A novel fluorescence assay for inorganic pyrophosphatase based on modulated aggregation of graphene quantum dots†
Xueli
Zhu
,
Jinwen
Liu
,
Haiyang
Peng
,
Jianhui
Jiang
* and
Ruqin
Yu
*
College of Chemistry and Chemical Engineering, Hunan University, State Key Laboratory for Chemo/Biosensing and Chemometrics, Changsha, Hunan, China. E-mail: rqyu@hnu.edu.cn; jianhuijiang@hnu.edu.cn
First published on 30th October 2015
Abstract
A simple and highly sensitive fluorometric method has been developed for inorganic pyrophosphatase (PPase) activity detection based on the disaggregation and aggregation of graphene quantum dots (GQDs). Copper ions can trigger the severe aggregation of GQDs with rich carboxyl groups, which results in effective fluorescence quenching. While, with the addition of pyrophosphate (PPi), the quenched fluorescence is effectively recovered owing to the strong interaction between PPi and Cu2+. Furthermore, under the catalytic hydrolysis of PPase, the complex of PPi–Cu2+–PPi is rapidly disassembled, and the fluorescence is re-quenched. This method is highly sensitive and selective for PPase detection, with a linear correlation between the fluorescence intensity and the PPase concentration in the range from 1 to 200 mU mL−1 with a detection limit down to 1 mU mL−1 (S/N = 3). Additionally, the inhibition effect of NaF on the PPase activity is also studied. Thus, the proposed method may hold a potential application in the diagnosis of PPase-related diseases and screening of PPase inhibitors, to evaluate the function and inhibition of PPase in biological systems.
Introduction
Pyrophosphate (PPi) is one of the most important biological anions, which is the product of adenosine triphosphate (ATP) hydrolysis under cellular conditions,1 playing an important role in pathological, energetic and metabolic progress.2 Soluble inorganic pyrophosphatase (EC 3.6.1.1; PPase) is a ubiquitous enzyme that can provide phosphate substrates and energy for biosynthesis reactions such as RNA, DNA and protein syntheses by hydrolyzing PPi,3 which also plays a vital role in controlling the level of PPi in cells.4 PPase has been proved to be relevant to evolutionary events, phosphorus metabolism and carbohydrate metabolism, which is of vital importance for cell growth and differentiation.5 Furthermore, the level of PPase is connected to several diseases, such as hyperthyroidism, lung adenocarcinomas and colorectal cancers.6 Thus, the analysis of PPase activity is of fundamental importance and significance. Additionally, PPases are strong metal-dependent enzymes,7 they need divalent ions such as Mg2+, Mn2+, Zn2+, Co2+ and Cd2+ for catalytic reactions, with Mg2+ conferring the highest activity.4a,c,5c,8 Up to now, several assays such as colorimetric,9 enzymatic10 and fluorometric11 have been developed for the detection of PPase activity. However, it has shown that these methods suffer from their own drawbacks such as time-consuming, high cost, sophisticated instrumentation, demand of specialized skills, high detection limit and poor stability in aqueous solution for noble metal nanoclusters etc. Thus, a convenient, simple yet effective method for monitoring the PPase activity is still urgently needed.In theoretical and experimental studies, graphene quantum dots (GQDs) whose diameters are less than 10 nm can exhibit quantum-confinement effects, and thus, wavelength-dependent and photobleaching-resistant