Detection of the tuberculosis antigenic marker mannose-capped lipoarabinomannan in pretreated serum by surface-enhanced Raman scattering
Alexis C.
Crawford
ab,
Lars B.
Laurentius
b,
Timothy S.
Mulvihill
a,
Jennifer H.
Granger
b,
John S.
Spencer
c,
Delphi
Chatterjee
c,
Kimberly E.
Hanson
d and
Marc D.
Porter
*abefg
aDepartment of Chemistry, University of Utah, Salt Lake City, UT 84112, USA
bNano Institute of Utah, University of Utah, Salt Lake City, UT 84112, USA. E-mail: marc.porter@utah.edu
cDepartment of Microbiology, Immunology, and Pathology, Colorado State University, Fort Collins, CO 80523, USA
dDepartments of Internal Medicine and Pathology, University of Utah School of Medicine, Salt Lake City, UT 84112, USA
eDepartment of Chemical Engineering, University of Utah, Salt Lake City, UT 84112, USA
fDepartment of Bioengineering, University of Utah, Salt Lake City, UT 84112, USA
gDepartment of Pathology, University of Utah, Salt Lake City, UT 84112, USA
First published on 28th November 2016
Abstract
The ability to detect tuberculosis (TB) continues to be a global health care priority. This paper describes the development and preliminary assessment of the clinical accuracy of a heterogeneous immunoassay that integrates a serum pretreatment process with readout by surface-enhanced Raman scattering (SERS) for the low-level detection of mannose-capped lipoarabinomannan (ManLAM). ManLAM is a major virulence factor in the infectious pathology of Mycobacterium tuberculosis (Mtb) that has been found in the serum and other body fluids of infected patients. The effectiveness of ManLAM as a TB diagnostic marker, however, remains unproven for reasons not yet well understood. As reported herein, we have found that (1) ManLAM complexes with proteins and possibly other components in serum; (2) these complexes have a strongly detrimental impact on the ability to detect ManLAM using an immunoassay; (3) a simple pretreatment step can disrupt this complexation; and (4) disruption by pretreatment improves detection by 250×. We also describe the results from a preliminary assessment on the utility of serum pretreatment by running immunoassays on archived specimens from 24 TB-positive patients and 10 healthy controls. ManLAM was measurable in 21 of the 24 TB-positive specimens, but not in any of the 10 control specimens. These findings, albeit for a very small specimen set, translate to a clinical sensitivity of 87.5% and a clinical specificity of 100%. Together, these results both provide much needed evidence for the clinical utility of ManLAM as a TB marker, and demonstrate the potential utility of our overall approach to serve as a new strategy for the development of diagnostic tests for this disease.
1. Introduction
Advances in tuberculosis (TB) diagnostics remain a global health priority.1–3 TB is one of the world's deadliest infectious diseases and has a severity amplified by the emergence of drug-resistant strains of Mycobacterium tuberculosis (Mtb) and by co-infection with human immunodeficiency virus (HIV).4 The World Health Organization (WHO) estimates that there were 10.4 M active cases of TB in 2015 and 1.4 M associated deaths.4Early detection of active infection is vital to treating and containing the spread of TB.5 However, the majority (∼80%) of TB cases occur in low- to middle-income countries, where resource limitations complicate the ability to fight the disease. Because of its low cost, sputum smear microscopy (SSM) is the most widely applied test in these areas of the world, but it cannot be used for the reliable identification of early-stage infection.6 Serological diagnostics, which use the detection of secondary markers of the disease (e.g., antibodies), have proven ineffective in patients with immune systems compromised by HIV co-infection.7–9 Nucleic acid amplification tests (NAATs) can be of immense value in early diagnosis9,10 and are becoming an important addition to the TB diagnostics toolbox. The adoption of this technology in limited resource regions of the world, however, is deterred by concerns over cost and ease of use.10–14
In recognition of these challenges, there has been a refocus in TB diagnostics towards the direct detection of primary antigenic markers for Mtb in serum and other body fluids.15–17 This strategy parallels a proven approach for the early-stage diagnosis of malaria and other diseases.18 The potential merits of applying this approach to TB include: (1) high clinical accuracy (i.e., high clinical sensitivity and clinical specificity); (2) direct quantifiable evidence of the disease and the response to treatment; (3) diagnosis of smear-negative pulmonary infection; and (4) lack of dependence on a functioning immune system.15,16 Assays using body fluids other than sputum may also be useful in diagnosing extrapulmonary TB,7,19 a form of TB that accounts for ∼10% of the globally infected population. Extrapulmonary TB is often found in children and in adults co-infected with HIV and cannot be effectively diagnosed by analyzing sputum samples.19,20
Several mycobacterial antigens have been found in the serum and other body fluids of TB-infected patients.15 The most widely investigated antigen for potential use as a TB marker is mannose-capped lipoarabinomannan (ManLAM), a highly branched lipoglycan (17.3 ± 5 kDa)21 unique to mycobacteria.8,9,16,22–36 The strong interest in ManLAM reflects several factors. First, ManLAM is a major virulence factor in the infectious pathology of Mtb.37 ManLAM suppresses immunological responses to bacterial infection, including phagosome maturation, apoptosis, and interferon-gamma signaling in macrophages.38 Second, ManLAM is a large, loosely associated fractional component (∼40%) of the mycobacterial cell wall.23,39,40 It is therefore easily shed, potentially in large amounts, by metabolic active or degrading organisms into the circulatory system of an infected patient. Nonetheless, the effectiveness of ManLAM as a TB marker remains unproven due to inconsistencies in clinical sensitivity.3,30
Lipoarabinomannan has three unique structural components.41 Two of these components, the mannosyl phosphate inositol anchor and the mannan backbone, are common to all mycobacteria. The third component, the capping region, is species dependent [e.g., the phosphoinositol-capped LAM (PILAM) of the nonpathogenic M. smegmatis and the mannose-capped ManLAM of pathogenic M. leprae, M. bovis, and M. tuberculosis]. The underlying basis for the use of ManLAM therefore rests on the simple logic that if ManLAM is found in an infected individual in a TB-endemic region of the world, the likelihood of this antigen originating from Mtb is extremely high.4 There is one particularly important, but not well recognized, obstacle to the detection of ManLAM: its presence in the circulatory system in both its free and complexed forms.23,26,42–44 These complexes, referred to hereafter as complexed ManLAM, arises from its binding to proteins and potentially other components in body fluids. It is therefore reasonable to expect, as demonstrated in the companion paper for PILAM,45 that the detection of ManLAM may be compromised due to the impact of steric effects on the binding and/or labeling of ManLAM in heterogeneous immunoassays.30,42
In building on our sample pretreatment work using PILAM,45 this paper tests the hypothesis that the diagnostic strength of ManLAM for TB can be significantly improved by applying a simple but vital sample pretreatment step to disrupt ManLAM complexation before performing the immunoassay. For this, we used a sandwich immunoassay for the detection of ManLAM that combines gold nanoparticle (AuNP) labels, ManLAM monoclonal antibodies (mAbs), and surface-enhanced Raman scattering (SERS) detection. This approach draws on the continued focus of our46–48 and other laboratories49–58 on exploiting the strengths of SERS for low-level quantification of disease markers.59–62 Herein, we show that the application of a simple pretreatment (acidification) process can improve the limit of detection (LoD) of ManLAM in serum by ∼250× compared to untreated serum. We also carried out a preliminary assessment of the clinical accuracy of this approach in tests of 24 TB-positive patients (culture-confirmed) and 10 healthy controls. As detailed, these findings begin to demonstrate the potential of our approach to serve as an important addition to the TB diagnostics toolbox. Prospects and challenges in extending this approach to clinical and other point-of-need settings, along with possible applications to other TB markers and different types of patient specimens and diseases, are also briefly discussed.
