Open Access Article
Open Access ArticleRegulating C4-dicarboxylate transporters for improving fumaric acid production
Ting Zhang†
 ac,
Ruirui Song†a,
Meng Wanga,
Li Deng*ab,
Lihai Fana and
Fang Wanga
ac,
Ruirui Song†a,
Meng Wanga,
Li Deng*ab,
Lihai Fana and
Fang Wanga
aNational Energy R&D Center for Biorefinery, Beijing University of Chemical Technology, Beijing, People's Republic of China. E-mail: dengli@mail.buct.edu.cn
bAmoy—BUCT Industrial Bio-technovation Institute, Amoy, People's Republic of China
cThe 302 Hospital of PLA, China
First published on 12th January 2017
Abstract
Although many efforts have been made to engineer Escherichia coli for fumaric acid production, the fumarate efflux system has not been investigated as an engineering target to improve fumaric acid production. In this work, we cloned and expressed C4-dicarboxylate transporters of different sources in a previously constructed fumaric-acid-producing strain to study their effects on the production of fumaric acid. In addition, each native C4-dicarboxylate transporter was deleted in separate experiments to investigate their individual effects on fumaric acid production. The results showed that the expression of the genes dcuB-Ec and dcuC-Ec can increase the fumaric acid yield by 48.5% and 53.1%, respectively. Fed-batch cultivations in a 5 L bioreactor of strain A-dcuB-Ec produced 9.42 g L−1 of fumaric acid after 50 hours.
Introduction
Fumaric acid (FA) as a C4-dicarboxylic acid is an important raw material and is widely applied.1–4 FA is usually obtained from petroleum-derived chemicals, but limited petroleum resources, rising oil prices, and concerns of environmental pollution caused by chemical synthesis have prompted renewed interest in and study of fumaric acid production from sugars by fermentation using microorganisms.5–11 These investigations have used many microorganisms such as Rhizopus oryzae,5–7 Saccharomyces cerevisiae,8 Escherichia coli9,10 and Candida glabrata.11Making use of the transport of molecular species, such as product efflux and substrate uptake, could increase the efficiency of target compound production. Expression of the genes galP and glk improved alternative glucose utilization under anaerobic conditions.12 Modulating the transporter genes dcuB and dcuC in combination led to a 34% increase of succinic acid (SA) titer.13 For malic acid (MA) production, expression of the malate export gene SpMAE1 in S. cerevisiae resulted in a >10-fold increase of the malate titer.14 Overproduction of organic acids requires the use of a proper exporter to increase the productivity.15 Employing the two dicarboxylic acid transporter genes SFC1 and SpMAE1 facilitated an improvement of fumarate production in C. glabrata.11 However, there has been to our knowledge no report about engineering an export system to improve FA production in E. coli.
The C4-dicarboxylate carriers from bacteria can be classified according to their protein sequences into five coherent groups: the dicarboxylate uptake DcuAB, DcuC, CitT families of proteins, the DctA-type dicarboxylate transporters, and the tripartite ATP-independent periplasmic transporter families.16,17 In E. coli, DctA has been mainly expressed under aerobic conditions, which catalyze the transport of C4-dicarboxylic acids or other amino acids together with the transport of H+/Na+.17,18 The DcuAB family, which could be divided into two subclasses, DcuA and DcuB, has been shown to be mainly involved in fumarate respiration under anaerobic conditions.19–21 The expression of the anaerobic fumarate–succinate antiporter DcuB has been shown to be regulated by DcuSR, a two-component regulatory system.22,23 The DcuC family has been shown to have functions (uptake, exchange and efflux) similar to those of DcuAB.19,24–26 The sequences of DcuD and DcuC proteins have been found to be similar, but the function of DcuD remains unclear.27
In the current work, strain ABCDIA10 was used as a model system to study the C4-dicarboxylate carriers for FA production. Two complementary types of experiments were carried out: various C4-dicarboxylate carriers from different species were expressed in this strain; and the native C4-dicarboxylate carriers were deleted separately to investigate their effects on FA efflux. In addition, the production of other C4-dicarboxylic acids, specifically MA and SA, was monitored to study the functions of the respective C4-dicarboxylate carriers.