photo-luminescence can be observed.12 GQDs, as a class of zero-dimensional graphitic nanomaterials, display special optical and electrical phenomena compared with other kinds of quantum dots.13 With regard to conventional dye molecules and semiconduction quantum dots, they have many advantages such as good photostability, low toxicity, excellent water solubility, easy modulation, excellent biocompatibility, and high electrical and thermal conductivity, which make them promising candidates for future applications in such fields like fluorescent probes, optoelectronic devices, bioimaging and photocatalysts.13a–c,14 Based on these superiorities, a multi-function fluorescence sensing platform was developed to achieve selective detection of DNA targets,14g as well as H2O2, glucose,14i,k metal ions,14a,e and proteases.14f
Herein, we report the development of a novel fluorometric method for highly sensitive detection of PPase using the GOD nanoprobe. This method relies on our new finding that carboxyl-modified GQDs can be assembled by coordination with Cu2+ into large aggregates, which can be reversely disaggregated into GQDs by the addition of PPi. The reversible modulation of fluorescence signal by Cu2+ and PPi in the aggregation and disaggregation enables the sensitive detection of PPase, as illustrated in Scheme 1. In the presence of Cu2+, GQDs are assembled into aggregates due to the crosslinked coordination between Cu2+ and the carboxyl group, which results in the substantial fluorescence quenching because of self-quenching between the GQDs. After the addition of PPi, the competition of PPi with GODs for Cu2+ due to the strong interaction between PPi and Cu2+ induces disaggregation of GODs, recovering the fluorescence of GODs. The presence of active PPase catalyzes the hydrolysis of PPi into phosphate, which releases Cu2+ and, in turn, mediates re-aggregation of GQDs with a quenched fluorescence signal. Therefore, the modulation of fluorescence in response to PPase gives a quantitative measure for the assay of activity or inhibition of PPase.
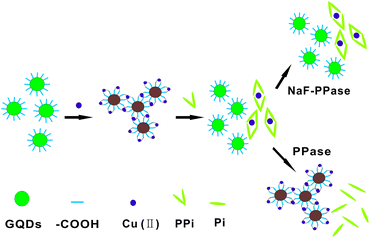 | ||
| Scheme 1 Illustration of the biosensing strategy for the detection of PPase activity based on aggregation and disaggregation of graphene quantum dots. | ||
Experimental
Chemicals and materials
Baker's yeast inorganic PPase (EC 3.6.1.1, 100 U) was purchased from Sigma-Aldrich (Shanghai). One unit of Baker's yeast inorganic PPase can liberate 1.0 μmol of inorganic orthophosphate per minute at pH = 7.2 at 25 °C. Sodium pyrophosphate (PPi) was acquired from Shanghai Sangon Biotechnology Co. Ltd (Shanghai). HEPES was obtained from HeFei BoMei Biotechnology Co. Ltd (Hefei). MgCl2·6H2O, Cu2SO4·5H2O and NaF were all bought from Sinopharm Chemical Reagent Co. Ltd (Shanghai). All other regents used in this work were at least analytical grade and directly used without any further purification. The solutions were prepared with ultrapure water (resistance >18.2 MΩ) that was obtained from a Milli-Q water purification system (Billerica, MA).The fluorescence emission spectra of GQDs were obtained from F-7000 and Fluoromax-4 spectrofluorometers. The UV-vis absorption was recorded on a UV-2450 spectrometer. Transmission electron microscopy (TEM) images were obtained from JEM-2100F with an accelerating voltage of 200 kV. Zeta potential and dynamic light scattering (DLS) analysis were performed by Malvern Zetasizer Nano ZS90.
