
- S. Dallongeville, N. Garnier, D. Bernal Casasola, M. Bonifay, C. Rolando and C. Tokarski, Anal. Bioanal. Chem., 2011, 399, 3053–3063 CrossRef CAS PubMed.
- W. Fremout, M. Dhaenens, S. Saverwyns, J. Sanyvova, P. Vandenabeele, D. Deforce and L. Moens, Anal. Chim. Acta, 2010, 658, 156–162 CrossRef CAS PubMed.
- G. Leo, L. Cartechini, P. Pucci, A. Sgamellotti, G. Marino and L. Birolo, Anal. Bioanal. Chem., 2009, 395, 2269–2280 CrossRef CAS PubMed.
- C. Tokarski, E. Martin, C. Rolando and C. Cren-Olivé, Anal. Chem., 2006, 78, 1494–1502 CrossRef CAS PubMed.
- S. Kuckova, M. Crhova, L. Vankova, A. Hnizda, R. Hynek and M. Kodicek, Int. J. Mass Spectrom., 2009, 284, 42–46 CrossRef CAS PubMed.
- M. Buckley, M. Collins, J. Thomas-Oates and J. C. Wilson, Rapid Commun. Mass Spectrom., 2009, 23, 3843–3854 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ay01766h |
| This journal is © The Royal Society of Chemistry 2015 |