Materials and methods
Strains, plasmids and medium
The strains and plasmids used in this study are listed in Table 1. The FA producer strain ABCDIA was used as the original strain. For routine cultures during plasmid construction and strain development, cells were grown in a Luria–Bertani (LB) broth or on LB plates (1.5%, w/v, agar) containing appropriate antibiotics: 50 μg mL−1 of kanamycin (Km), 100 μg mL−1 of ampicillin (Ap). All flask cultures were performed in M9 medium28 supplemented with 15 g L−1 glucose, 3 g L−1 yeast extract, 1 g L−1 citrate, and 3 g L−1 NaHCO3.| Strains | Phenotype | Source |
|---|---|---|
| Klebsiella pneumoniae | KCTC2242, without capsule | Lab collection |
| Lactobacillus rhamnosus 705 | Lactobacillus rhamnosus | Lab collection |
| Clostridium acetobutylicum | Clostridium acetobutylicum 824 | Lab collection |
| Strain ABCDIA | JM109(DE3)-fumA−fumB−fumC−frdABCD−iclR−arcA− | 10 |
| A-dcuA-Ec | Strain ABCDIA overexpressed dcuA-Ec, KanR | This study |
| A-dcuB-Ec | Strain ABCDIA overexpressed dcuB-Ec, KanR | This study |
| A-dcuC-Ec | Strain ABCDIA overexpressed dcuC-Ec, KanR | This study |
| A-dcuD-Ec | Strain ABCDIA overexpressed dcuD-Ec, KanR | This study |
| A-dctA-Ec | Strain ABCDIA overexpressed dctA-Ec, KanR | This study |
| A-dcuA-kp | Strain ABCDIA overexpressed dcuA-kp, KanR | This study |
| A-dcuB-kp | Strain ABCDIA overexpressed dcuB-kp, KanR | This study |
| A-dcuC-kp | Strain ABCDIA overexpressed dcuC-kp, KanR | This study |
| A-dcuD-kp | Strain ABCDIA overexpressed dctA-kp, KanR | This study |
| A-dauA-Ec | Strain ABCDIA overexpressed dauA-Ec, KanR | This study |
| A-00582-Lr | Strain ABCDIA overexpressed 00582-Lr, KanR | This study |
| A-Sdc-Lr | Strain ABCDIA overexpressed Sdc-Lr, KanR | This study |
| A-dcuC-Lr | Strain ABCDIA overexpressed dcuC-Lr, KanR | This study |
| A-dcuB-Ca | Strain ABCDIA overexpressed dcuB-ca, KanR | This study |
| ABCDIA-dcuA | Deleted the dcuA gene in strain ABCDIA | This study |
| ABCDIA-dcuB | Deleted the dcuB gene in strain ABCDIA | This study |
| ABCDIA-dcuC | Deleted the dcuC gene in strain ABCDIA | This study |
| ABCDIA-dcuD | Deleted the dcuD gene in strain ABCDIA | This study |
| ABCDIA-dctA | Deleted the dctA gene in strain ABCDIA | This study |
| Plasmids | Description | Source |
| pET28a-GFP | pET28a+ contained GFP, T7 promoters, KanR | Lab collection |
| pET28a-dcuA-Ec-GFP | pET28a contained gene dcuA-Ec, fusion with GFP, KanR | This study |
| pET28a-dcuB-Ec-GFP | pET28a contained gene dcuB-Ec, fusion with GFP, KanR | This study |
| pET28a-dcuC-Ec-GFP | pET28a contained gene dcuC-Ec, fusion with GFP, KanR | This study |
| pET28a-dcuD-Ec-GFP | pET28a contained gene dcuD-Ec, fusion with GFP, KanR | This study |
| pET28a-dctA-Ec-GFP | pET28a contained gene dctA-Ec, fusion with GFP, KanR | This study |
| pET28a-dcuA-kp-GFP | pET28a contained gene dcuA-kp, fusion with GFP, KanR | This study |
| pET28a-dcuB-kp-GFP | pET28a contained gene dcuB-kp, fusion with GFP, KanR | This study |
| pET28a-dcuC-kp-GFP | pET28a contained gene dcuC-kp, fusion with GFP, KanR | This study |
| pET28a-dcuD-kp-GFP | pET28a contained gene dcuD-kp, fusion with GFP, KanR | This study |
| pET28a-dauA-Ec-GFP | pET28a contained gene dauA-Ec, fusion with GFP, KanR | This study |
| pET28a-00582-Lr-GFP | pET28a contained gene 00582-Lr, fusion with GFP, KanR | This study |
| pET28a-Sdc-Lr-GFP | pET28a contained gene Sdc-Lr, fusion with GFP, KanR | This study |
| pET28a-dcuC-Lr-GFP | pET28a contained gene dcuC-Lr, fusion with GFP, KanR | This study |
| pET28a-dcuB-Ca-GFP | pET28a contained gene dcuB-Ca, fusion with GFP, KanR | This study |
Gene expression and inactivation methods
For the expression of the C4-dicarboxylate transporters, the C4-dicarboxylate transporters genes were respectively cloned from the genome DNA of Escherichia coli, Klebsiella pneumoniae, Lactobacillus rhamnosus and Clostridium acetobutylicum. Each of these genes was fusion expressed in respective experiments with eGFP using the GS linker.For the deletion of the C4-dicarboxylate transporters, the one-step inactivation of chromosomal genes method29 was used. In respective experiments, each one of the genes dcuA, dcuB, dcuC, dcuD and dctA was deleted respectively. All primers used in this study are listed in Table 2.