Fluorescence PPase activity assay
Different activity units of PPase, ranging from 0 to 600 mU mL−1, were separately added to the mixed solution that contains GQDs, Cu2+ (100 μM), Mg2+ (0.1 μM), PPi (160 μM) and HEPES buffer (10 mM), the final volume was 100 μL. After an incubation time of 60 min in a water bath at 37 °C, the fluorescence spectra of the mixture were recorded.Inhibition efficiency of F− on PPase activity
For evaluating the inhibition efficiency of F− on PPase activity, we first separately added PPase (200 mU mL−1) to the aqueous solution of NaF with different concentrations. After 15 min at room temperature, the NaF-treated PPase was added to the mixed solution containing GODs, Cu2+ (100 μM), Mg2+ (0.1 μM), PPi (160 μM) and HEPES buffer (10 mM), the final volume was 100 μL. After another 60 min in a water bath at 37 °C, the fluorescence spectra of the mixture were obtained.Results and discussion
Mechanism of PPase activity detection
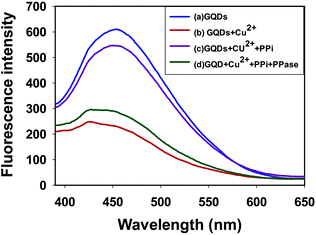
Fluorescent carboxyl-modified GQDs can be facilely synthesized and purified based on the previous procedure.13e The fluorescence emission spectrum of the GQDs showed a peak at 449 nm, while the excitation spectrum exhibited a peak at 368 nm (Fig. 1, blue curve, and Fig. S1 in the ESI†). The fluorescence quantum yield of GQDs was determined to be 5.58% by comparing the wavelength integrated intensity of the samples to that of standard quinine sulfate (ΦST = 0.577) (Fig. S2 in the ESI†). After the addition of Cu2+, appreciable quenching of the fluorescence was observed (Fig. 1, red curve). It is reasonable to assume that the quenching mechanism should be the coordination of Cu2+ with the carboxyl group on the surface of GQDs, which eventually leads to the aggregation of GQDs. We can also prove the proposed mechanism of aggregation by comparing the TEM image of Cu2+–GQD complex with that of GQDs. We observed that the GQDs were well-dispersed and gave a diameter of ∼3 nm. In contrast, the Cu2+–GQD complex was found to form clear aggregates with a diameter of ∼20 nm (Fig. 2A and B). This observation gave an immediate evidence for the induced assembly of GQDs into large aggregates triggered by Cu2+. Further evidence from zeta potential analysis revealed that the zeta potential of the Cu2+–GQD complex showed a positive shift in comparison with that of GQDs, indicating the formation of Cu2+–GQD complex (Fig. S3 in the ESI†).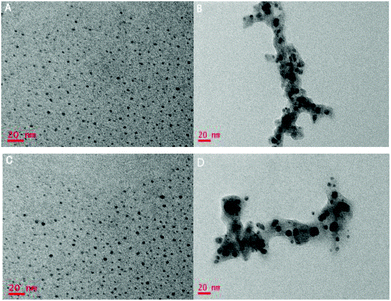 | ||
| Fig. 2 (A)TEM images of single GQDs, (B) Cu2+–GQD complex, (C) re-disaggregated GQDs and PPi–Cu2+ complexes, and (D) re-aggregated GQDs with copper ion complexes in the presence of PPi and PPase. | ||
On adding PPi into the solution of Cu2+–GQD complex, the fluorescence spectrum was found to be recovered up to 90% (Fig. 1, purple curve). This was ascribed to the disaggregation of the Cu2+–GQD complex into free GQDs, because the binding affinity between Cu2+ and PPi was reported to be higher than that between Cu2+ and the carboxyl group.11b,15 The TEM image in Fig. 2C manifests that most of the GODs were restored to monodispersity in this case, confirming that PPi mediated disaggregation of Cu2+–GQD complex into GQDs. After PPase was incubated for 30 minutes with the solution containing Cu2+, PPi and GQDs, the fluorescence was quenched again (Fig. 1, green curve). This result was attributed to the hydrolysis of PPi into Pi catalyzed by PPase. Because Pi had a much weaker interaction with Cu2+, the coordination between Cu2+ and GQDs became dominant, which caused re-aggregation of GQDs with quenched fluorescence. The TEM image in Fig. 2D also manifests that most of the GODs were restored to monodispersity in this case, testifying the presumed mechanism for our assay.
