Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Towards on-site testing of Phytophthora species†
Lydia
Schwenkbier
abc,
Sibyll
Pollok
abc,
Stephan
König
d,
Matthias
Urban
a,
Sabine
Werres
*d,
Dana
Cialla-May
abc,
Karina
Weber
*abc and
Jürgen
Popp
abc
aLeibniz Institute of Photonic Technology Jena (IPHT), Jenaer BioChip Initiative, Albert-Einstein-Straße 9, 07745 Jena, Germany. E-mail: karina.weber@ipht-jena.de; Fax: +49 3641 206399; Tel: +49 3641 206309/948390
bFriedrich Schiller University Jena, Institute of Physical Chemistry and Abbe Center of Photonics, Helmholtzweg 4, 07745 Jena, Germany
cInfectoGnostics Forschungscampus Jena, Zentrum für Angewandte Forschung, Philosophenweg 7, 07743 Jena, Germany
dJulius Kuehn Institute-Federal Research Centre for Cultivated Plants (JKI), Institute for Plant Protection in Horticulture and Forests, Messeweg 11/12, 38104 Braunschweig, Germany. E-mail: sabine.werres@jki.bund.de; Fax: +49 531 2993009; Tel: +49 531 2994407
First published on 11th November 2014
Abstract
Rapid detection and accurate identification of plant pathogens in the field is an ongoing challenge. In this study, we report for the first time on the development of a helicase-dependent isothermal amplification (HDA) in combination with on-chip hybridization for the detection of selected Phytophthora species. The HDA approach allows efficient amplification of the yeast GTP-binding protein (Ypt1) target gene region at one constant temperature in a miniaturized heating device. The assay's specificity was determined by on-chip DNA hybridization and subsequent silver nanoparticle deposition. The silver deposits serve as stable endpoint signals that enable the visual as well as the electrical readout. Our promising results point to the direction of a near future on-site application of the combined techniques for a reliable detection of Phytophthora species.
Introduction
Fungal-like organisms of the genus Phytophthora belong to some of the world's most devastating plant pathogens.1Phytophthora fragariae (P. fragariae), Phytophthora kernoviae (P. kernoviae), Phytophthora ramorum (P. ramorum) and Phytophthora rubi (P. rubi) are on the EPPO A2 list of the European and Mediterranean Plant Protection Organization (EPPO) (https://www.eppo.int/QUARANTINE/listA2.htm). These species are sufficiently dangerous to recommend a regulation as quarantine organisms. The pathogen P. ramorum2 for instance is the causal agent of Sudden Oak Death in the forests of the west coast of the United States (http://www.suddenoakdeath.org) and Larix decline in the United Kingdom.3 A wide range of trees, shrubs and plants in natural and landscaped environments as well as in nursery industries can be affected by Phytophthora species. Therefore the prevention of the worldwide spread of these fungus-like plant pathogens due to an increasing trade between countries is of great importance. Thus, suitable detection methods are mandatory in order to facilitate effective screening to control and eradicate Phytophthora. In particular, rapid and reliable approaches which are inexpensive plus field applicable are needed to significantly minimize the delay between sampling and diagnosis.Common techniques for routine diagnosis of Phytophthora on the species level rely upon molecular biological, immunological or microbiological approaches.4–6 The last two methods are time consuming, laborious, and require extensive knowledge of classical taxonomy. An accurate discrimination between various Phytophthora species was successfully realized by the polymerase chain reaction (PCR).5 Nevertheless, their application in the field is hampered due to the need for thermal cycling instruments.7–12
An important step towards on-site detection of regulated Phytophthora species is provided by isothermal nucleic acid amplification techniques.7–12 Recently, several articles highlighted the loop-mediated amplification (LAMP) for a DNA-based Phytophthora specification.13–15 Here the Bst DNA polymerase amplifies the target gene region under conditions that omit the use of a thermal cycler.16–18 Although LAMP allows a convenient usage and is highly sensitive, primer designing is arduous and requires dedicated software. Moreover, an initial heat denaturation of the double-stranded template DNA prior to the isothermal amplification is often mandatory. Thus, LAMP needs a two temperature profile and cannot be claimed as really isothermal.