| Primers | Sequences (5′ to 3′) |
|---|---|
| dcuA-F-NdeI-Ec | CGCCATATGCTAGTTGTAGAACTCA |
| dcuA-R-BamHI-Ec | CGGGATCCAGAACCACCACCACCAGAACCACCACCACCAGAACCACCACCACCAGTAGCCAGCATGAAGCTACCCAG |
| dcuA-F-NdeI-kp | CGCCATATGTTTGGTGCAGAACTGG |
| dcuA-R-BamHI-kp | CGGGATCCAGAACCACCACCACCAGAACCACCACCACCAGAACCACCACCACCAGTAGCCAGCATAATCCCGCC |
| dcuB-F-NheI-Ec | CGGCTAGCATGTTATTTACTATCCAAC |
| dcuB-R-BamHI-Ec | CGGGATCCAGAACCACCACCACCAGAACCACCACCACCAGAACCACCACCACCAGTAGCTAAGAACCCGTACATCG |
| dcuB-F-NdeI-kp | CGCCATATGGAATTCGCGATACAAC |
| dcuB-R-BamHI-kp | CGGGATCCAGAACCACCACCACCAGAACCACCACCACCAGAACCACCACCACCAGTAGCCAGAAAGCCGTACATCG |
| dcuC-F-NdeI-Ec | CGCCATATGCTGACATTCATTGAGC |
| dcuC-R-BamHI-Ec | CGGGATCCAGAACCACCACCACCAGAACCACCACCACCAGAACCACCACCACCAGTAGCCTTGCCTGTGACCGCTGC |
| dcuC-F-NdeI-kp | CGCCATATGCTGACGT TTATTGAGA |
| dcuC-R-BamHI-kp | CGGGATCCAGAACCACCACCACCAGAACCACCACCACCAGAACCACCACCACCAGTAGCGTGGAGCGCGCTGCCGGGC |
| dcuD-F-NdeI-Ec | CGCCATATGTTCGGCATAATTATATC |
| dcuD-R-BamHI-Ec | CGGGATCCAGAACCACCACCACCAGAACCACCACCACCAGAACCACCACCACCAGTAGCATAAAATAGCGTGATTGTGG |
| dctA-F-NdeI-Ec | CGCCATATGAAAACCTCTCTGTTTAAAAGCC |
| dctA-R-BamHI-Ec | CGGGATCCAGAACCACCACCACCAGAACCACCACCACCAGAACCACCACCACCAGTAGCAGAGGATAATTCGTGCGTT |
| dctA-F-NheI-kp | CGGCTAGCATGAAAACCTCTATCTTTAAAAGC |
| dctA-R-BamHI-kp | CGGGATCCAGAACCACCACCACCAGAACCACCACCACCAGAACCACCACCACCAGTAGCAGAGGATAATTCGTGCGTT |
| dauA-F-NdeI-Ec | CGCCATGTGAACAAAATATTTTCCTCAC |
| dauA-R-BamHI-Ec | CGGGATCCAGAACCACCACCACCAGAACCACCACCACCAGAACCACCACCACCAGTAGCTAAATCCGCCATCGCCGC |
| dcuB-F-NdeI-Ca | CGCCATATGTTTTGGGTAGAGTTAG |
| dcuB-R-BamHI-Ca | CGGGATCCAGAACCACCACCACCAGAACCACCACCACCAGAACCACCACCACCAGTAGCGAAAACGATTGCACCAAGTA |
| sdcS-F-NdeI-Lr705 | CGCCATATGCAAACTACGGCAAAAGCTTTTAG |
| sdcS-R-BamHI-Lr705 | CGGGATCCAGAACCACCACCACCAGAACCACCACCACCAGAACCACCACCACCAGTAGCATTAAAGATCAGTGACGCA |
| dcuC-F-NdeI-Lr705 | CGCCATATGAACGCTATTTTGATCTC |
| dcuC-R-BamHI-Lr705 | CGGGATCCAGAACCACCACCACCAGAACCACCACCACCAGAACCACCACCACCAGTAGCAAAGAGTATCAGTGAC |
| L00582-F-NdeI-Lr705 | CGCCATATGTATAAACGGTACAAAGATG |
| L00582-R-BamHI-Lr705 | CGGGATCCAGAACCACCACCACCAGAACCACCACCACCAGAACCACCACCACCAGTAGCCCACTTTTTGGTCGTGGCAC |
| dcuA-F1 | AACAAGGAAGGCTAATATGC |
| dcuB-F1 | AATAGGATTATCGCGAGG |
| dcuB-R1 | TTATTCGAAAATGGCGTG |
| dcuA-R1 | TGTTAGTCGGCGGGAAAG |
| dcuC-F1 | TTACTTTGGGGCCTGGAGAC |
| dcuC-R1 | GCATTACTATTTACTTGCCTGT |
| dcuD-F1 | GGATTGACCGTTTTCTTTTGA |
| dcuD-R1 | GCCCAGGCGGCCTGTTTTG |
| dctA-F1 | ACTAAAGGACACCCTATG |
| dctA-R1 | CGGGCATAAGTGATGAGA |
| dcuA-ko-F | TAGTTGTAGAACTCATCATAGTTTTGCTGGCGATCTTCTTGGGCGCCAGCTGTCAAACATGAGAATTAA |
| dcuA-ko-R | GCGTGAACGCCCCGCGATTGATCATTACAGCATGAAGCTACCCAGCACGAGTGTAGGCTGGAGCTGCTTC |
| dcuB-ko-F | GTTCACACATGTTATTTACTATCCAACTTATCATAATACTGATATGTCTGCTGTCAAACATGAGAATTAA |
| dcuB-ko-R | CGCGTAGTCACGGGCGGCACGCAAAGTGCATTTATAAGAACCCGTACATGTGTAGGCTGGAGCTGCTTC |
| dcuC-ko-F | AGGAAAAATTATGCTGACATTCATTGAGCTCCTTATTGGGGTTGTGGTTACTGTCAAACATGAGAATTAA |
| dcuC-ko-R | GACCGCTGCTGCCGTTCCTGGCACCATCAGCTCTGTAGCAACGATAACAGTGTAGGCTGGAGCTGCTTC |
| dcuD-ko-F | CACGGAGTTCAACAATGTTCGGCATAATTATATCTGTCATCGTATTAACTGTCAAACATGAGAATTAA |
| dcuD-ko-R | TCTTTTTAATGACTTAATAAAATAGCGTGATTGTGGCAATCATATTCACCGTGTAGGCTGGAGCTGCTTC |
| dctA-ko-F | AACCTCTCTGTTTAAAAGCCTTTACTTTCAGGTCCTGACAGCGATAGCCCTGTCAAACATGAGAATTAA |
| dctA-ko-R | TTAAGAGGATAATTCGTGCGTTTTGCCATCCGGCGCACGATTATTCAGCGTGTAGGCTGGAGCTGCTTC |
Cultivation