2. Materials and methods
2.1 Assay format
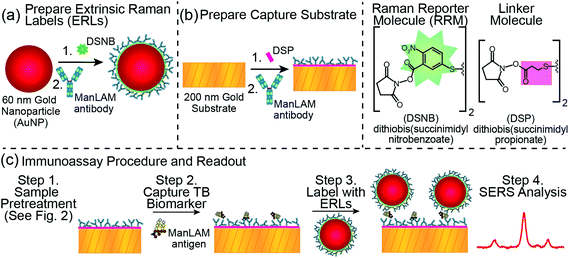
Fig. 1 overviews the details of our SERS-based immunoassay,61 which sandwiches ManLAM (isolated from the CDC-1551 strain of Mtb and obtained from Colorado State University)63,64 between an extrinsic Raman label (ERL) and a capture substrate. ERLs are prepared by modifying 60 nm AuNPs with a thiolate monolayer that forms by the spontaneous adsorption of the disulfide-bearing Raman reporter molecule (RRM) 5,5′-dithiobis(succinimidyl-2-nitrobenzoate) (DSNB). This step is followed by the adsorption of a layer of ManLAM mAbs on the RRM.65 This construction places the Raman scattering centers of the RRM monolayer in close proximity to the AuNP surface in order to maximize the SERS signal.66,67 The smooth, glass-supported gold (∼200 nm thick) capture substrate is also coated with ManLAM mAbs adsorbed on the thiolate monolayer formed from the spontaneous adsorption of dithiobis(succinimidyl propionate) (DSP).65 As a result, the presence of captured ManLAM is indirectly signaled by the characteristic Raman spectrum of the RRM, and the amount of ManLAM is indirectly quantified by the strength of its most intense spectral feature (i.e., the symmetric nitro stretch, νs(NO2), centered at 1336 cm−1 of the DSNB-derived monolayer). The selection of the ManLAM antibody for this work evaluated the same panel of three mAbs as used in the companion paper for PILAM for their effectiveness in capturing and labeling ManLAM.45,68 We found that the mAb used for PILAM, CS906.7 mAb (obtained from Colorado State University), also worked best with ManLAM and was therefore employed in this work; we note, however, that the slope of the dose–response plot for ManLAM is not as steep as for PILAM, indicating a higher affinity for PILAM by CS906.7.It is worthwhile to point out a few of the considerations adopted in moving SERS forward as a reliable analytical measurement tool in diagnostics. These relate, in large part, to designing an assay in which the response from SERS is reproducibly managed.69,70 We control reproducibility by (1) the size and shape distribution of the gold nanoparticles that constitute the ERL core; (2) the ability to form a monomolecular layer of RRMs and mAbs on the ERLs; and (3) the use of a smooth gold capture substrate. The latter component is particularly important due to the plasmonic coupling between the gold core of the ERL and the gold support of the capture substrate.71 We also use UV-Vis spectrophotometry per the method by Haiss et al.,72 to assure a fixed concentration of ERLs in the suspension used to tag the captured antigen.
2.2 Extrinsic Raman labels (ERLs)
The preparation61 and plasmonic signal optimization71 of ERLs and the synthesis of DSNB48 have appeared elsewhere. The preparative steps are depicted in Fig. 1A. First, an aqueous suspension of 60 nm AuNPs (NanoPartz) in 2.0 mM borate buffer (BB, pH 8.5, Thermo Fisher Scientific) was mixed with 1.0 mM DSNB in acetonitrile (Sigma-Aldrich) for 1.5 h at 4 °C. This step yielded a DSNB-derived thiolate monolayer on the AuNP surface. Next, a 10.0 μL at 100 μg mL−1 aliquot of the ManLAM mAb CS906.768 was added to the suspension and incubated for 1 h at 4 °C. This step was followed by pipetting 100 μL of 10% (w/v) bovine serum albumin (BSA, Sigma-Aldrich) in 2.0 mM BB into the suspension and incubating the resulting suspension at room temperature for 1 h. BSA serves as a blocking agent and a stabilizer of the colloidal suspension. This suspension was subsequently centrifuged at ∼2000g for 10 min, and the clear supernatant was carefully removed. The ERL pellet was resuspended in 1.0 mL of 1% BSA in 2.0 mM BB, and the centrifugation and resuspension steps were repeated two more times. The final resuspension used 0.25 mL of 2% BSA in 2.0 mM BB containing 150 mM NaCl (Thermo Fisher Scientific) to achieve an ERL concentration of 8.0 × 1010 particles per mL. Spectrophotometric measurements of the amounts of DSNB and mAbs coated on the ERLs varied by ±5.0% and ±10.2%, respectively.732.3 Capture substrate
Capture substrates (Fig. 1B) were prepared at 21 °C on 1 × 1 cm glass squares that supported a 200 nm layer of template stripped gold (TSG).74 A 2 mm diameter address was created in the center of the substrate by octadecanethiol (ODT, FLUKA) microprinting with polydimethylsiloxane (PDMS, Dow Corning Sylgard).75 The ODT layer produced a hydrophobic boundary around an uncoated 2 mm address, the latter of which was modified with an ethanolic solution of DSP (1.0 mM, Thermo Fisher Scientific) for 1 h. Next, the DSP-derived monolayer was reacted with a 20.0 μL drop of capture antibody (2.5 μg mL−1, CS906.7 ManLAM mAbs) for 1 h to form a layer of ManLAM mAbs on the hydrolyzed surface.65 As overviewed in Fig. 1C, the substrate was then rinsed three times with phosphate-buffered saline containing 0.1% Tween 20 (PBST, pH 7.4, Thermo Fisher Scientific); blocked with 20.0 μL of StartingBlock® (Thermo Fisher Scientific) for 1 h; rinsed three more times with PBST; and exposed to 20.0 μL of a ManLAM-containing sample. After 1 h, the sample was rinsed three times with 2 mL of 2.0 mM BB containing 150 mM NaCl and 0.1% Tween 20, exposed to 20.0 μL of the ERL suspension, and incubated for 16 h. Finally, the sample was rinsed three times with 2 mL of 2.0 mM BB containing 10 mM NaCl and 0.1% Tween 20, and dried under ambient conditions (21 °C) for ∼1 h prior to SERS interrogation.2.4 Instrumentation and sample readout
A modified NanoRaman spectrometer (Concurrent Analytical) was used to collect Raman spectra.76 This instrument has three primary components: laser excitation source, fiber optic probe, and spectrograph. The light source is a 22 mW HeNe laser (632.8 nm) with a spectrograph consisting of an f/2.0 Czerny Turner imaging spectrometer with 6–8 cm−1 resolution and a Kodak 0401E charged coupled device (CCD) thermoelectrically cooled to 0 °C.Raman spectra were collected at a 1 s integration time by irradiating a 20 μm spot on the sample surface at a laser power of 3.0 mW. The laser power was checked periodically in each run and varied by no more than 0.1 mW. The samples were analyzed at 10 different locations to account for surface heterogeneity.70 Spectra were baseline corrected by applying a 6th order polynomial fit, with the height of the νs(NO2) peak at 1336 cm−1 of the DSNB adlayer on the ERLs used for the construction of the dose–response plot.