A further improvement of isothermal amplification that mimics in vivo DNA replication is introduced by the helicase-dependent amplification (HDA).19–22 Similar to the common PCR, the target gene region, which is enclosed by two primers, is selectively amplified. In more detail, a DNA helicase separates the double-stranded DNA and the resulting strands are immediately coated by single-stranded binding proteins (SSBs). Two sequence-specific primers bind to the template and get extended by the DNA polymerase. The newly synthesized DNA strands serve as matrices for a new amplification cycle which allows an exponential amplification.19 This approach possesses several advantages compared to other isothermal amplification methods. Firstly, a helicase unwinds and separates the double-stranded DNA; a prior heat denaturation step and subsequent thermal cycling are unnecessary. Thus, HDA can be referred to as a real isothermal technique with performance at one constant temperature for the entire process. Secondly, only one specific primer pair has to be designed. Last but not least kits are commercially available that enable a more convenient usage.22,23 These depicted properties offer promising potential towards the development of on-site detection systems for plant pathogens.19,24–26 Optimized HDA protocols have already been adapted for the detection of several bacterial pathogens like Clostridium difficile,27,28Staphylococcus aureus,23,29,30Neisseria gonorrhoeae,20,23,31,32Mycobacterium tuberculosis;33,34 as well as different viruses.35–39
Within this context, we adapted the HDA approach for Phytophthora pathogen detection for the first time. The DNA for this isothermal amplification was isolated from cultivated Phytophtora species or infected rhododendron leaves. A subsequent precise specification of the phylogenetically closely related Phytophthora species was realized by on-chip DNA hybridization.
Experimental
Phytophthora cultivation, plant samples and DNA extraction
Isolates of P. ramorum (BBA9/95), P. fragariae (BBA L1) and P. kernoviae (JKI 080-09-00-00-00-03) (culture collection of the Julius Kuehn Institute, Braunschweig, Germany) were cultivated on carrot piece agar2 with the exception of P. fragariae, which was cultivated on red kidney bean agar.40 50 mg mycelia per isolate were harvested by scraping them from the agar surface. Furthermore, the DNA of P. ramorum BBA9/95 and P. kernoviae JKI-080-09-00-00-00-03 was extracted from 100 mg of artificially infected rhododendron leaves. The mycelium and infected plant material were frozen in liquid nitrogen and ground twice for 30 s at 70% speed in a mill (Retsch, Haan, Germany).DNA extraction was performed by using the InviMag Plant DNA Mini Kit according to the recommendation of the manufacturer (Invitek, Berlin, Germany).
Thermophilic helicase-dependent amplification
We decided to exploit an asymmetric thermophilic HDA (tHDA) approach that amplifies target DNA efficiently at 65 °C and requires less protein components than the ambient temperature platform.22,35 This second-generation HDA approach led to higher specificity and sensitivity.20 For the adaption of the Phytophthora target DNA amplification41 to the HDA system we used the commercially available IsoAmp II tHDA kit from BioHelix. The IsoAmp® II Universal tHDA kit (BioHelix, Beverly, MA, USA) was utilized according to the manufacturer's recommendations. The reaction mix contained 1x annealing buffer II, 4 mM MgSO4, 40 mM NaCl, 1 μM BSA, 3.5 μl dNTPs, 75 nM biotin-labeled reverse primer, 25 nM forward primer and 3.5 μl of IsoAmp enzyme mix in a final volume of 50 μl. The primers and capture probes (Table 1; Fig. 1) were designed within the yeast GTP-binding protein (Ypt1) target region,42,43 using the program Sequencher 5.1. Primers fulfill the following criteria: (i) a length of 29–34 bp, (ii) an optimized melting temperature of 64 °C (±2 °C) and a G/C base content between 45 and 55%. Capture probes were constructed to achieve (i) the highest discrimination of target sequences in relation to non-corresponding sequences, (ii) a length of 30–35 bp and (iii) a melting temperature between 62 and 65 °C. A low tendency for sequence secondary structure formation is expressed in delta G values44 between −1 and 1.5. The primer positions are close to the capture probes because the complete amplicon should not extend more than 110 bp to ensure amplification by the Bst polymerase. The tHDA reaction was conducted asymmetrically whereas the ratio between the forward and biotin-labeled reverse primer was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.