For FA fermentation, the cells were inoculated into 4 mL LB medium and cultivated overnight in a rotary shaker at 37 °C, 180 rpm. Then 500 μL cultures were transferred into a 50 mL Erlenmeyer flask containing 20 mL LB medium for the seed cultures. The intensity of the absorption at a wavelength of 600 nm (OD600) can allow for an approximation of the biomass concentration in the culture. When the OD600 reached ca. 0.6, the cells were transferred into a 500 mL Erlenmeyer flask containing 50 mL of fermentation culture, making sure the initial OD600 was 0.1. Use of the same initial inoculum OD allowed for a convenient comparison of the growth rates of the strains. The specific use of the initial inoculum OD600 of 0.1 for all fermentation culture experiments resulted in the strains reaching log phase relatively easily and rapidly. All of the flasks were cultivated at 37 °C, 220 rpm in a rotary shaker. For overexpression of the C4-dicarboxylate transporter, 1 mM IPTG was added when the OD600 was about 0.6 (i.e., at approximately three hours of being cultured). After the IPTG was added, the contents of the flasks were cultured at the 30 °C, 220 rpm condition.9,10Fed-batch fermentations were carried out in a 5 L jar fermentor containing 3 L of M9 medium supplemented with 15 g L−1 of glucose, 3 g L−1 of yeast extract and 1 g L−1 of citrate. Seed cultures were prepared by transferring samples of 4 mL of overnight-grown culture in LB medium into 150 mL of the same medium in 500 mL Erlenmeyer flasks and incubating them in a rotary shaker for 3 h at 220 rpm and 37 °C. The seed culture (300 mL) was added to the fermentor to give the initial OD600 of ca. 0.1. The culture pH was maintained at 7.0 using 30% (w/v) Na2CO3. The amount of dissolved oxygen was kept at 40% (v/v) by flowing 6 L min−1 of air, and the agitation speed was 300 rpm. The feeding solution contained 700 g L−1 of glucose and 8 g L−1 of MgSO4·7H2O. When the glucose concentration of the culture broth decreased to below 5 g L−1, feeding solution was added to adjust the glucose concentration in the fermentor to ca. 20 g L−1.9,10
Expression analyses and confocal laser scanning microscopy (CLSM) observations
Strains were grown overnight in 4 mL LB medium, then diluted to OD600 0.1 in the same medium, and after three hours of cultivation, 0.1% of IPTG was added and cultured for six hours at 30 °C. The eGFP fluorescence intensity of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells was measured using flow cytometry.
000 cells was measured using flow cytometry.
The induced recombinant strains were harvested by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and washed two times with PBS. Images were acquired on a Lecia TCS SP2/AOBS (Germany). Excitation light at a wavelength of 488 nm was used to excite the eGFP.
000g and washed two times with PBS. Images were acquired on a Lecia TCS SP2/AOBS (Germany). Excitation light at a wavelength of 488 nm was used to excite the eGFP.
Dry cell weight (DCW) determination
Cell concentrations were measured using a spectrophotometer at a wavelength of 600 nm after an appropriate dilution. The optical density at 600 nm (OD600) was converted to DCW according to the calibration curve DCW![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OD600 = 0.38
OD600 = 0.38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (g L−1), which showed an R2 = 0.99.
1 (g L−1), which showed an R2 = 0.99.