2.5 Control and TB patient sera
Pooled human AB serum (Innovative research, lot# IPLA-SERAB-13458) was used for the development of the assay and the generation of calibration curves (i.e., serum spiked with ManLAM). The vendor prepares this product, referred to hereafter as negative human serum, by pooling and processing healthy male donor plasma collected at centers across the USA. This serum was stored at −30 °C and allowed to thaw to ambient temperature (21 °C) immediately prior to use. All calibration data are presented as the average and standard deviation of the collected spectra (i.e., a spectrum from 10 different locations per sample from triplicate samples).The TB-positive sera were collected from patients in South Africa who were enrolled in the Tuberculosis Trials Consortium Study Group 22 (TBTC-22) with culture-confirmed cavitary TB.77 This study group participated in a randomized clinical trial that was designed to test the effectiveness of the anti-TB drugs rifapentine and isoniazid in treating pulmonary TB in adult, HIV-negative patients. These deidentified samples were procured by Colorado State University from the Centers for Disease Control and Prevention (CDC) after TBTC-22 approval. This specimen set consisted of 24 different serum samples, each at a volume of ∼100 μL. We do not have information with regard to treatment status (e.g., drug regimen or time course of treatment) for any of these specimens. However, tests for immunoblot reactivity confirmed the presence of ManLAM antibodies in all TB-positive specimens, but not in any of the healthy controls (data not shown). These results suggest that there was a high likelihood that ManLAM antigen will also be present in the TB-positive serum specimens.
Healthy, nonendemic control sera, referred to hereafter as healthy controls, were obtained from U.S.-born residents of Colorado. These non-Bacillus Calmette–Guérin (BCG)-vaccinated residents gave informed consent to participate in a study of reactivity to M. leprae and M. tuberculosis antigens.78 These residents had no known exposure to TB or leprosy and did not work in a mycobacterial laboratory. The assay of all patient specimens and healthy controls were performed at the University of Utah. All assays were carried out in a biosafety cabinet contained in a Biosafety Level (BSL) 2, which had been enhanced for handling some risk group 3 pathogens.
2.6 Ethical statement
The anonymized archived serum TB-patient samples used in this study were provided by the CDC Repository. These samples were collected as part of the TBTC Study 22 clinical trial,77 with patient informed consent in accordance with the guidelines of the Declaration of Helsinki. As a pre-existing collection, the studies herein were considered exempt by our Institutional Review Board. The designated sample numbers used herein are arbitrary and cannot be traced back to the patients. All subjects providing healthy control samples signed an informed consent at the time of enrollment and before sample collection. The study was approved by the Institutional Review Board at Colorado State University. The participants were informed that the samples were going to be stored in a repository. None of the authors have access to any identifying patient information.2.7 Serum pretreatment
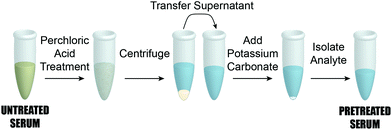
As detailed in the companion paper,45 the ability to detect PILAM directly from serum was significantly impaired by complex formation.42–44 We therefore adapted the same classic pretreatment procedure79 designed to disrupt complexation via protein denaturation by acidification with HClO4 for our work with ManLAM. This procedure consists of the five steps outlined in Fig. 2. It begins by adding 2.0 μL of HClO4 (70%, Sigma-Aldrich) to 100.0 μL of a calibration/patient sample in a small centrifuge tube, which brings the pH to ∼2 and forms a milky suspension. After vortexing for 10 s and centrifuging at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min, a 75.0 μL aliquot of the resulting supernatant was transferred to a second centrifuge tube and neutralized to pH 7.5 with 6.0 μL of 2.0 mM aqueous solution of K2CO3 (Thermo Fisher Scientific). The samples were then cooled to 4 °C for 30 min to accelerate KClO4 precipitation, and subsequently warmed to room temperature (21 °C, ∼20 min) before being pipetted (20.0 μL) onto the capture substrate. We add that ManLAM is acid-labile.80–82 We have found, however, that such degradation does not appear to have an effect on the reproducibility of the dose–response plots presented in Fig. 4. Nonetheless, experiments to quantify the potential impact of the serum pretreatment process on the recovery of ManLAM are underway.
000g for 5 min, a 75.0 μL aliquot of the resulting supernatant was transferred to a second centrifuge tube and neutralized to pH 7.5 with 6.0 μL of 2.0 mM aqueous solution of K2CO3 (Thermo Fisher Scientific). The samples were then cooled to 4 °C for 30 min to accelerate KClO4 precipitation, and subsequently warmed to room temperature (21 °C, ∼20 min) before being pipetted (20.0 μL) onto the capture substrate. We add that ManLAM is acid-labile.80–82 We have found, however, that such degradation does not appear to have an effect on the reproducibility of the dose–response plots presented in Fig. 4. Nonetheless, experiments to quantify the potential impact of the serum pretreatment process on the recovery of ManLAM are underway.
2.8 Data analysis
Due to the small volumes of the TB-positive and healthy control serum specimens (∼100 μL), assays for each sample were only run in duplicate. The levels of ManLAM in all patient samples are therefore reported as averages, with the uncertainties reflecting the range of measurements from reading 10 sites per sample from the duplicate substrates prepared from a single specimen.The serum blank specimens had an average response of 64 cts s−1, with a standard deviation of 11 cts s−1. We employed a confidence interval of >99% to define the LoD, which yielded a signal on the calibration curve that matched the blank response (64 cts s−1) plus three times its standard deviation (3 × 11 cts s−1) for a cutoff of 97 cts s−1. Patients were identified using our SERS-based assay as “TB-positive” if the level of ManLAM was statistically measurable above the LoD (i.e., the average response including the standard deviation of the patient sample is statistically different from the cutoff of 97 cts s−1). The fit of the data for the pretreated calibration samples used a linear least squares approach for data that ranged from the blank serum samples to serum samples spiked at 100 ng mL−1 (y = 12.9x + 64.1). For samples with signals beyond the linear portion of the dose–response plot (i.e., ManLAM concentrations above ∼100 ng mL−1), a second order polynomial fit (y = −0.00727x2 + 13.5x + 64.1) was used for quantification.
Clinical sensitivity and clinical specificity measure diagnostic test accuracy. The former is defined as the percentage of infected individuals correctly identified by the test as infected; it is expressed as: [TP/(TP + FN)] × 100, where TP is the number of true positive tests and FN is the number of false negative tests. The latter is defined as the percentage of uninfected subjects correctly identified by the test as being uninfected; it is given as: [TN/(FP + TN)] × 100, where TN is the number of true negatives and FP is the number of false positives.