4.
| DNA | Sequence 5′-3′ | Modification |
|---|---|---|
| HDA_frag.F | GAC CAT TGG CGT CGA CTT TGT GAG TGC TA | |
| HDA_frag.R | GCA CGA TAA CGT CAG CAA TCG GAG AGC AAA TC | 5′-Biotin |
| HDA_ram.F | CCA TCA AGC TCC AGA TTG TAC GTC TGC | |
| HDA_ram.R | GAG TAA AAT ATA GAT GTT AGC TGC ATG TCG TTG C | 5′-Biotin |
| HDA_ker.F | GGC TGC ACG AGA TCG ATA GGT GAG TTC TAC | |
| HDA_ker.R | TCT CMC AGG CGT ATC TGA TTT AAC ACG TGT TCC | 5′-Biotin |
| P. kernoviae | CAC CAC ATG AAT ACC TGC CAG GCG AGA TGC | 5′-NH2–C6 |
| P. lateralis | CGG GAG ATT TTT TCC CGC TTT CCT TGG GGT AAG | 5′-NH2–C6 |
| P. ramorum | CCC CCC ACT TTC CGT GGG TGA GTT TCC TTT | 5′-NH2–C6 |
| P. pinifolia | CCG CGG ACG AAA ACT AAC TCT CTT GTG TAG TG | 5′-NH2–C6 |
| P. fragariae | CTA GCC TTG CCA TTT CTA GGT CCA AAA AGG C | 5′-NH2–C6 |
| P. rubi | CTA GCC TTG CCA TTC CTA GGT CCA AAA AGG C | 5′-NH2–C6 |
| P. austrocedrae | CCT CCG TGG TTC ATG TAC AAA ACG TGC AGC | 5′-NH2–C6 |
| P. cambivora | GTC CAC CAT GGC TAA GTT TTG ACC TCC AGG | 5′-NH2–C6 |
| P. cinnamomi | CTG TCT GCC CCA TTC AAC AGA CGC TAA CGT C | 5′-NH2–C6 |
| Negative control I | GGA CAG GAG CGA TTC AGG ACY ATA ACA AGC AG | 5′-NH2–C6 |
| Negative control II | ATC GAG CTG GAC GGC AAG ACC ATC AAG CT | 5′-NH2–C6 |
| Positive control | AGA ATC AAG GAG CAG ATG CTG AAA AAA | 5′-NH2, 3′-biotin |
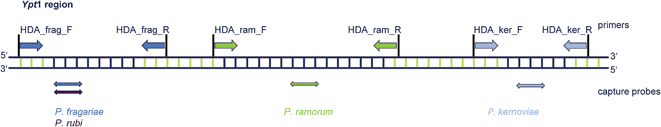 | ||
| Fig. 1 This draft shows the positions of the various primer (upper part) and capture probes (lower part) within the Ypt1 target gene sequence. | ||
The HDA reaction mixture was incubated for 90 min at 65 °C (thermophilic) in a miniaturized heating module allowing simple temperature management (Fig. 2). A Peltier heat pump element covered with a copper plate and a heat sink at the other side was used to create isothermal temperature conditions. A polycarbonate plate (thickness 4 mm) with drill-holes (diameter 4 mm, reaction volume 50 μl) was placed between these elements and sealed with a thin foil for incubating the HDA reaction mixture. The size of the heating element is 15 × 15 mm with an electric power of 8.5 Watt. The temperature was measured with a PT1000 platinum resistor thermometer pasted within the copper plate. An electronic controller used this temperature signal to generate a pulse-width modulation (PWM) signal to switch the Peltier element for holding a constant temperature of 65 °C. The operating points for this controller were set by USB connection from a PC.
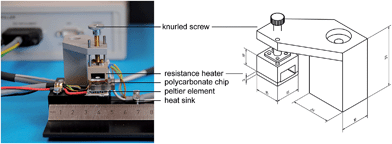 | ||
| Fig. 2 HDA reaction module consisting of the polycarbonate reaction chamber and the electronically controllable heating device. The scale bars within the technical drawing are given in mm. | ||
Agarose gel electrophoresis
Successful DNA amplification was verified on a 2% (w/v) agarose gel. For visualization the DNA was stained with GelRed (VWR International GmbH, Darmstadt, Germany) according to the recommendations of the manufacturer. The molecular weight marker ‘GeneRuler 100 bp DNA Ladder’ was purchased from Thermo Scientific.On-chip DNA hybridization
The preparation of the chips was performed as previously described.45,46 The Ypt1 region was chosen to design the species-specific capture probes41 (Fig. 1). These capture probes (Eurofins MWG Operon, Ebersberg, Germany; Table 1) were dissolved in spotting buffer (160 mM Na2SO4, 130 mM Na2HPO4) to a final concentration of 20 μM and spotted within the electrode gaps of the chip platform (Nanoplotter 2.1 GeSim, Germany; spotting layout see ESI†). A biotin-labeled non-complementary probe was immobilized as a positive control to verify successful enzyme binding via biotin–streptavidin interaction and subsequent silver deposition. After UV-linking at 254 nm for 5 min the chips were washed with 0.1 × saline-sodium citrate (SSC)/0.5% sodium dodecyl sulphate (SDS).The specific detection of Phytophthora species was performed in a microfluidic device as previously described.41,46,47 20 μl of the HDA products were dissolved in 50 μl buffer (5 × SSC/0.1% SDS) and applied on the chips for 15 min at 58 °C using an interval flow and further processed.