Analysis of C4-dicarboxylic acids and glucose
The quantification of organic acids and glucose in the culture supernatant was performed using HPLC analysis. The organic acids were detected at a wavelength of 210 nm with an Aminex HPX-87H column (25 cm, 0.4 cm i.d., Bio-Rad, USA) on an UltiMate 3000 HPLC system (Dionex, USA). The glucose was detected using an Refractive Index detector connected serially to the same HPLC system. The mobile phase consisted of 5 mmol L−1 H2SO4; the flow rate was 0.6 mL min−1 and the column temperature was 50 °C.Results and discussion
The effect of deleted transporters
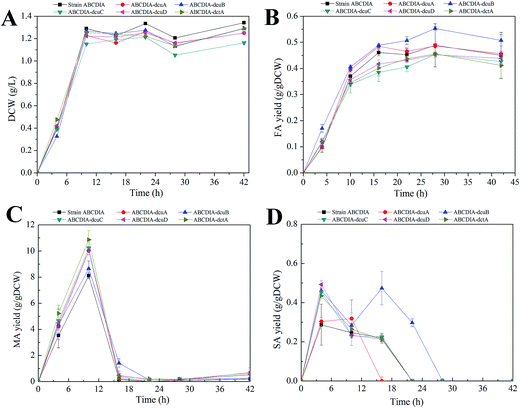
The effect of the transporters was investigated in a genetically engineered strain called strain ABCDIA, which has been shown to be able to produce an appreciable amount of FA through the deletion of the genes fumABC, frdABCD, iclR and arcA.10In order to investigate the effect of the C4-dicarboxylate transporters on FA production, the C4-dicarboxylate transporter genes (dcuA, dcuB, dcuC, dcuD, dctA) were individually deleted in separate experiments. The cell growth and C4-dicarboxylic acid production of the strains with these deletions are shown in Fig. 1. The dry cell weight was chosen to assess strain growth. As shown in Fig. 1A, the deletion of each transporter led to a decrease of the biomass. Deletion of the dcuC gene yielded an especially significant inhibition of the cell growth. The cell growth rate and the maximal biomass of strain ABCDIA-dcuC were decreased by 11.5% and 9.1%, respectively, relative to those of the control strain (Fig. 1A).
At the same time, the three kinds of C4-dicarboxylic acids (FA, MA and SA) produced in the fermentation broth by the recombinant strains were detected (Fig. 1B–D). As shown in Fig. 1B, the deletion of the dcuB gene increased the FA yield from 0.488 g gDCW−1 to 0.55 g gDCW−1. The FA efflux rate of strain ABCDIA-dcuB was 0.042 g (gDCW h)−1, which was 1.75-fold higher than that of the control strain ABCDIA. The deletions of the dcuB and dcuC genes seemed, according the data, to each have resulted in significant changes in FA yield. The deletion of dcuA had little effect on the FA yield, and the deletions of dcuC, dcuD and dctA decreased the FA yield by 16.49%, 9.5% and 12.8%, respectively, in 16 hours (Fig. 1B). As shown in Fig. 1B, the FA yield was decreased after 28 hours of fermentation. We believe that at the later period of fermentation, the main carbon source glucose was exhausted, and the organic acids were used as a kind of substitute carbon.
As shown in Fig. 1C, the MA yield was increased as a result of the deletions of the transporter genes. The MA yield of strain ABCDIA-dctA was increased by 42.8% relative to that of the control strain. Moreover, the MA yields of strains ABCDIA-dcuA, ABCDIA-dcuB, ABCDIA-dcuC and ABCDIA-dcuD were increased by 22.1%, 13.1%, 27% and 22.4%, respectively, relative to that of the control.
The SA yields produced by the recombinant strains are shown in Fig. 1D. These recombinant strains produced little SA, which was taken up and reused at the later period of the fermentation (Fig. 1D). The SA yields reached their maxima in 6 hours, except for strain ABCDIA-dcuB, and the deletion of dcuA led to an approximately 50% decrease in SA production. The produced SA was taken up at 22 hours of fermentation, except for the ABCDIA-dcuB strain, which showed a significant increase at 16 hours. Moreover, the time it took for SA in the fermentation broth to be completely taken up by the ABCDIA-dcuA strain was 16 hours; but in the ABCDIA-dcuB strain, the time it took for SA to become exhausted was delayed to 28 hours (Fig. 1D). These results indicated that the uptake rate of SA was speeded up by the deletion of dcuA but slowed down by the deletion of dcuB.
In most conditions, changes of biomass significantly affect the production of the aimed for product. Generally, the increase of biomass means more strain to produce this product. Sometimes the biomass has been observed to decrease after the genetic modification, yet the yield of product increased. For example, in our current work, the biomass decreased when the dcuB gene was deleted, but the yield of FA increased. When we expanded the culture to increase the biomass, the FA production increased significantly (Fig. 4).