3. Results and discussion
3.1 Preliminary findings
As described in the companion paper,45 this work began by conducting a set of studies to assess the effectiveness of our assay strategy by spiking PILAM (M. smegmatis) into negative human serum, as this fast growing and nonpathogenic mycobacterium is often used as a model for Mtb research.83–85 The companion paper demonstrated the severe detrimental impact of complex formation on the detection of PILAM directly from spiked human serum, and devised an approach to disrupt complexation and thereby improve detection.3.2 ManLAM measurements in untreated and pretreated human serum
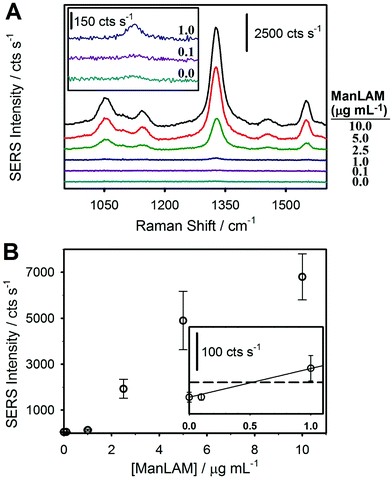
The first assessment of the potential merits of our integrated approach focused on a comparison of assays that analyzed ManLAM after spiking into untreated, negative human serum and then subjecting the spiked serum samples to the HClO4 pretreatment protocol shown in Fig. 2. The SERS spectra and dose–response plot for the untreated samples are shown in Fig. 3. These samples included serum blanks and serum spiked with ManLAM at levels up to 10 μg mL−1. There are two important points to draw from these spectra (Fig. 3A). First, all of the observable spectral features can be assigned to functional groups of the monomolecular coating of the RRM on the ERLs (e.g., νs(NO2) at 1336 cm−1 and aromatic ring mode at 1558 cm−1 of the DSNB-derived coating).61 None of the vibrational modes of the mAb layer on the capture substrate or ERLs are detectably enhanced. Second, there is evidence for a small, but measureable, level of ERL adsorption in the spectrum for the negative control (48 ± 14 cts s−1). However, the response is barely above the instrument noise level of 39 ± 7 cts s−1. We attribute this observation to the effectiveness of the blocking agent and other preparative procedures in reducing nonspecific adsorption.The dose–response plot for ManLAM in untreated serum is shown in Fig. 3B. It was constructed from the average signal for the strongest feature in the SERS spectrum, νs(NO2), from 10 different locations per sample and triplicate samples for each calibration point. The response at low levels of ManLAM follows a linear dependence. Though not shown, the response at higher ManLAM concentrations begins to level off (∼5 μg mL−1) as binding sites on the capture substrate approach saturation, plateauing at ∼10 μg mL−1. The LoD, calculated as the response on the calibration line that matches the signal for the blank sample plus three times its standard deviation, is ∼500 ng mL−1. This value is ∼30× greater than the LoD found for PILAM from M. smegmatis, which is phosphoinositol-capped, using the same set of assay conditions.45 This difference is attributed to the lower affinity of the CS906.7 mAb for ManLAM versus that for PILAM.
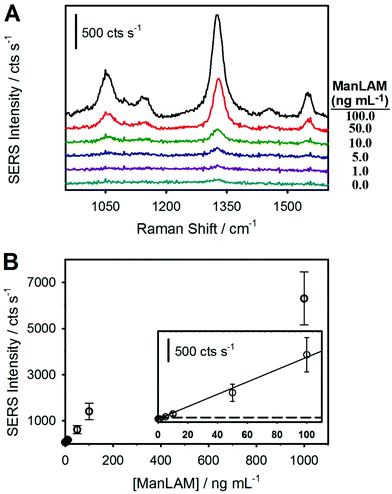
Following the hypothesis that the complexes formed between ManLAM and serum constituents sterically inhibit binding to the capture mAb, the next set of assays analyzed the ManLAM-spiked sera after completing the HClO4 pretreatment procedure illustrated in Fig. 2. Indeed, this assertion is in accord with a growing body of evidence for the presence of complexes for ManLAM in human serum,23,26,27,42–44,86,87 the most recent of these studies pointing to the strong association of ManLAM with high density lipoproteins (HDLs).42 The impact of pretreatment is immediately evident in the results presented in Fig. 4, which shows that the spectral responses for the pretreated samples are much stronger than those of the untreated samples. This difference is particularly evident when comparing the responses for a ManLAM concentration of 100 ng mL−1. With the untreated sample, the response at 100 ng mL−1 is indistinguishable from that of the serum blank, whereas the signal for the acid-pretreated specimen is slightly above 1400 cts s−1. Another comparison shows that the signal (∼130 cts s−1) for the untreated ManLAM sample at 1 μg mL−1 is close to the response (∼170 cts s−1) for the pretreated sample at a ManLAM level that is lower by a factor of 100 (0.010 μg mL−1).
The dose–response plot, obtained from triplicate samples for each calibration point for ManLAM spiked into negative human serum followed by acid pretreatment, is shown in Fig. 4B. Similar to the data for ManLAM spiked into human serum without pretreatment (Fig. 3), the response at low ManLAM levels follows a linear dependence and plateaus (data not shown) at ∼1 μg mL−1 of ManLAM as mAb binding sites on the capture substrate begin to saturate. The LoD from an analysis of these data is ∼2 ng mL−1 or ∼250× below that in serum prior to pretreatment.