Optimization of the experimental conditions
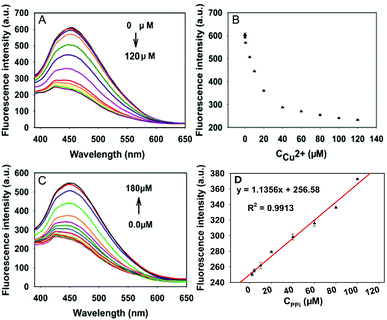
We first studied the effect of Cu2+ concentration on the sensing system. As shown in Fig. 3A and B, different concentrations of Cu2+ were mixed into the GQD aqueous solution with the incubation time of 30 min, leading to gradual quenching of the GQD fluorescence. When it reached 100 μM, the fluorescence of the mixture solution changed little with further increase of Cu2+ concentration, thus, the Cu2+ concentration of 100 μM was adopted in the following experiments.Fig. 3C exhibits the fluorescence spectra of the GQD solution containing 100 μM Cu2+ and different concentrations of PPi from 0 to 180 μM, as we can see in Fig. S4 (ESI†), 30 min later, the addition of 160 μM PPi made the fluorescence intensity close to the initial intensity, and further addition caused a little change in it. Moreover, the fitted linear data between fluorescence intensity and concentrations of PPi in the range of 2–100 μM (Fig. 3D), and the equation can be expressed as y = 1.1356x + 256.58, R2 = 0.9913. This strategy can also be used to detect PPi.
Sensitivity and selectivity
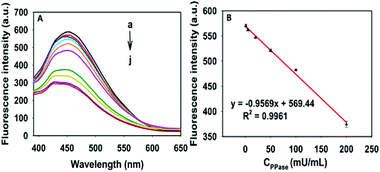
Under the optimized experimental conditions, the activity of PPase was tested in the GQD aqueous solution containing Cu2+ (100 μM), Mg2+ (0.1 μM), PPi (160 μM) and HEPES buffer (10 mM). As displayed in Fig. 4A and S5 (ESI†), the mixed solution was respectively added to different activity units of PPase in the range of 0–600 mU mL−1, 60 min later in a water bath at 37 °C, the decrease of fluorescence intensity was recorded, and there is a linear correlation between the fluorescence intensity and the concentration of PPase in the range of 1–200 mU mL−1. The linear equation can be expressed as y = −0.9569x + 569.44, R2 = 0.9961 (Fig. 5B), and the limit of detection (LOD) based on 3σ/s was calculated to be about 1 mU mL−1.The developed assay was compared with literature reports. From Table S1 (ESI†), it can be seen that our method exhibits comparable or even better performances than the published methods.
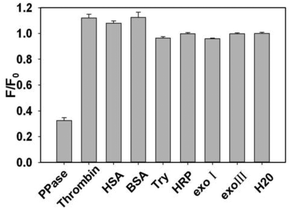
Before applying the proposed biosensor for real sample analysis, we investigated its selectivity comparing with several representative interfering proteins such as thrombin, human serum albumin (HAS), bovine serum albumin (BSA), trypsin (Try), horseradish peroxidase (HRP), exonuclease I and exonuclease II. As displayed in Fig. 5, the relative fluorescence intensities of these proteins did not decrease evidently in the absence of PPase, although their concentrations are much higher than PPase. The result indicated that these representative interfering proteins could not influence the quenching effect of PPase assay, that is to say, the proposed assay shows superior selectivity for PPase activity detection, which is promising to be applied in real biological samples.