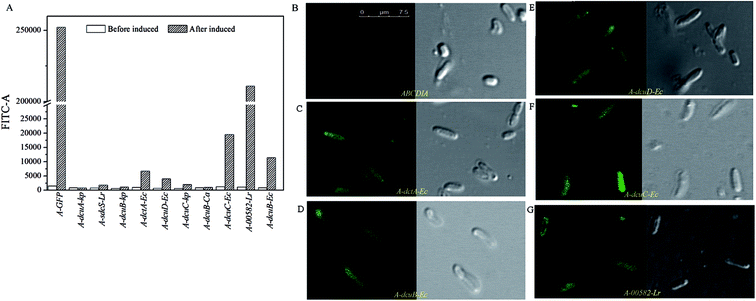 | ||
| Fig. 2 (A) The fluorescence intensity levels of eGFP of the recombinant strains. (B–G) The results of the CLSM observations of the recombinant strains. | ||
The expression of C4-dicarboxylate transporters
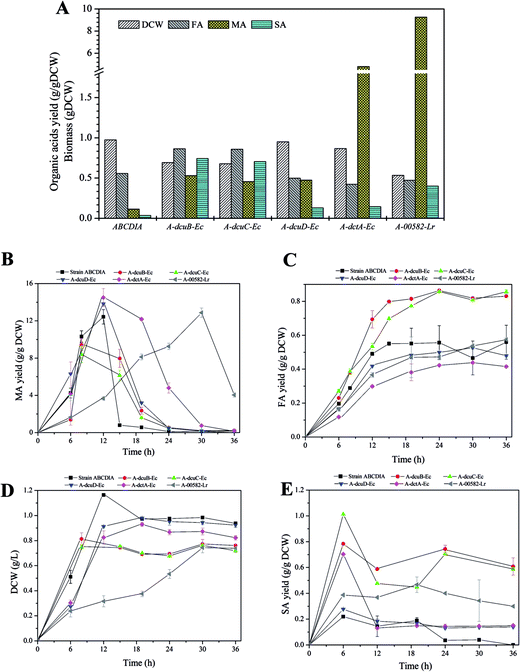
The C4-dicarboxylate transporters genes dcuA-Ec, dcuB-Ec, dcuC-Ec and dctA-Ec were cloned from the genome DNA of E. coli, and the genes dcuA-Kp, dcuB-kp and dcuC-kp were cloned from the K. pneumoniae genome DNA. The C4-dicarboxylate transporters genes 00582-Lr and sdcS-Lr were cloned from the genome DNA of L. rhamnosus 705 and gene dcuB-Ca was cloned from C. acetobutylicum genome DNA.The recombinant plasmid containing the transporter gene fused with eGFP was transformed into strain ABCDIA. After induction with IPTG, the expression level of the transporters could be estimated by the fluorescence intensity of eGFP. We used flow cytometry and CLSM to verify the expression and location of the eGFP fusion. As shown in Fig. 2A, the eGFP fluorescence of the recombinant strains was detected using flow cytometry. Here, after induction, the highest fluorescence intensity was observed for the positive control strain A-GFP. The fluorescence intensities of strains A-dcuB-Ec, A-dcuC-Ec, A-dcuD-Ec, A-dctA-Ec and A-00582-Lr were observed to increase after induction. We believe that the dcuB-Ec, dcuC-Ec, dcuD-Ec, dctA-Ec and 00582-Lr C4-dicarboxylate transporters were correctly folded. However, the fluorescence intensities of other strains after induction did not show a noticeable increase, indicating that the C4-dicarboxylate transporters of K. pneumoniae and C. acetobutylicum were either not expressed or not correctly folded in E. coli. In these cases, we chose strains A-dcuB-Ec, A-dcuC-Ec, A-dcuD-Ec, A-dctA-Ec and A-00582-Lr as samples to investigate the effects of these C4-dicarboxylate transporters on the yields of fumaric acid and other C4-dicarboxylic acids.
The effect of the overexpressed transporters
The overexpression of the transporters was observed to make a difference in DCW and FA yield (Fig. 3A). The cell growth rates (gDCW h−1) of strains A-dcuB-Ec and A-dcuC-Ec were 0.102 and 0.094, and that of the control strain ABCDIA was 0.097. However, the biomass levels of strains with overexpressed dcuB-Ec and dcuC-Ec were decreased, respectively, by 28.8% and 30.4% in 24 hours relative to that of the control strain ABCDIA (Fig. 3D), which was consistent with previous results.30 Moreover, the FA yields of strains A-dcuB-Ec and A-dcuC-Ec were increased by 48.5% and 53.1%, respectively, in 36 hours relative to that of the control strain ABCDIA (Fig. 3C). In a previous experiment (Fig. 1), after the gene dcuB was deleted, the FA yield of the recombinant strain was increased by 12.7%. To the best of our knowledge, the main function of dcuB is external C4-dicarboxylate absorption.22,23 For this reason, deletion of dcuB resulted in the enhancement of FA yield. However, the FA yield was increased to a greater extent when the dcuB was overexpressed under aerobic conditions. The fumarate channel of DcuB has been suggested to allow for transport in both directions under aerobic conditions. To confirm this view, we propose that the recombinant strain with 14C-FA should, in a laboratory with appropriate qualifications, be cultured to determine the transport direction of the DcuB channel. (Please note that our laboratory does not have such qualifications.)In contrast to the control strain ABCDIA, the increase of the FA yield in both strains A-dcuB-Ec and A-dcuC-Ec coincided with a significant decrease of MA yield (Fig. 3B). Here it was demonstrated for the first time that, under aerobic conditions, transporters DcuB and DcuC were able to facilitate fumarate excretion accompanied with malate absorption. However, previous reports only indicated the function of DcuB and DcuC under anaerobic conditions being involved in succinate efflux during glucose fermentation.13,19,20,24 Meanwhile, compared to the control strain ABCDIA, the overexpression of genes dcuD-Ec and dctA-Ec resulted in lower cell growth rates (0.076 and 0.069 gDCW h−1), lower FA yields (decreased by 14.3% and 25.9%) and higher MA yields (increased by 11% and 17%) (Fig. 3B–D). As shown in Fig. 3E, the strains A-dcuB-Ec and A-dcuC-Ec could yield 0.78 and 1.01 gSA gDCW−1 respectively, while the control strain ABCDIA only produced a small amount of SA (∼0.2 g gDCW−1).