Optical and electrical signal readout
The amount of silver deposits was measured optically and electrically. The optical readout was realized by scanning with a reflecta ProScan 7200 slide scanner (reflecta GmbH, Rottenburg, Germany) with a 8 bit grey value and a resolution of 3600 dpi and the subsequent analysis of grey values was performed with ImageJ software (National Institutes of Health, USA). The grey value is calculated by mean grey value calculation, subtracting the measured background value from the sample values and setting the positive control to 100%. The mean grey value of the internal hybridization control (negative control I) of all experiments was used to set the threshold which is three times the standard deviation (6.41% ± 6.46%).For conductance measurement, the DC resistance is computed using an in-house developed portable chip-reader48 and converted to electrical conductance.
Results and discussion
Optimization of helicase-dependent amplification for Phytophthora species
Within the present study we explored for the first time HDA as an attractive alternative amplification method for plant pathogens. The HDA reaction was established and optimized for selected Phytophthora species (Table 1) by using a robust, miniaturized heating module combined with an existent detection platform.First, specific primers and capture probes were designed. The capture probes for P. fragariae, P. ramorum and P. kernoviae were recently published by Schwenkbier et al.41 The positions of primers and capture probes were set to amplify a region within the yeast GTP-binding protein 1 (Ypt1) gene (Fig. 1). Established isothermal LAMP-based Phytophthora detection systems used capture probes that hybridize with the internal transcribed spacer 1 region (ITS1). Due to the fact that by targeting the Ypt1 instead of the ITS1 region a higher specificity is achievable concomitant with easier adaptability to other regulated Phytophthora species, we addressed this molecular target with our chip-based amplification and detection assay. Extensive studies with the Ypt1 region from Phytophthora species showed that it is the best region to get species-specific base pairs within a length of 30–40 bp.49
Isolated genomic DNA from various Phytophthora cultures was used as a template for the amplification of specific Ypt1 target gene regions via the tHDA approach (Fig. 1). The primer pair HDA_frag. allowed the amplification of P. fragariae and P. rubi Ypt1 DNA parts. These two species differ in only one single base. And as both P. fragariae and P. rubi are on the EPPO A2 list, it is not stringently required to discriminate between those plant pathogens. The amplification of P. ramorum was realizable with the primers HDA_ram.F/R. Lastly, the primer set HDA_ker. was used to amplify a fragment of P. kernoviae within the Ypt1 gene region. A further HDA approach was conducted with genomic DNA isolated from rhododendron leaves infected with P. ramorum/kernoviae. Additionally, an asymmetric amplification strategy was chosen to generate ssDNA, which facilitates the subsequent hybridization.
The isothermal amplification was performed in a miniaturized HDA reaction module consisting of a polycarbonate plate providing reaction cavities of 50 μl and a heating device to ensure a constant temperature of 65 °C (Fig. 2). It offers several advantages including an accurate temperature control and a small size that ensure its portability. As there is a heater for both, bottom and top plus another cooling from the bottom, we achieve efficient heat conduction. Furthermore the system can be easily adapted to various chip formats and reaction volumes since the height is changeable.