3.3 TB patient assay
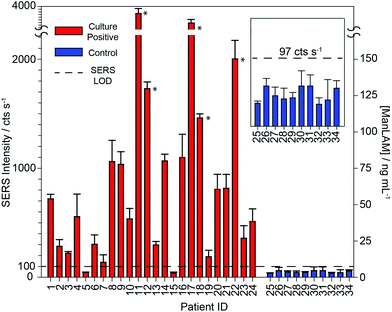
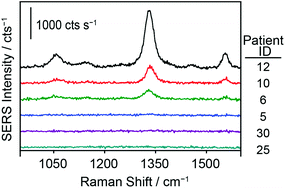
To further test the hypothesis that an effective pretreatment method significantly improves the utility of ManLAM as a diagnostic marker for TB, we analyzed 24 TB-positive (identifiers #1 to #24) and 10 healthy control (identifiers #25 to #34) serum specimens. The post pretreatment results for the assays of these specimens are presented in Fig. 5 and 6 and Table 1. The small specimen volumes precluded the opportunity to compare the responses of these samples before and after pretreatment. Fig. 5 summarizes these measurements by using histograms to represent the average signal strength of the νs(NO2) spectral feature measured for each sample, and includes a delimiter (dashed black line) for the LoD (2 ng mL−1) of the SERS assay. Fig. 6 consists of a small set of specimen spectra for illustrative purposes; it includes spectra for two of the healthy control samples (#25 and #30) and four of the TB-positive samples (#5, #6, #10, and #12). These spectra were selected to demonstrate the range of measured responses, which for the TB-positive samples, includes a non-detectable response (specimen #5) and a response outside the linear range of the dose–response plot (specimen #12).| Sample ID | SERS/cts s−1 | ManLAM/ng mL−1 |
|---|---|---|
| The calculated ManLAM levels are derived from the linear fit equation from the calibration curve (y = 12.9x + 64.1) for ManLAM ranging from 0–100 ng mL−1 in pretreated human serum. SERS responses corresponding to ManLAM > 100 ng mL−1 are calculated from a second order polynomial fit (y = −0.00727x2 + 13.5x + 64.1). ND (not detected) represents ManLAM below the LoD in human serum of 2 ng mL−1. The Raman instrument noise level is 39 ± 7 cts s−1. | ||
| 1 | 728 ± 41 | 52 ± 3 |
| 2 | 282 ± 61 | 17 ± 5 |
| 3 | 219 ± 16 | 12 ± 1 |
| 4 | 553 ± 208 | 38 ± 16 |
| 5 | 43 ± 3 | ND |
| 6 | 301 ± 86 | 18 ± 7 |
| 7 | 131 ± 67 | ND |
| 8 | 1059 ± 194 | 77 ± 15 |
| 9 | 1034 ± 116 | 75 ± 9 |
| 10 | 533 ± 96 | 36 ± 7 |
| 11 | 3710 ± 208 | 328 ± 16 |
| 12 | 1728 ± 61 | 129 ± 5 |
| 13 | 295 ± 31 | 18 ± 2 |
| 14 | 1065 ± 61 | 78 ± 5 |
| 15 | 37 ± 9 | ND |
| 16 | 1096 ± 214 | 80 ± 17 |
| 17 | 3349 ± 173 | 288 ± 13 |
| 18 | 1460 ± 36 | 108 ± 3 |
| 19 | 187 ± 60 | 10 ± 5 |
| 20 | 803 ± 139 | 57 ± 11 |
| 21 | 814 ± 129 | 58 ± 10 |
| 22 | 2001 ± 170 | 157 ± 13 |
| 23 | 356 ± 115 | 23 ± 9 |
| 24 | 509 ± 146 | 35 ± 11 |
| 25 | 36 ± 4 | ND |
| 26 | 60 ± 15 | ND |
| 27 | 47 ± 11 | ND |
| 28 | 42 ± 14 | ND |
| 29 | 44 ± 7 | ND |
| 30 | 59 ± 27 | ND |
| 31 | 60 ± 21 | ND |
| 32 | 35 ± 8 | ND |
| 33 | 41 ± 27 | ND |
| 34 | 56 ± 11 | ND |
As evident in Fig. 5, ManLAM was detectable in 21 of the 24 TB-positive samples. ManLAM was not found in three of the TB-positive samples (i.e., #5, #7, and #15) or in any of the 10 healthy control specimens (i.e., ManLAM < 2 ng mL−1). Importantly, these results yield a clinical sensitivity of 87.5% and a clinical specificity of 100% for this limited data set.
Further inspection of Table 1 points to three ranges for the ManLAM levels found in the patient serum specimens. First, five of the TB-positive samples (#11, #12, #17, #18, and #22) have ManLAM concentrations greater than 100 ng mL−1. Second, several other samples (#2, #3, #6, #13, and #19) have ManLAM levels above the LoD but below 20 ng mL−1. Third, ManLAM levels in the majority of samples (#1, #4, #8, #9, #10, #14, #16, #20, #21, #23, and #24) are between 20 and 100 ng mL−1. While we are not able to offer any correlations between ManLAM levels and the stage of disease progression, the ability to quantify small differences in ManLAM levels in TB-patient sera suggests that our approach may be useful in monitoring treatment response, determining the optimal duration of therapy, and assessing new drug treatment regimens and vaccines.88–90
These data also show that the responses for 3 of the TB-positive patient samples (#5, #7, and #15) were not distinguishable from those of the calibration blank or any of the controls. Even though the average of patient sample #7 lies above the cutoff, its uncertainty makes it indistinguishable from the blank. The inability to detect ManLAM in these specimens is attributed to one or a combination of four possibilities: (1) ManLAM was present in these three specimens at levels below the LoD of the assay; (2) these patients may have had a positive response to one of the drug treatments used in the TBTC-22 clinical trial; (3) these specimens may have degraded during storage and/or shipment prior to receipt or from freeze/thaw cycling when realiquoted for distribution; or (4) not all patients with cavitary TB have circulating ManLAM antigenemia.
Finally, these data show that the responses for all 10 healthy control samples were commensurate with that of the serum blank used in the construction of the calibration plot. The spectra for sample #25 and #30 in Fig. 6 are representative of those for all of the healthy control samples. Moreover, these responses could not be differentiated from instrument noise even after increasing the signal acquisition time from 1 to 60 s (data not shown).
Taken together, these results support the potential value of our integrated approach for the detection of TB. Experiments to more fully validate the performance of this methodology by analyzing a much larger specimen sample set and to evaluate other types of specimens (e.g., patients with non-cavitary lung disease, HIV co-infection, and extrapulmonary infection) are being planned. We also believe that one of the obstacles in the detection of ManLAM in other body fluids (e.g., urine and cerebral spinal fluid) is likely to be a consequence of very low concentrations of unbound antigen due to complexation.
4. Conclusions and prospectus
These results begin to demonstrate the potential of ManLAM to serve as a long sought-after antigenic marker for TB and of our technological approach to become an exciting addition to the TB diagnostics toolbox. It is important, however, to ground these preliminary results with a reality check in terms of the many-faceted obstacles faced by TB diagnostics in meeting the global challenges in fighting this disease.First and foremost is the challenge to prove the effectiveness of our ManLAM assay through an extensive series of validation studies. While the results herein point to a potential technical solution for a test for active infection, an extensive series of validation studies must be carried out. Both the number of specimens and types of specimens must be markedly expanded. Examples of the types of specimens include those from patients who (1) are both TB-positive and HIV-positive; (2) have cavitary, non-cavitary, and extrapulmonary TB; or (3) have been BCG (bacilli Calmette-Guerin) vaccinated. Moreover, since ManLAM can potentially be linked to the Mtb burden of a patient, longitudinal studies could be used to test the effectiveness of the test in determining the stage of disease progression and in providing a means to monitor the response to treatment. The overall goal of these validation studies is to quantify the clinical sensitivities and clinical specificities of ManLAM for each of these manifestations of TB, which requires specimens that are well characterized in terms of clinical symptoms, comparative data from other TB tests, (e.g., culture and molecular tests), and detailed patient health histories. Interestingly, the 2006 Global Health Diagnostics Forum, which was convened by the Bill and Melinda Gates Foundation, estimated that a rapid and globally accessible diagnostic test for active TB infection, which has a clinical sensitivity of ≥85% and a clinical specificity of ≥97%, could help save ∼400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 lives each year.91 Our small data set suggests that we may be on a path that could reach these metrics, but it remains to be seen if this level of effectiveness can be realized with ManLAM alone or if a multiplexed test to include other antigenic markers for TB will be required. There have also been recent efforts aimed at improving ManLAM detection by developing more specific and higher affinity antibodies,87 which may again lower the LoD, and therefore further improve clinical accuracy. There are other promising approaches for ManLAM detection,92 but it is difficult to compare the utility of the different platforms as the capture and labeling reagents are not the same.