With respect to the overexpression of the heterogeneous transporter gene 00582-Lr, the cell growth and C4-dicarboxylic acid yield of strain A-00582-Lr showed significantly different trends than did the other strains (Fig. 3B–D). The cell growth rate was only 0.025 gDCW h−1, and after 24 hours the DCW was 45.3% less than that of the control strain (Fig. 3B). In addition, the FA yield of strain A-00582-Lr was observed to continuously increase to 0.57 g gDCW−1 over the course of 36 hours, and this yield was 3.24% greater than that of the control (Fig. 3B). In addition, at the earlier stage of the fermentation, the MA yield of strain A-00582-Lr was quite low and reached its maximum in 30 hours (Fig. 3D). As for the production of SA, the expression of the 00582-Lr gene increased the yield of this C4-dicarboxylic acid (0.47 g gDCW−1 vs. 0.19 g gDCW−1).
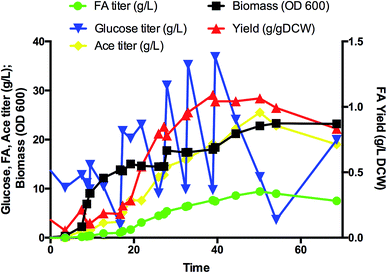
The fed-batch fermentation
Flask shaking yielded greater FA yields for both strains ABCDIA-dcuB (in which dcuB was deleted) and A-dcuB-Ec (in which dcuB was overexpressed) than for the original strain ABCDIA. In addition, the function of ducB under anaerobic conditions was mainly fumarate uptake.19–21 We suggest that a laboratory with appropriate qualifications deploy isotope labelling of FA to verify the transport direction in DcuB channel. (Note that we do not have such qualifications in our laboratory.) The increased FA yield of strain A-dcuB-Ec was higher than that of strain ABCDIA-dcuB. It was suggested that the efflux effect was greater than the absorption. In addition, the other strains constructed did not show any greater positive effects on the FA production than strain A-dcuB-Ec. Thus, we further studied the best FA-producing strain, i.e., A-dcuB-Ec, in a 5 L aerobic fed-batch fermentor (Fig. 4). The culture produced over 9.4 g L−1 FA from glucose in 50 hours, and the yield was 1.07 g gDCW−1. Although the maximum yield of FA was 1.09 g gDCW−1 at 39 hours, the FA titer was 7.62 g L−1. At the later of the fermentation, after the supplementation of glucose, the strains preferred to produce more acetate than FA. How to avoid the overproduction of acetate will be the main focus of further studies. The titer of the major acetate byproduct was approximately 25 g L−1. We recognize that commercialization will require another three- to five-fold improvement in the FA production, and to this end we are taking steps to raise the rates of key steps in the pathway, to remove metabolic inefficiencies, and to substantially reduce the amounts of byproducts. Fermentation process engineering can also optimize the feeding and aeration strategy to achieve higher cell densities and improve the concentration of specific products.Conclusions
In order to increase the efficiency of the production of target compounds, optimizing the product efflux system is an effective strategy. In this work, the direction of the C4-dicarboxylate carrier exchange was investigated. The fumarate channel of DcuB was found to be a two-way channel, and the efflux effect was found to be greater than the uptake effect. In the end, the FA titer and yield were 9.42 g L−1 and 1.07 g gDCW−1 when dcuB-Ec was overexpressed in a 5 L fermentor. To the best of our knowledge, there have been no previous reports on FA production under aerobic conditions involving an engineered C4-dicarboxylate transportation system in E. coli.Acknowledgements
This work was supported by the National High Technology Research and Development Program and Development Program of China (863 program, Grant No. 2014AA020522), Amoy Industrial Biotechnology R&D and Pilot Conversion Platform (No. 3502Z20121009), and the National Nature Science Foundation of China (21390202, 81372248).References
- I. Goldberg, J. S. Rokem and O. Pines, J. Chem. Technol. Biotechnol., 2006, 81, 1601–1611 CrossRef CAS.
- R. C. A. Engel, A. J. J. Straathof, T. W. Zijlmans, W. M. Gulik and L. A. M. Wielen, Appl. Microbiol. Biotechnol., 2008, 78, 379–389 CrossRef PubMed.