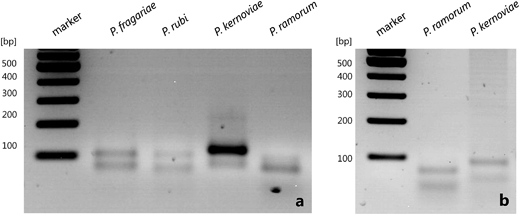
The resulting HDA products were analyzed by agarose gel electrophoresis (Fig. 3). In general the asymmetric tHDA approach led to two distinct bands in the analytical gel. The faster migrating DNA emerged single-stranded and the higher molecular weight band represented double-stranded DNA. The asymmetric tHDA reaction was successfully realized with genomic DNA isolated from cultures (Fig. 3a) as well as from infected rhododendron leaves (Fig. 3b). Thus, the newly designed HDA primer pairs allow for the successful amplification of Ypt1 target gene regions of regulated Phytophthora species by asymmetric isothermal HDA.
Phytophthora specification by on-chip hybridization
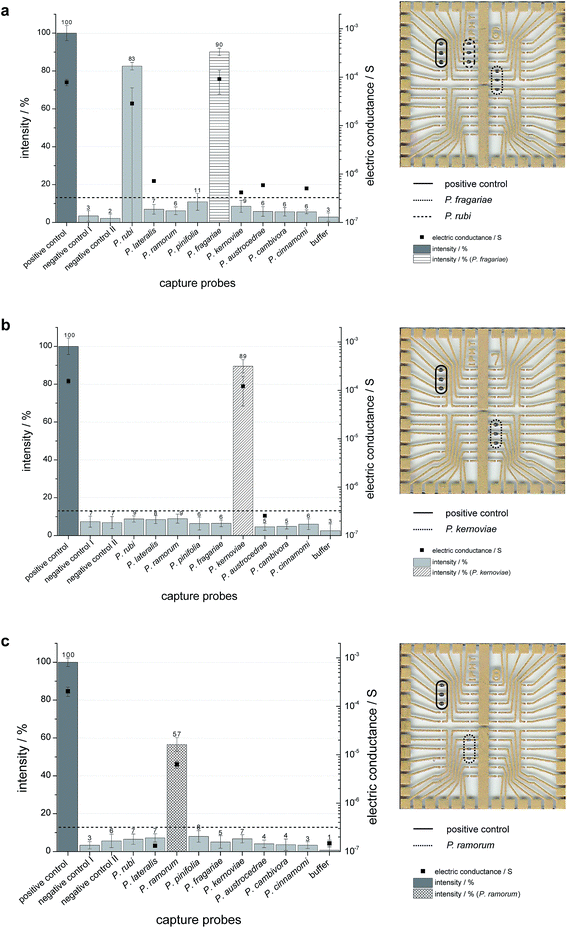
A subsequent on-chip hybridization step for proper discrimination of selected Phytophthora species concerning their Ypt1 gene region was performed. The resulting hybridization signals were detectable by the naked eye due to the formed silver deposits in the case of matching the capture and target probe. We used HDA-generated DNA from P. fragariae, P. kernoviae, P. ramorum (template DNA isolated from cultures) and P. ramorum or P. kernoviae-infected plant samples (template DNA isolated from infected rhododendron leaves) to verify the functionality of on-chip hybridization. The results of five independent experiments for each of the Phytophthora isolates are displayed in Fig. 4 (left panel: diagrams with grey values for the spotted capture probes; right panel: chips with silver deposits). Specific signals were obtained for tHDA amplicons of P. fragariae, which indicated that a sufficient amount of ssDNA specifically bound to the matching capture probes (Fig. 4a). Besides the specific signals for P. fragariae, signals for P. rubi can also be detected. This can be explained by the fact that the amplified sequence of both Phytophthora species only differs in one single base. In the case of P. kernoviae the tHDA amplicons also yielded high and specific hybridization signals (Fig. 4b). In contrast, a slightly lower but still very distinct hybridization signal was detectable for P. ramorum (Fig. 4c). These experiments were repeated independently for at least five times per species to ensure the reproducibility. Due to the necessity of short fragments for the HDA, the hybridization efficiency is great and the signals are still specific. The target DNA binds selectively to the capture probes without showing any false-positive signals. Thus, the specificity of the capture probes is illustrated.In addition to the grey value analysis an electrical detection was performed. A matching hybridization and subsequent enzyme binding result in the deposition of silver between the electrode gaps of the chip. The metallic silver enables the closure of the gap and the electric resistance on each individual spot can be measured. The conductivity signals of P. fragariae, P. kernoviae and P. ramorum reflect the results obtained by grey value analysis (Fig. 4).