Inhibition study
According to recent crystallographic results, the fluoride ion is a forceful and specific inhibitor of cytoplasmic pyrophosphatase due to the formation of the F−–PPase intermediate which results in an instant decrease in enzyme activity.7a,10b,11a,16 To attest this result further, PPase with a final concentration of 200 mU mL−1 was separately added into the aqueous solution of NaF with different concentrations (i.e., 0, 0.1, 0.5, 1, 5, 50, 200, 400, 500 μM), and incubated for 15 min at room temperature. Then different concentrations of NaF-treated PPase were respectively mixed into the solution containing diluted GQDs, Cu2+ (100 μM), Mg2+ (0.1 μM), PPi (160 μM) and HEPES buffer (10 mM), and incubated for another 60 min at 37 °C. As shown in Fig. 6, the fluorescence intensity of the system increased gradually with the increase of NaF concentrations. Furthermore, the IC50 (the half maximal inhibitory concentration) value of 200 mU mL−1 PPase was calculated to be about 7.730 μM, which is in accord with the previous report.9a These results suggest that the proposed method can also be applied to evaluate the inhibition efficiency of PPase.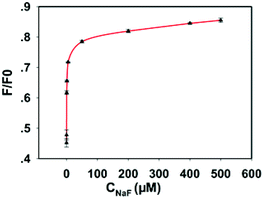 | ||
| Fig. 6 Effect of NaF inhibition of PPase activity by measuring the fluorescence intensity of the GQDs in 10 mM HEPES buffer at pH 7.2. | ||
Conclusions
In summary, we have reported a new finding on the aggregation and disaggregation of GQDs mediated by Cu2+ and its interacting anion PPi, which enables the development of a simple and highly sensitive fluorometric method for PPase activity and inhibition assay. To our knowledge, the proposed method is the first analytical system based on a fluorescence GQD sensor that can quantitatively, selectively and sensitively detect PPase activity with a detection limit of 1 mU mL−1. This method can also be applied to screening inhibitors of PPase. In virtue of these advantages, the developed method may have great potential for the ascertainment of PPase activity and its potential inhibitors for basic biology and clinical diagnostics for PPase-related diseases and PPase-targeted drug discovery.Acknowledgements
This work was supported by NSFC (21527810, 21190041, 21221003) and National Key Basic Research Program (2011CB911000).Notes and references
- (a) S. J. Yang, G. Q. Feng and N. H. Williams, Org. Biomol. Chem., 2012, 10, 5606 RSC; (b) B. B. Liu, X. M. Li, R. J. Gao, W. H. Zhou, G. Q. Xie, M. Bartlam, H. Pang, Y. Feng and Z. H. Rao, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 577 CrossRef PubMed.
- (a) J. J. Deng, P. Yu, L. F. Yang and L. Q. Mao, Anal. Chem., 2013, 85, 2516 CrossRef CAS PubMed; (b) W. X. Yu, J. Qiang, J. Yin, S. Kambam, F. Wang, Y. Wang and X. Q. Chen, Org. Lett., 2014, 16, 2220 CrossRef CAS PubMed; (c) R. Selegård, K. Enander and D. Aili, Nanoscale, 2014, 6, 14204 RSC.
- (a) L. Wang, L. Yuan, H. W. Wang, X. L. Liu, X. M. Li and H. Chen, Bioconjugate Chem., 2014, 25, 1252 CrossRef PubMed; (b) X. Li, L. Wang, G. J. Chen, D. M. Haddleton and H. Chen, Chem. Commun., 2014, 50, 6506 RSC; (c) M. K. Islam, T. Miyoshi, H. K. Aoki, T. Isibe, T. Arakawa, Y. Matsumoto and N. Tsuji, Eur. J. Biochem., 2003, 270, 2814 CrossRef CAS PubMed; (d) A. Ramos, S. A. I. Adham and J. A. Gil, FEMS Microbiol. Lett., 2003, 225, 85 CrossRef CAS PubMed.
- (a) T. C. Chao, H. M. Huang, J. Y. Tsai, C. Y. Huang and Y. J. Sun, Proteins: Struct., Funct., Bioinf., 2006, 65, 670 CrossRef CAS PubMed; (b) H. S. Lee, Y. Cho, Y. J. Kim, T. O. Lho, S. S. Cha, J. H. Lee and S. G. Kang, FEMS Microbiol. Lett., 2009, 300, 68 CrossRef CAS; (c) E. H. Harutyunyan, I. P. Kuranova, B. K. Vainshtein, W. E. HÖHNE, V. S. Lamzin, Z. Dauter, A. V. Teplyakov and K. S. Wilson, Eur. J. Biochem., 1996, 239, 220 CAS.