- W. L. Araújo, A. Nunes-Nesi and A. R. Fernie, Phytochemistry, 2011, 72, 838–843 CrossRef PubMed.
- T. Werpy, G. Petersen, A. Aden, J. Bozell, J. Holladay, J. White, A. Manheim, D. Eliot, L. Lasure and S. Jones, Synthetic Fuels, 2004 Search PubMed.
- B. Zhang, C. D. Skory and S.-T. Yang, Metab. Eng., 2012, 14, 512–520 CrossRef CAS PubMed.
- C. Gu, Y. Zhou, L. Liu, T. Tan and L. Deng, Bioresour. Technol., 2013, 131, 303–307 CrossRef CAS PubMed.
- Q. Xu, S. Li, Y. Fu, C. Tai and H. Huang, Bioresour. Technol., 2010, 101, 6262–6264 CrossRef CAS PubMed.
- G. Xu, L. Liu and J. Chen, Microb. Cell Fact., 2012, 11, 24 CrossRef CAS PubMed.
- C. W. Song, D. I. Kim, S. Choi, J. W. Jang and S. Y. Lee, Biotechnol. Bioeng., 2013, 110, 2025–2034 CrossRef CAS PubMed.
- T. Zhang, Z. Wang, L. Deng, T. Tan, F. Wang and Y. Yan, Appl. Microbiol. Biotechnol., 2015, 99, 5033–5044 CrossRef CAS PubMed.
- X. Chen, X. Dong, Y. Wang, Z. Zhao and L. Liu, Metab. Eng., 2015, 31, 62–73 CrossRef CAS PubMed.
- J. Lu, J. Tang, Y. Liu, X. Zhu, T. Zhang and X. Zhang, Appl. Microbiol. Biotechnol., 2012, 93, 2455–2462 CrossRef CAS PubMed.
- J. Chen, X. Zhu, Z. Tan, H. Xu, J. Tang, D. Xiao and X. Zhang, Appl. Microbiol. Biotechnol., 2014, 98, 2197–2205 CrossRef CAS PubMed.
- M. R. Zelle, E. d. Hulster, W. A. van Winden, P. d. Waard, C. Dijkema, A. A. Winkler, J.-M. A. Geertman, J. P. v. Dijken, J. T. Pronk and A. J. A. v. Maris, Appl. Environ. Microbiol., 2008, 74, 2766–2777 CrossRef PubMed.
- E. Jamalzadeh, P. J. T. Verheijen, J. J. Heijnen and W. M. van Gulik, Appl. Environ. Microbiol., 2011, 78, 705–716 CrossRef PubMed.
- I. Janausch, E. Zientz, Q. H. Tran, A. Kroger and G. Unden, Biochim. Biophys. Acta, Mol. Cell Res., 2002, 1553, 39–56 CrossRef CAS.
- I. Janausch, O. Kim and G. Unden, Arch. Microbiol., 2001, 176, 224–230 CrossRef CAS PubMed.
- I. T. Paulsen, M. K. Sliwinski and M. H. Saier Jr, J. Mol. Biol., 1998, 277, 573–592 CrossRef CAS PubMed.
- E. Zientz, S. Six and G. Unden, J. Bacteriol., 1996, 178, 7241–7247 CrossRef CAS PubMed.
- S. Six, S. C. Andrews, G. Unden and J. R. Guest, J. Bacteriol., 1994, 176, 6470–6478 CrossRef CAS PubMed.
- P. Golby, D. J. Kelly, J. R. Guest and S. C. Andrews, J. Bacteriol., 1998, 180, 6586–6596 CAS.
- E. Zientz, J. Bongaerts and G. Unden, J. Bacteriol., 1998, 180, 5421–5425 CAS.
- A. Kleefeld, B. Ackermann, J. Bauer, J. Krämer and G. Unden, J. Biol. Chem., 2008, 284, 265–275 CrossRef PubMed.
- E. Zientz, I. Janausch, S. Six and G. Unden, J. Bacteriol., 1999, 181, 3716–3720 CAS.
- E. Zientz, I. G. Janausch, S. Six and G. Unden, J. Bacteriol., 1999, 181, 3716–3720 CAS.
- E. Zientz, S. Six and G. Unden, J. Bacteriol., 1997, 178, 7241–7247 CrossRef.
- I. G. Janausch and G. Unden, Arch. Microbiol., 1999, 172, 219–226 CrossRef CAS PubMed.
- T. Maniatis, E. F. Fritsch and J. Sambrook, Molecular cloning: a laboratory manual, CSHL Press, 1982 Search PubMed.
- K. A. Datsenko and B. L. Wanner, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 6640–6645 CrossRef CAS PubMed.
- J. J. Beauprez, M. R. Foulquie-Moreno, J. Maertens, E. van Horen, K. Bekers, G. J. E. Baart, R. M. Cunin, D. Charlier, J. J. Heijnen and W. Soetaert, Green Chem., 2011, 13, 2179–2186 RSC.
Footnote |
| † These authors contributed equally to this paper. |
| This journal is © The Royal Society of Chemistry 2017 |