The results for the infected plant samples are illustrated in Fig. 5 (left panel: diagrams with grey values for the spotted capture probes; right panel: chips with silver deposits). The hybridization signals of amplified target DNA, which was isolated from P. ramorum or P. kernoviae infected rhododendron leaves appear significantly. Thus, also the combined technology of tHDA and on-chip hybridization allowed a discrimination of Phytophthora species in real plant samples.
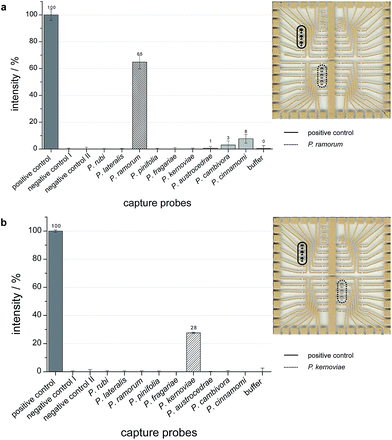 | ||
| Fig. 5 Grey values as endpoint signals for successful on-chip hybridization of HDA products from rhododendron leaves infected with P. ramorum (a) or P. kernoviae (b). | ||
Conclusions
Within this contribution, the identification of some EPPO-listed Phytophthora species by combining isothermal asymmetric tHDA with on-chip hybridization is introduced for the first time. The optical as well as electrical detection provided valid results for the analysis of the regulated plant pathogens.Isothermal amplification techniques were developed as an alternative to PCR for target gene amplification omitting the use of a thermocycler. In particular, HDA enables truly isothermal amplification without the need for prior heat denaturation or elaborated primer design, which is mandatory for LAMP. To date, no report addresses the HDA technique for the amplification of plant pathogens, in particular Phytophthora. Our developed asymmetric tHDA approach was successfully applied to amplify isolated template DNA from Phytophthora cultures and infected plant material. For an effective discrimination of several regulated Phytophthora species the Ypt1 region was chosen to design species-specific capture probes. These probes are located within a 450 bp region of the Ypt1 gene. Current HDA protocols allow the amplification of DNA fragments with a maximum length of 120 bp, hence three different primer pairs had to be designed to cover all species investigated in this study. For the establishment of our HDA experiments we started with only one primer pair per reaction.
HDA combined with chip-based detection of regulated Phytophthora species offers great potential for on-site detection. Significant improvements can allow the use of portable testing devices directly in the field or at the location, where a suspicious plant has to be investigated. This can concentrate sampling, detection as well as intervention and, thereby, reduce the delay between taking a sample of infected plants and obtaining a valid result. In order to realize a putative field application, isothermal nucleic acid amplification was optimized to substitute PCR, which requires a cost-intensive thermocycler. We demonstrated that the tHDA-based amplification as well as on-chip detection can be conducted in miniaturized and portable devices that enable on-site operating performance. The tHDA performance omits the need for thermal cycling and laborious technical requirements. Additionally, the development of disposable, low-cost chips can facilitate the near future availability of portable devices for chip-based DNA analytics. Further, the generated silver spots on the chips represent robust and long-lasting endpoint signals, which are already detectable by the naked eye. In contrast to a recently reported study based on HDA and fluorescence detection of bacterial pathogens, our colorimetric approach eliminated signal loss due to fading or expensive detection equipment. Also a conductance measurement is realizable via metallic silver, bridging the electrode gaps in the case of a matching DNA hybridization. The resulting decreased electrical resistance can be readout with our proprietary portable chip reader. Taken together, the presented results concerning an isothermal amplification and subsequent on-chip detection of Phytophthora pathogens in plant samples, realized in simple, modular, miniaturized devices, display great potential for upcoming on-site applications.
Acknowledgements
The project “PhytoChip-Validierung” (28-1-54.081-10) is supported by the Federal Ministry of Food and Agriculture (BMEL) based on a decision of the Parliament of the Federal Republic of Germany via the Federal Office for Agriculture and Food (BLE) under the innovation support program. The heating system was provided by the LungCARD project. The project “JBCI 2.0” (InnoProfile-Transfer, Unternehmen Region), supported by the Federal Ministry of Education and Research (BMBF) Germany, is gratefully acknowledged.References
- D. C. Erwin and O. K. Ribeiro, Phytophthora Diseases Worldwide, APS Press, St. Paul, Minn., 1996 Search PubMed.