000 lives each year.91 Our small data set suggests that we may be on a path that could reach these metrics, but it remains to be seen if this level of effectiveness can be realized with ManLAM alone or if a multiplexed test to include other antigenic markers for TB will be required. There have also been recent efforts aimed at improving ManLAM detection by developing more specific and higher affinity antibodies,87 which may again lower the LoD, and therefore further improve clinical accuracy. There are other promising approaches for ManLAM detection,92 but it is difficult to compare the utility of the different platforms as the capture and labeling reagents are not the same.
Another challenge revolves around accessibility in the regions of the world where such a test is needed the most.12,13 The only platforms today that have a high clinical accuracy are microbial culturing and NAATs,10 but both have a cost, complexity, and/or specimen type limitation that impedes widespread use in resource-limited settings.7 Within this context, there are several issues that need to be addressed with respect to instrumentation, sample processing, and reagent packaging and stability before potentially transitioning our approach from the research laboratory to field settings.13 More so, issues differ depending on whether the test will be conducted in settings with reliable infrastructure (e.g., hospital clinic) in terms of power, sample refrigeration/freezing, and technical skills, or if the intent is to create a field-deployable (e.g., point-of-need) test, in which case the infrastructure and technical capabilities may be limited. In either case, approaches to streamline sample preparation and readout, instrument and test standardization and calibration, reagent packaging and handling, and overall ease of use must be addressed.
These results have another potentially important implication. If we assume that the complexed form of ManLAM has a molecular weight greater than 100 kDa as discussed in the companion paper,45 then the ∼30 kDa cutoff by the filtration mechanisms of the kidneys would preclude passage of the complex into urine. ManLAM would then only be a viable diagnostic marker in urine specimens from patients with nephropathy. This interpretation is consistent with the increased accuracy reported in tests for ManLAM in the urine of HIV co-infected patients, and may aid in understanding the origins of the poor clinical accuracies of such tests.16,23–26,34,35,44,93,94
In closing, we would like to briefly comment on the potential impact of our approach to pretreatment beyond that for ManLAM. There are a number of other serum-based disease markers that could be prone to complexation, which may interfere with detection when using a capture-styled assay. Examples include the carbohydrate antigen CA 19-9 (210 kDa), a marker for pancreas adenocarcinoma, and the polysaccharide galactomannan, a marker for invasive aspergillosis infection. Future studies to examine the effects of our pretreatment approach on these and other markers are planned.
Acknowledgements
Discussions with Nicholas Schlotter (Department of Chemistry, Hamline University) are gratefully acknowledged. This work was supported by the Critical Path Initiative of the U.S. Food and Drug Administration (contract U18 FD004034), the Bill and Melinda Gates Foundation (OOP10396210), and the Kerith Foundation.References
- L. Phillips, Nature, 2013, 493, 14–16 CrossRef PubMed.
- M. Pai, S. Kalantri and K. Dheda, Expert Rev. Mol. Diagn., 2006, 6, 423–432 CrossRef CAS PubMed.
- S. Dorman, Int. J. Tuberc. Lung Dis., 2015, 19, 504–516 CrossRef CAS PubMed.
- Global Tuberculosis Report, World Health Organization, Geneva, 2016.
- A. Zumla, M. Raviglione, R. Hafner and C. Fordham von Reyn, N. Engl. J. Med., 2013, 368, 745–755 CrossRef CAS PubMed.
- K. R. Steingart, V. Ng, M. Henry, P. C. Hopewell, A. Ramsay, J. Cunningham, R. Urbanczik, M. D. Perkins, M. A. Aziz and M. Pai, Lancet Infect. Dis., 2006, 6, 664–674 CrossRef PubMed.
- K. R. Steingart, L. L. Flores, N. Dendukuri, I. Schiller, S. Laal, A. Ramsay, P. C. Hopewell and M. Pai, PLoS Med., 2011, 8, e1001062 Search PubMed.
- V. B. Patel, R. Singh, C. Connolly, V. Kasprowicz, A. Zumla, T. Ndungu and K. Dheda, PLoS One, 2010, 5, e15664 CAS.
- M. D. Perkins and J. Cunningham, J. Infect. Dis., 2007, 196, S15–S27 CrossRef PubMed.
- G. Theron, A. Pooran, J. Peter, R. van Zyl-Smit, H. K. Mishra, R. Meldau, G. Calligaro, B. Allwood, S. K. Sharma, R. Dawson and K. Dheda, Eur. Respir. J., 2012, 40, 161–168 CrossRef PubMed.
- M. Liong, A. N. Hoang, J. Chung, N. Gural, C. B. Ford, C. Min, R. R. Shah, R. Ahmad, M. Fernandez-Suarez, S. M. Fortune, M. Toner, H. Lee and R. Weissleder, Nat. Commun., 2013, 4, 1752 CrossRef PubMed.
- S. Wang, M. A. Lifson, F. Inci, L.-G. Liang, Y.-F. Sheng and U. Demirci, Expert Rev. Mol. Diagn., 2016, 16, 449–459 CrossRef CAS PubMed.
- R. McNerney and P. Daley, Nat. Rev. Microbiol., 2011, 9, 204–213 CrossRef CAS PubMed.
- S. K. Srivastava, C. J. M. van Rijn and M. A. Jongsma, RSC Adv., 2016, 6, 17759–17771 RSC.
- F. Abebe, C. Holm-Hansen, H. Wiker and G. Bjune, Scand. J. Immunol., 2007, 66, 176–191 CrossRef CAS PubMed.
- K. Reither, E. Saathoff, J. Jung, L. T. Minja, I. Kroidl, E. Saad, J. F. Huggett, E. N. Ntinginya, L. Maganga, L. Maboko and M. Hoelscher, BMC Infect. Dis., 2009, 9, 141 CrossRef PubMed.
- P. Tucci, G. González-Sapienza and M. Marin, Front. Microbiol., 2014, 5, 549 CrossRef PubMed.
- WHO Malaria Rapid Diagnostic Test Performance: Results of WHO Product Testing of Malaria RDTs: Round 3 (2010–2011), World Health Organization, Geneva, 2011 Search PubMed.
- L. Norbis, R. Alagna, E. Tortoli, L. R. Codecasa, G. B. Migliori and D. M. Cirillo, Expert Rev. Anti-Infect. Ther., 2014, 12, 633–647 CrossRef CAS PubMed.
- P. Piccini, E. Chiappini, E. Tortoli, M. de Martino and L. Galli, BMC Infect. Dis., 2014, 14, S4 CrossRef PubMed.
- A. Venisse, J. M. Berjeaud, P. Chaurand, M. Gilleron and G. Puzo, J. Biol. Chem., 1993, 268, 12401–12411 CAS.
- D. Chatterjee and K. H. Khoo, Glycobiology, 1998, 8, 113–120 CrossRef CAS PubMed.
- S. D. Lawn, BMC Infect. Dis., 2012, 12, 103 CrossRef PubMed.
- S. Sarkar, X. Tang, D. Das, J. S. Spencer, T. L. Lowary and M. R. Suresh, PLoS One, 2012, 7, e32340 CAS.
- B. Hamasura, J. Bruchfeld, M. Haile, A. Pawlowski, B. Bjorvatn, G. Källenius and S. B. Svenson, J. Microbiol. Methods, 2001, 45, 41–52 CrossRef.