- (a) M. Ilias and T. W. Young, Biochim. Biophys. Acta, 2006, 1764, 1299 CrossRef CAS; (b) E. Kukko and J. Heinonen, Eur. J. Biochem., 1982, 127, 347 CrossRef CAS; (c) A. Teplyakov, G. Obmolova, K. S. Wilson, K. Ishii, H. Kaji, T. Samejima and I. Kuranova, Protein Sci., 1994, 3, 1098 CrossRef CAS; (d) T. S. Sitnik, J. P. Vainonen, E. V. Rodina, T. I. Nazarova, S. A. Kurilova, N. N. Vorobyeva and S. M. Avaeva, IUBMB Life, 2003, 55, 37 CrossRef CAS.
- (a) E. Koike, S. Toda, F. Yokoi, K. Izuhara, N. Koike, K. Itoh, K. Miyazaki and H. Sugihara, Biochem. Biophys. Res. Commun., 2006, 341, 691 CrossRef CAS PubMed; (b) Z. H. Lu, L. P. Hu, S. Evers, J. Chen and Y. Shen, Proteomics, 2004, 4, 3975 CrossRef CAS PubMed; (c) D. B. Fredman, S. Hill, J. W. Keller, N. B. Merchant, S. E. Levy, R. J. Coffey and R. M. Caprioli, Proteomics, 2004, 4, 793 CrossRef PubMed.
- (a) A. A. Baykov, I. P. Fabrichniy, P. Pohjanjoki, A. B. Zyryanov and R. Lahti, Biochemistry, 2000, 39, 11939 CrossRef CAS PubMed; (b) C. A. Wu, N. K. Lokanath, D. Y. Kim, H. J. Park, H. Y. Hwang, S. T. Kim, S. W. Suh and K. K. Kim, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2005, 61, 1459 CrossRef PubMed; (c) V. A. Sklyankina and S. M. Avaeva, Eur. J. Biochem., 1990, 191, 195 CrossRef CAS PubMed.
- (a) D. S. Cooerman, Phosphorus Chem., 1981, 23, 119 Search PubMed; (b) P. Heikinheimo, P. Pohjanjoki, A. Helminen, M. Tasanen, B. S. Copperman, A. Goldman, A. Baykov and R. Lahti, Eur. J. Biochem., 1996, 239, 138 CAS; (c) T. H. Liu, S. H. Jsu, Y. T. Huang, S. M. Lin, T. W. Huang, T. H. Chuang, S. K. Fan, C. C. Fu, F. G. Tseng and R. L. Pan, FEBS J., 2009, 276, 4381 CrossRef CAS PubMed; (d) E. V. Rodina, Y. P. Vainonen, N. N. Vorobyeva, S. A. Kurilova, T. I. Nazarova and S. M. Avaeva, Eur. J. Biochem., 2001, 268, 3851 CrossRef CAS PubMed.
- (a) J. J. Deng, Q. Jiang, Y. X. Wang, L. F. Yang, P. Yu and L. Q. Mao, Anal. Chem., 2013, 85, 9409 CrossRef CAS; (b) L. L. Zhang, M. Li, Y. F. Qin, Z. D. Chu and S. L. Zhao, Analyst, 2014, 139, 6298 RSC; (c) J. K. Heinonen and R. J. Lahti, Anal. Biochem., 1981, 113, 313 CrossRef CAS PubMed; (d) A. A. Baykov, V. N. Kasho and S. M. Avaea, Anal. Biochem., 1988, 171, 271 CrossRef CAS PubMed.
- (a) T. Biswas, E. Resto-Roldán, S. K. Sawyer, I. Artsimovitch and O. V. Tsodikov, Nucleic Acids Res., 2013, 41, 1 CrossRef PubMed; (b) J. Eriksson, S. Karamohamed and P. Nyrén, Anal. Biochem., 2001, 293, 67 CrossRef CAS PubMed.
- (a) K. F. Xu, Z. G. Chen, L. Zhou, O. Zheng, X. P. Wu, L. H. Guo, B. Qiu, Z. Y. Lin and G. N. Chen, Anal. Chem., 2015, 87, 816 CrossRef CAS PubMed; (b) J. Sun, F. Yang, D. Zhao and X. R. Yang, Anal. Chem., 2014, 86, 7883 CrossRef CAS PubMed.
- (a) Y. Wang, L. Zhang, R. P. Liang, J. M. Bai and J. D. Qiu, Anal. Chem., 2013, 85, 9148 CrossRef CAS.
- (a) L. P. Lin, M. C. Rong, F. Luo, D. M. Chen, Y. R. Wang and X. Chen, Trends Anal. Chem., 2014, 54, 83 CrossRef CAS; (b) X. T. Zheng, A. Ananthanarayanan, K. Q. Luo and P. Chen, Small, 2015, 11, 1620 CrossRef CAS PubMed; (c) C. Y. Wu, C. Wang, T. Han, X. J. Zhou, S. W. Guo and J. Y. Zhang, Adv. Healthcare Mater., 2013, 2, 1613 CrossRef CAS PubMed; (d) Z. K. Huang, J. F. Qu, X. G. Peng, W. L. Liu, K. W. Zhang, X. L. Wei and J. X. Zhong, Phys. Status Solidi RRL, 2014, 8, 436 CrossRef CAS; (e) Y. Q. Dong, G. L. Li, N. N. Zhou, R. X. Wang, Y. C. Wu and G. N. Chen, Anal. Chem., 2012, 84, 8378 CrossRef CAS PubMed.
- (a) F. X. Wang, Z. Y. Gu, W. Lei, W. J. Wang, X. F. Xia and Q. L. Hao, Sens. Actuator, B, 2014, 190, 516 CrossRef CAS; (b) Z. S. Qian, X. Y. Shan, L. J. Chai, J. J. Ma, J. R. Chen and H. Feng, Biosens. Bioelectron., 2014, 60, 64 CrossRef CAS PubMed; (c) X. M. Li, M. C. Rui, J. Z. Song, Z. C. Shen and H. B. Zeng, Adv. Funct. Mater., 2015, 25, 4929 CrossRef CAS; (d) H. J. Sun, A. D. Zhao, N. Gao, K. Li, J. S. Ren and X. G. Qu, Angew. Chem., Int. Ed., 2015, 54, 7176 CrossRef CAS PubMed; (e) Z. B. Qu, M. Zhang, T. S. Zhou and G. Y. Shi, Chem. – Eur. J., 2014, 20, 13777 CrossRef CAS PubMed; (f) Y. Y. Lin, R. Chapman and M. M. Stevens, Adv. Funct. Mater., 2015, 25, 3183 CrossRef CAS; (g) Z. S. Qian, X. Y. Shan, L. J. Chai, J. R. Chen and H. Feng, Chem. – Eur. J., 2014, 20, 16065 CrossRef CAS; (h) X. H. Zhu, X. Xiao, X. X. Zuo, Y. Liang and J. M. Nan, Part. Part. Syst. Char., 2014, 31, 801 CrossRef CAS; (i) L. Zhang, D. Peng, R. P. Liang and J. D. Qiu, Chem. – Eur. J., 2015, 21, 9343 CrossRef CAS PubMed; (j) D. Wang, J. F. Chen and L. M. Dai, Part. Part. Syst. Char., 2015, 32, 515 CrossRef CAS; (k) L. Zhou, J. L. Geng and B. Liu, Part. Part. Syst. Char., 2013, 30, 1086 CrossRef CAS; (l) D. Chakravarty, M. B. Erande and D. J. Late, J. Sci. Food Agric., 2015, 20, 46 Search PubMed.
- (a) J. B. English, A. E. Martell, R. J. Motekaitis and I. Murase, Inorg. Chim. Acta, 1997, 258, 183 CrossRef CAS.
- (a) P. Halonen, A. A. Baykov, A. Goldman, R. Lahti and B. S. Cooperman, Biochemistry, 2002, 41, 12025 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5an01937k |
| This journal is © The Royal Society of Chemistry 2016 |