- S. Werres, R. Marwitz, W. Veld, A. De Cock, P. J. M. Bonants, M. De Weerdt, K. Themann, E. Ilieva and R. P. Baayen, Mycol. Res., 2001, 105, 1155–1165 CrossRef CAS.
- C. Brasier and J. Webber, Nature, 2010, 466, 824–825 CrossRef CAS PubMed.
- O. Lazcka, F. J. Del Campo and F. X. Munoz, Biosens. Bioelectron., 2007, 22, 1205–1217 CrossRef CAS PubMed.
- P. A. O'Brien, N. Williams and G. E. S. Hardy, Crit. Rev. Microbiol., 2009, 35, 169–181 CrossRef PubMed.
- F. N. Martin, Z. G. Abed, Y. Baldi and K. Ivors, Plant Dis., 2012, 96, 1080–1103 CrossRef.
- P. Craw and W. Balachandran, Lab Chip, 2012, 12, 2469–2486 RSC.
- C.-C. Chang, C.-C. Chen, S.-C. Wei, H.-H. Lu, Y.-H. Liang and C.-W. Lin, Sensors, 2012, 12, 8319–8337 CrossRef CAS PubMed.
- P. J. Asiello and A. J. Baeumner, Lab Chip, 2011, 11, 1420–1430 RSC.
- J. Kim and C. J. Easley, Bioanalysis, 2011, 3, 227–239 CrossRef CAS PubMed.
- F. Sidoti, M. Bergallo, C. Costa and R. Cavallo, Mol. Biotechnol., 2013, 53, 352–362 CrossRef CAS PubMed.
- P. Gill and A. Ghaemi, Nucleosides, Nucleotides Nucleic Acids, 2008, 27, 224–243 CAS.
- J. A. Tomlinson, I. Barker and N. Boonham, Appl. Environ. Microbiol., 2007, 73, 4040–4047 CrossRef CAS PubMed.
- J. A. Tomlinson, M. J. Dickinson and N. Boonham, Phytopathology, 2010, 100, 143–149 CrossRef CAS PubMed.
- T.-T. Dai, C.-C. Lu, J. Lu, S. Dong, W. Ye, Y. Wang and X. Zheng, FEMS Microbiol. Lett., 2012, 334, 27–34 CrossRef CAS PubMed.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, e63 CrossRef CAS PubMed.
- Y. Mori, K. Nagamine, N. Tomita and T. Notomi, Biochem. Biophys. Res. Commun., 2001, 289, 150–154 CrossRef CAS PubMed.
- Y. Mori, M. Kitao, N. Tomita and T. Notomi, J. Biochem. Biophys. Methods, 2004, 59, 145–157 CrossRef CAS PubMed.
- M. Vincent, Y. Xu and H. M. Kong, EMBO Rep., 2004, 5, 795–800 CrossRef CAS PubMed.
- L. X. An, W. Tang, T. A. Ranalli, H. J. Kim, J. Wytiaz and H. M. Kong, J. Biol. Chem., 2005, 280, 28952–28958 CrossRef CAS PubMed.
- A. Motre, Y. Li and H. Kong, Gene, 2008, 420, 17–22 CrossRef CAS PubMed.
- Y.-J. Jeong, K. Park and D.-E. Kim, Cell. Mol. Life Sci., 2009, 66, 3325–3336 CrossRef CAS PubMed.
- D. Andresen, M. von Nickisch-Rosenegk and F. F. Bier, Clin. Chim. Acta, 2009, 403, 244–248 CrossRef CAS PubMed.
- D. Andresen, M. von Nickisch-Rosenegk and F. F. Bier, Expert Rev. Mol. Diagn., 2009, 9, 645–650 CrossRef CAS PubMed.
- M. Mahalanabis, J. do, H. Almuayad, J. Y. Zhang and C. M. Klapperich, Biomed. Microdevices, 2010, 12, 353–359 CrossRef CAS PubMed.
- N. Ramalingam, T. C. San, T. J. Kai, M. Y. M. Mak and H.-Q. Gong, Microfluid. Nanofluid., 2009, 7, 325–336 CrossRef CAS.
- W. H. A. Chow, C. McCloskey, Y. Tong, L. Hu, Q. You, C. P. Kelly, H. Kong, Y.-W. Tang and W. Tang, J. Mol. Diagn., 2008, 10, 452–458 CrossRef CAS PubMed.