- R. Wood, K. Racow, L.-G. Bekker, K. Middelkoop, M. Vogt, B. N. Kreiswirth and S. D. Lawn, BMC Infect. Dis., 2012, 12, 47 CrossRef PubMed.
- E. Sada, D. Aguilar, M. Torres and T. Herrera, J. Clin. Microbiol., 1992, 30, 2415–2418 CAS.
- E. Sada, P. Brennan, T. Herrera and M. Torres, J. Clin. Microbiol., 1990, 28, 2587–2590 CAS.
- S. D. Lawn, D. J. Edwards, K. Kranzer, M. Vogt, L.-G. Bekker and R. Wood, AIDS, 2009, 23, 1875–1880 CrossRef PubMed.
- P. Sarkar, D. Biswas, G. Sindhwani, J. Rawat, A. Kotwal and B. Kakati, Postgrad. Med. J., 2014, 90, 155–163 CrossRef CAS PubMed.
- P. Drain, E. Losina, S. M. Coleman, J. Giddy, D. Ross, J. N. Katz, R. P. Walensky, K. A. Freedberg and I. V. Bassett, BMC Infect. Dis., 2014, 14, 110 CrossRef PubMed.
- T. A. Tessema, B. Hamasur, G. Bjune, S. Svenson and B. Bjorvatn, Scand. J. Infect. Dis., 2001, 33, 279–284 CrossRef CAS PubMed.
- A. D. Kerkhoff, R. Wood, M. Vogt and S. D. Lawn, PLoS One, 2014, 9, e103285 Search PubMed.
- M. P. Nicol, V. Allen, L. Workman, W. Isaacs, J. Munro, S. Pienaar, F. Black, L. Adonis, W. Zemanay, Y. Ghebrekristos and H. J. Zar, Lancet Glob. Health, 2014, 2, e278–e284 CrossRef PubMed.
- K. Dheda, V. Davids, L. Lenders, T. Roberts, R. Meldau, D. Ling, L. Brunet, R. van Zyl Smit, J. Peter, C. Green, M. Badri, L. Sechi, S. Sharma, M. Hoelscher, R. Dawson, A. Whitelaw, J. Blackburn, M. Pai and A. Zumla, PLoS One, 2010, 5, e9848 Search PubMed.
- C. Boehme, E. Molokova, F. Minja, S. Geis, T. Loscher, L. Maboko, V. Koulchin and M. Hoelscher, Trans. R. Soc. Trop. Med. Hyg., 2005, 99, 893–900 CrossRef CAS PubMed.
- J. Chan, X. D. Fan, S. W. Hunter, P. J. Brennan and B. R. Bloom, Infect. Immun., 1991, 59, 1755–1761 CAS.
- V. Briken, S. A. Porcelli, G. S. Besra and L. Kremer, Mol. Microbiol., 2004, 53, 391–403 CrossRef CAS PubMed.
- L. M. Pereira Arias-Bouda, L. N. Nguyen, L. M. Ho, S. Kuijper, H. M. Jansen and A. H. J. Kolk, J. Clin. Microbiol., 2000, 38, 2278–2283 CAS.
- B. Al-Sayyed, S. Piperdi, X. Yuan, A. Li, G. S. Besra, W. R. Jacobs, A. Casadevall and A. Glatman-Freedman, Tuberculosis, 2007, 87, 489–497 CrossRef CAS PubMed.
- A. K. Mishra, N. N. Driessen, B. J. Appelmelk and G. S. Besra, FEMS Microbiol. Rev., 2011, 35, 1126–1157 CrossRef CAS PubMed.
- R. Sakamuri, D. Price, M. Lee, S. Cho, C. Barry, L. Via, B. Swanson and H. Mukundan, Tuberculosis, 2013, 93, 301–307 CrossRef CAS PubMed.
- T. A. Tessema, G. Bjune, B. Hamasur, S. Svenson, H. Syre and B. Bjorvatn, Scand. J. Infect. Dis., 2002, 34, 97–103 CrossRef PubMed.
- J. A. Cox, R. L. Lukande, S. Kalungi, E. Van Marck, K. Van de Vijver, A. Kambugu, A. M. Nelson, R. Colebunders and Y. C. Manabe, PLoS One, 2015, 10, e0123323 Search PubMed.
- L. B. Laurentius, A. C. Crawford, T. S. Mulvihill, J. H. Granger, R. Robinson, J. S. Spencer, D. Chatterjee, K. E. Hanson and M. D. Porter, Analyst, 2016 10.1039/c6an02109c.
- J. H. Granger, M. C. Granger, M. A. Firpo, S. J. Mulvihill and M. D. Porter, Analyst, 2013, 138, 410–416 RSC.
- J. D. Driskell, K. M. Kwarta, R. J. Lipert and M. D. Porter, Anal. Chem., 2005, 77, 6147–6154 CrossRef CAS PubMed.
- D. S. Grubisha, R. J. Lipert, H.-Y. Park, J. D. Driskell and M. D. Porter, Anal. Chem., 2003, 75, 5936–5943 CrossRef CAS PubMed.
- D. A. Stuart, A. J. Haes, C. R. Yonzon, E. M. Hicks and R. P. Van Duyne, IEE Proc. Nanobiotechnol., 2005, 152, 13–32 CrossRef CAS PubMed.
- S. Xu, X. Ji, W. Xu, X. Li, L. Wang, Y. Bai, B. Zhao and Y. Ozaki, Analyst, 2004, 129, 63–68 RSC.
- H. Chon, S. Lee, S. W. Son, C. H. Oh and J. Choo, Anal. Chem., 2009, 81, 3029–3034 CrossRef CAS PubMed.
- A. Pallaoro, G. B. Braun and M. Moskovits, Nano Lett., 2015, 15, 6745–6750 CrossRef CAS PubMed.
- A.-I. Henry, B. Sharma, M. F. Cardinal, D. Kurouski and R. P. Van Duyne, Anal. Chem., 2016, 88, 6638–6647 CrossRef CAS PubMed.
- C. Westley, Y. Xu, A. J. Carnell, N. J. Turner and R. Goodacre, Anal. Chem., 2016, 88, 5898–5903 CrossRef CAS PubMed.
- H. Fisk, C. Westley, N. J. Turner and R. Goodacre, J. Raman Spectrosc., 2016, 47, 59–66 CrossRef CAS PubMed.
- C. P. Shaw, M. Fan, C. Lane, G. Barry, A. I. Jirasek and A. G. Brolo, J. Phys. Chem. C, 2013, 117, 16596–16605 CAS.
- L. Laurentius, S. R. Stoyanov, S. Gusarov, A. Kovalenko, R. Du, G. P. Lopinski and M. T. McDermott, ACS Nano, 2011, 5, 4219–4227 CrossRef CAS PubMed.
- S. Laing, K. Gracie and K. Faulds, Chem. Soc. Rev., 2016, 45, 1901–1918 RSC.
- K. C. Bantz, A. F. Meyer, N. J. Wittenberg, H. Im, Ö. Kurtuluş, S. H. Lee, N. C. Lindquist, S.-H. Oh and C. L. Haynes, Phys. Chem. Chem. Phys., 2011, 13, 11551–11567 RSC.
- D. Graham and R. Goodacre, Chem. Soc. Rev., 2008, 37, 883–884 RSC.