- S. Huang, J. Do, M. Mahalanabis, A. Fan, L. Zhao, L. Jepeal, S. K. Singh and C. M. Klapperich, PLoS One, 2013, 8, e60059 CAS.
- J. Goldmeyer, H. Li, M. McCormac, S. Cook, C. Stratton, B. Lemieux, F. Kong, W. Tang and Y.-W. Tang, J. Clin. Microbiol., 2008, 46, 1534–1536 CrossRef CAS PubMed.
- G. C. Frech, D. Munns, R. D. Jenison and B. J. Hicke, BMC Res. Notes, 2012, 5, 430 CrossRef PubMed.
- V. Doseeva, T. Forbes, J. Wolff, Y. Khripin, D. O'Neil, T. Rothmann and I. Nazarenko, Diagn. Microbiol. Infect. Dis., 2011, 71, 354–365 CrossRef CAS PubMed.
- Y. Tong, B. Lemieux and H. Kong, BMC Biotechnol., 2011, 11, 50 CrossRef CAS PubMed.
- E. Torres-Chavolla and E. C. Alocilja, Biosens. Bioelectron., 2011, 26, 4614–4618 CrossRef CAS PubMed.
- A. Motre, R. Kong and Y. Li, J. Microbiol. Methods, 2011, 84, 343–345 CrossRef CAS PubMed.
- J. Goldmeyer, H. Kong and W. Tang, J. Mol. Diagn., 2007, 9, 639–644 CrossRef CAS PubMed.
- J. A. Jordan, C. O. Ibe, M. S. Moore, C. Host and G. L. Simon, J. Clin. Virol., 2012, 54, 11–14 CrossRef CAS PubMed.
- C. Domingo, P. Patel, J. Yillah, M. Weidmann, J. A. Mendez, E. R. Nakoune and M. Niedrig, J. Clin. Microbiol., 2012, 50, 4054–4060 CrossRef PubMed.
- H.-J. Kim, Y. Tong, W. Tang, L. Quimson, V. A. Cope, X. Pan, A. Motre, R. Kong, J. Hong, D. Kohn, N. S. Miller, M. D. Poulter, H. Kong, Y.-W. Tang and B. Yen-Lieberman, J. Clin. Virol., 2011, 50, 26–30 CrossRef CAS PubMed.
- N. S. Miller, B. Yen-Lieberman, M. D. Poulter, Y.-W. Tang and P. A. Granato, J. Clin. Virol., 2012, 54, 355–358 CrossRef PubMed.
- S. Werres and R. Casper, J. Phytopathol., 1987, 118, 367–369 CrossRef.
- L. Schwenkbier, S. König, S. Wagner, S. Pollok, J. Weber, M. Hentschel, J. Popp, S. Werres and K. Weber, Microchim. Acta, 2013, 180, 15–16 CrossRef.
- L. Schena and D. E. L. Cooke, J. Microbiol. Methods, 2006, 67, 70–85 CrossRef CAS PubMed.
- L. Schena, J. M. Duncan and D. E. L. Cooke, Plant Pathol., 2008, 57, 64–75 CAS.
- M. Zuker, Nucleic Acids Res., 2003, 31, 3406–3415 CrossRef CAS PubMed.
- T. Schueler, R. Kretschmer, S. Jessing, M. Urban, W. Fritzsche, R. Moeller and J. Popp, Biosens. Bioelectron., 2009, 25, 15–21 CrossRef CAS PubMed.
- B. Seise, A. Brinker, R. Kretschmer, M. Schwarz, B. Rudolph, T. Kaulfuss, M. Urban, T. Henkel, J. Popp and R. Moeller, Eng. Life Sci., 2011, 11, 148–156 CrossRef CAS.
- S. Wuenscher, B. Seise, D. Pretzel, S. Pollok, J. Perelaer, K. Weber, J. Popp and U. S. Schubert, Lab Chip, 2014, 14, 392–401 RSC.
- M. Urban, R. Moller and W. Fritzsche, Rev. Sci. Instrum., 2003, 74, 1077–1081 CrossRef CAS.
- S. König, L. Schwenkbier, M. Riedel, S. Wagner, S. Pollok, J. Popp, K. Weber and S. Werres, Plant Pathol., 2014 Search PubMed , submitted.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ay02287d |
| This journal is © The Royal Society of Chemistry 2015 |