- M. D. Porter, R. J. Lipert, L. M. Siperko, G. Wang and R. Narayanan, Chem. Soc. Rev., 2008, 37, 1001–1011 RSC.
- B. Sharma, R. R. Frontiera, A.-I. Henry, E. Ringe and R. P. Van Duyne, Mater. Today, 2012, 15, 16–25 CrossRef CAS.
- L. Shi, R. Zhou, Z. Liu, T. L. Lowary, P. H. Seeberger, B. L. Stocker, D. C. Crick, K. H. Khoo and D. Chatterjee, J. Bacteriol., 2008, 190, 5248–5255 CrossRef CAS PubMed.
- J. B. Torrelles, P. A. Sieling, N. Zhang, M. A. Keen, M. R. McNeil, J. T. Belisle, R. L. Modlin, P. J. Brennan and D. Chatterjee, Glycobiology, 2012, 22, 1118–1127 CrossRef CAS PubMed.
- C. Y. Lim, N. A. Owens, R. D. Wampler, Y. Ying, J. H. Granger, M. D. Porter, M. Takahashi and K. Shimazu, Langmuir, 2014, 30, 12868–12878 CrossRef CAS PubMed.
- M. Moskovits, Rev. Mod. Phys., 1985, 57, 783–826 CrossRef CAS.
- R. L. McCreery, Raman Spectroscopy for Chemical Analysis, John Wiley & Sons, Inc., New York, 2000 Search PubMed.
- H. Gaylord, P. J. Brennan, D. B. Young and T. M. Buchanan, Infect. Immun., 1987, 55, 2860–2863 CAS.
- H.-Y. Park, R. J. Lipert and M. D. Porter, in Proc. SPIE-Int. Soc. Opt. Eng, ed. and M. S. Islam and A. K. Dutta, 2004, vol. 5593, pp. 464–477 Search PubMed.
- A. C. Crawford, A. Skuratovsky and M. D. Porter, Anal. Chem., 2016, 88, 6515–6522 CrossRef CAS PubMed.
- J. D. Driskell, R. J. Lipert and M. D. Porter, J. Phys. Chem. B, 2006, 110, 17444–17451 CrossRef CAS PubMed.
- W. Haiss, N. T. K. Thanh, J. Aveyard and D. G. Fernig, Anal. Chem., 2007, 79, 4215–4221 CrossRef CAS PubMed.
- M. Bradley, MSc thesis, University of Utah, Dec. 2010.
- M. Hegner, P. Wagner and G. Semenza, Surf. Sci., 1993, 291, 39–46 CrossRef CAS.
- A. Kumar, H. A. Biebuyck and G. M. Whitesides, Langmuir, 1994, 10, 1498–1511 CrossRef CAS.
- H.-Y. Park, J. D. Driskell, K. M. Kwarta, R. J. Lipert, M. D. Porter, C. Schoen, J. D. Neill and J. F. Ridpath, in Surface-Enhanced Raman Scattering - Physics and Applications, ed. K. Kneipp, M. Moskovits and H. Kneipp, Springer-Verlag Berlin, Heidelberg, 2006, pp. 427–446 Search PubMed.
- D. Benator, M. Bhattacharya, L. Bozeman, W. Burman, A. Cantazaro, R. Chaisson, F. Gordin, C. R. Horsburgh, J. Horton, A. Khan, C. Lahart, B. Metchock, C. Pachucki, L. Stanton, A. Vernon, M. E. Villarino, Y. C. Wang, M. Weiner and S. Weis, Lancet, 2002, 360, 528–534 CrossRef.
- J. S. Spencer, H. J. Kim, W. H. Wheat, D. Chatterjee, M. V. Balagon, R. V. Cellona, E. V. Tan, R. Gelber, P. Saunderson, M. S. Duthie, S. T. Reece, W. Burman, R. Belknap, W. R. Mac-Kenzie, A. Geluk, L. Oskam, H. M. Dockrell and P. J. Brennan, Clin. Vaccine Immunol., 2011, 18, 260–267 CrossRef CAS PubMed.
- A. J. Anderson, Nature, 1965, 208, 491–492 CrossRef CAS PubMed.
- D. Chatterjee, C. M. Bozic, M. McNeil and P. J. Brennan, J. Biol. Chem., 1991, 266, 9652–9660 CAS.
- A. Vercellone, J. Nigou and G. Puzo, Front. Biosci., 1998, 3, e149–e163 CrossRef CAS.
- P. Ludwiczak, T. Brando, B. Monsarrat and G. Puzo, Anal. Chem., 2001, 73, 2323–2330 CrossRef CAS PubMed.
- L. T. N. Thi, R. B. Maura, S. Férnandez, G. Reyes, J. L. Perez, F. Reyes, M. de los Angeles García, M. Fariñas, J. F. Infante, Y. Tirado, A. Puig, G. Sierra, N. Álvarez, J. C. Ramírez, M. E. Sarmiento, M.-N. Norazmi and A. Acosta, Vaccimonitor, 2010, 19, 20–26 Search PubMed.
- A. K. Singh and J. M. Reyrat, Curr. Protoc. Microbiol., 2009, 14:C:10C.1, 10C.1.1–10C.1.12 Search PubMed.
- J. M. Reyrat and D. Kahn, Trends Microbiol., 2001, 9, 472–474 CrossRef CAS PubMed.
- S. Patil, G. Ramu and R. Prasad, J. Neuroimmunol., 2000, 105, 64–68 CrossRef CAS PubMed.
- C. E. Chan, S. Gotze, G. T. Seah, P. H. Seeberger, N. Tukvadze, M. R. Wenk, B. J. Hanson and P. A. MacAry, Sci. Rep., 2015, 5, 1–12 Search PubMed.
- N. Mdivani, H. Li, M. Akhalaia, M. Gegia, L. Goginashvili, D. S. Kernodle, G. Khechinashvili and Y. W. Tang, Clin. Chem., 2009, 55, 1694–1700 CAS.
- L. J. Jiang, W. J. Wu, H. Wu, S. S. Ryang, J. Zhou, W. Wu, T. Li, J. Guo, H. H. Wang, S. H. Lu and Y. Li, J. Microbiol. Biotechnol., 2012, 22, 1301–1306 CrossRef PubMed.
- W. W. Yew, Clin. Chim. Acta, 2001, 313, 31–36 CrossRef CAS.
- E. Keeler, M. D. Perkins, P. Small, C. Hanson, S. Reed, J. Cunningham, J. E. Aledort, L. Hillborne, M. E. Rafael, F. Girosi and C. Dye, Nature, 2006, 444, 49–57 CrossRef PubMed.
- B. Hamasur, J. Bruchfeld, P. van Helden, G. Källenius and S. Svenson, PLoS One, 2015, 10, e0123457 Search PubMed.
- P. Daley, J. S. Michael, P. Hmar, A. Latha, P. Chordia, D. Mathai, K. R. John and M. Pai, Int. J. Tuberc. Lung Dis., 2009, 13, 989–995 CAS.
- J. Minion, E. Leung, E. Talbot, K. Dheda, M. Pai and D. Menzies, Eur. Respir. J., 2011, 38, 1398–1405 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |