Open Access Article
Open Access ArticleBifunctional cationic solid lipid nanoparticles of β-NaYF4:Yb,Er upconversion nanoparticles coated with a lipid for bioimaging and gene delivery
Chenxi Song ab,
Shubiao Zhang
ab,
Shubiao Zhang *b,
Quan Zhoub,
Lei Shic,
Linying Duc,
Defu Zhib,
Yinan Zhaob,
Yuhong Zhenc and
Defeng Zhao*ad
*b,
Quan Zhoub,
Lei Shic,
Linying Duc,
Defu Zhib,
Yinan Zhaob,
Yuhong Zhenc and
Defeng Zhao*ad
aState Key Laboratory of Fine Chemicals, Dalian University of Technology, Dalian 116012, Liaoning, China. E-mail: zhaodfg@chem.dlut.edu.cn
bKey Laboratory of Biotechnology and Bioresources Utilization, Ministry of Education, Dalian Minzu University, Dalian 116600, Liaoning, China. E-mail: zsb@dlnu.edu.cn
cCollege of Pharmacy, Dalian Medical University, 116044 Dalian, China
dZhejiang Jihua Group Ltd. Co., Hangzhou Qianjiang Specially Engaged Expert, Hangzhou, China
First published on 17th May 2017
Abstract
β-NaYF4:Yb,Er upconversion nanoparticles (UCNPs) were synthesized by a one-pot solvothermal method. The UCNPs were characterized through X-ray diffraction (XRD), scanning electron microscopy (SEM) and transmission electron microscopy (TEM) to prove that they possessed hexagonal crystal phase with uniform particle size distribution. The cationic solid lipid nanoparticles (CSLNs) were then obtained by coating the UCNPs with a lipid. Further analysis by FTIR, XRD and TEM indicated the lipid can successfully coat UCNPs, and the coating process had no obvious influences on morphology and crystallization of the UCNPs. Furthermore, the CSLNs were used for bioimaging in cells and delivering small interfering RNA (siRNA). Cellular bioimaging and upconversion luminescence studies clearly demonstrated that the CSLNs could effectively enter into cells with the help of the lipid. Silencing assays and cytotoxicity in A549 cells convincingly showed that the CSLNs could effectively deliver siRNA and possessed low cytotoxicity and good biocompatibility. Thus, the bifunctional CSLNs could be used as a potential candidate in bioimaging and therapeutic applications.
1. Introduction
Conventional downconversion fluorescent materials, including organic dyes,1 semiconductor nanoparticles (such as CdS, CdSe, CdTe)2 and dye-coupled hybrid materials (carbon nanotubes3 and mesoporous silica4) are commonly used fluorophores for biological studies and clinical application due to their unique features. However, they also have many intrinsic limitations, such as weak photostability, broad absorption and emission bands.1,5 In addition, these downconversion fluorescent materials usually emit one lower-energy photon after absorption of a higher-energy ultraviolet (UV) or visible photon. The use of higher-energy light is also associated with several significant disadvantages, such as low light-penetration depth, possible severe photodamage to living organisms, and the autofluorescence noise of biological samples.6To solve these problems, the development of alternative biological luminescent labels through the use of UCNPs has attracted tremendous attention due to the unique luminescence properties of rare-earth nanoparticles, such as sharp absorption and emission lines, high quantum yields, long lifetimes and superior photostability.7–11 Among all the UCNPs, fluorides are considered as a best host material while oxides have high vibration energies which tend to quench the quantum yield of the luminescence.12–14 In particular, upconversion is a process where low-energy light, usually near-infrared (NIR) or infrared (IR), is converted to higher-energy light (UV or visible), through multiple photon absorptions or energy transfers.15 In comparison with downconversion fluorescent materials, upconversion rare-earth materials have many advantages in biological applications, such as noninvasive and deep penetration of NIR radiation, the absence of autofluorescence of biological tissues and feasibility of multiple labeling by UCNPs with different emissions under same excitation.7
Unfortunately, despite recent advances in the synthetic methods for controlling the size and shape of UCNPs,16–18 the UCNPs with hydrophobic organic ligands (such as oleic acid) coating their surface cannot be used directly in biological applications because of very low solubility in water and unfavorable surface properties. So a prerequisite for the development of UCNP-based biological labels is to gain access to water-soluble nanoparticles bearing appropriate functional groups (such as –COOH, –NH2, or –SH) on their surface.19 But these methods are still limited by the biocompatibility and toxicity of the modified UCNPs.20
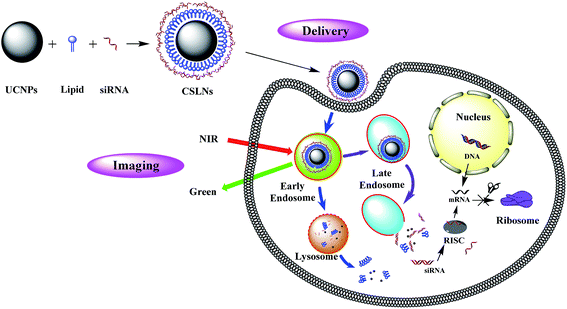
Cationic lipids for gene delivery could be used to coat UCNPs, because of their handy synthesis, potential to incorporate targeting ligands, low immune response, and good safety.21 As peptides based cationic lipids have shown more superiorities over other cationic lipids, such as good biodegradability, excellent biocompatibility and targeting ability to cells,22–24 the solid lipid nanoparticles (SLNs) obtained by coating UCNPs with peptides based lipids would show promising applications both for bioimaging and gene delivery. Therefore, we demonstrate the possibility of novel bifunctional CSLNs for bioimaging and gene delivery through peptide lipid coated UCNPs, the structure and gene delivery of CSLNs were illustrated in Fig. 1.
2. Results and discussion
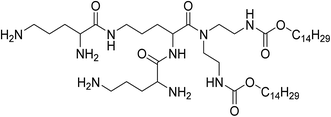
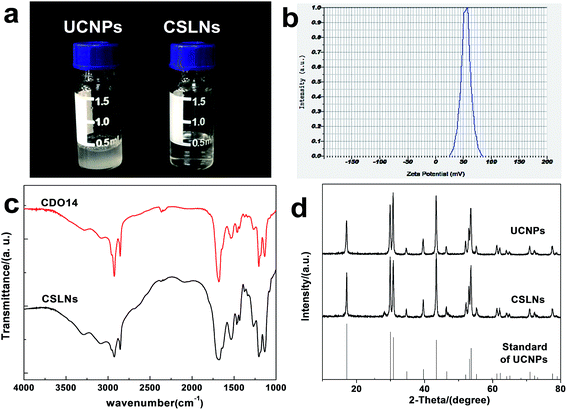
UCNPs were synthesized by a one-pot solvothermal method, as among all the methods for the synthesis of UCNPs solvothermal reaction with high-boiling solvent is an efficient way to obtain UCNPs with small size and high upconversion efficiency. CSLNs were then obtained through the simple and effective thin-film dispersion method, in which UCNPs were coated with a peptide cationic lipid CDO14, and the chemical structure shown in Fig. 2. Our previous study has proved that this lipid had superior gene delivery ability in vitro and in vivo.25As shown in Fig. 3a, the CLSNs formed a clearer aqueous solution compared with UCNPs at the same concentration (1 mg mL−1). The zeta-potential of CSLNs was also measured with a Zeta Sizer at room temperature (Fig. 3b). The zeta-potential was +54.4 mV and show that the particles were positively charged. The photos of the aqueous dispersions and zeta-potential measurement revealed the obvious phase-transfer indeed took place resulting in stable dispersions in water. Moreover, the FTIR spectrum (Fig. 3c) of CLSNs was nearly consistent with that of CDO14, which proved the successful coating of UCNPs with the lipid. The structure of the UCNPs and CSLNs was determined by XRD. Fig. 3d showed the XRD patterns of the UCNPs and the CSLNs. All the peaks in XRD of the products were indexed well with the standard XRD pattern (JCPDS no. 28-1192) of β-NaYF4:Yb,Er. The XRD pattern confirmed the hexagonal crystal phase of β-NaYF4:Yb,Er, which is very beneficial for obtaining bright luminescence. Moreover, no YbF4 and ErF4 were observed to indicate that during solvothermal synthesis process, YCl3·6H2O, YbCl3·6H2O and ErCl3·6H2O were completely dissolved and the dopants were successfully inserted into the host. And the size of the UCNPs was 38 nm calculated using Scherrer formula, D = nλ/β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θB (n = 1). Where D represents the crystallite size (nm), λ is the wavelength of the target used, θB is half the diffraction angle and β is full-width at half maximum for this angle.26–29
θB (n = 1). Where D represents the crystallite size (nm), λ is the wavelength of the target used, θB is half the diffraction angle and β is full-width at half maximum for this angle.26–29
The morphology of the UCNPs and CSLNs was investigated by SEM as shown in Fig. 4(a and b). The particle size was about 40 nm, which was consistent well with the XRD result. It can be clearly seen that the UCNPs were in interparticle agglomeration state, whereas CSLNs particles did not show obvious aggregation to indicate that they could be dispersed better in water. Typical TEM image given in Fig. 4c confirmed the uniform distribution of the hexagonal phase of β-NaYF4:Yb,Er nanoparticles and the size (from 38 to 44 nm) conformed to the XRD and SEM results. After coating with the lipid, the size of CSLNs increased to 45–50 nm and the membrane could be observed as indicated with the arrow in Fig. 4d. During the preparation of UCNPs, oleic acid was used to cap their surface; therefore, after the addition of the lipid they interact through the hydrophobic forces to cause the formation of a lipid membrane with the headgroups towards outside making the particles hydrophilic.
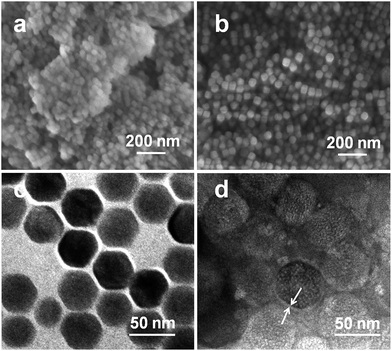 | ||
| Fig. 4 SEM images of (a) UCNPs and (b) CSLNs and TEM images of (c) UCNPs and (d) CSLNs. Scale bars (a = 500 nm, b = 500 nm, c = 50 nm, d = 50 nm). | ||
The electrostatic binding interaction between CSLNs and siRNA is a key factor for gene transfection.30–32 It is known that once the negatively charged siRNA complexed with the positively charged vector, the migration of siRNA in electric field toward the anode will be retarded due to the charge neutralization. Therefore, the electrostatic binding interactions between CSLNs and siRNA were confirmed by the gel retardation assay. The results of the gel retardation assay indicated that the binding ability between CSLNs and siRNA increased with the increasing mass ratio of CSLNs to siRNA. As shown in Fig. 5, the mobility of siRNA was not observed when CSLNs were complexed with siRNA at the ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. No obvious bands were visualized with the CSLNs/siRNA ratios of 6
1. No obvious bands were visualized with the CSLNs/siRNA ratios of 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 or more, because CSLNs/siRNA had more positive charges leading to the movement of CSLNs/siRNA to the opposite side in the gel. These results demonstrated that the CSLNs can easily bind with the siRNA, due to the cationic nature of the lipid. The binding ability is a key factor for the vectors as this can facilitate siRNA entry into cells and protect siRNA from degradation and this can subsequently affect water dispersibility and transfection efficiency of the vectors.33–36
1 or more, because CSLNs/siRNA had more positive charges leading to the movement of CSLNs/siRNA to the opposite side in the gel. These results demonstrated that the CSLNs can easily bind with the siRNA, due to the cationic nature of the lipid. The binding ability is a key factor for the vectors as this can facilitate siRNA entry into cells and protect siRNA from degradation and this can subsequently affect water dispersibility and transfection efficiency of the vectors.33–36
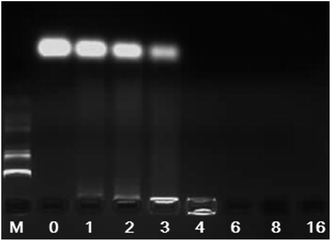 | ||
| Fig. 5 Gel electrophoresis of CSLNs/siRNA complexes at various mass ratios. (M: marker; CSLNs/siRNA ratio: 0, 1, 2, 3, 4, 6, 8, 16. The amount of siRNA was 0.5 μg). | ||
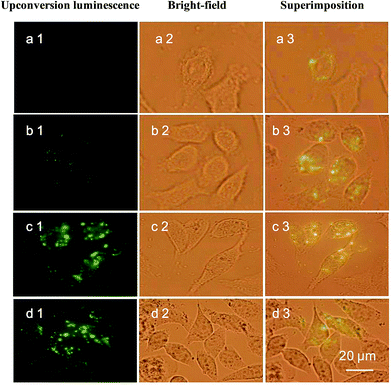
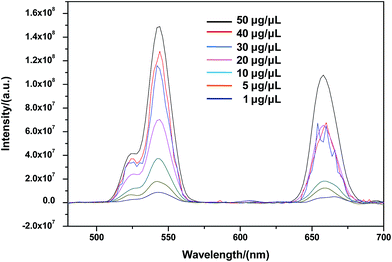
To study whether CSLNs could effectively enter into A549 cells, the uptake of CSLNs into A549 cells was experimented by fluorescence microscopy and fluorescence spectrometry. After the addition of the CSLNs, the cells were cultured in 6-well plates for 4 h at 37 °C and 5% CO2, monitored by the microscopy with 980 nm exciting light source. As shown in Fig. 6, the CSLNs could evenly spread throughout the cytoplasm, though a small amount of punctate distribution in the extracellular region was also observed, which was consistent with the results reported in the literature. It can be found that the fluorescence intensity increased with CSLNs ranging from 1 μg μL−1 to 20 μg μL−1, while the fluorescence intensity reached the maximum at 15 μg μL−1. Clear fluorescence of the CSLNs obviously demonstrated that the CSLNs have the potential to be used as probes for cell imaging. We deduced that a passive endocytosis process may be involved in the uptake of CSLNs for the lipid characteristics and thereof readily across cell membranes. Furthermore, peptide cationic lipids can significantly improve the adhesion of CSLNs to cell surfaces.37 The results of cellular uptake by fluorescence microscopy clearly indicated that the lipid can effectively deliver UCNPs into cells, though the specific details of the uptake of CSLNs in cells need to be further investigated. To quantify the cell uptake of CSLNs, the upconversion luminescence was measured as shown in Fig. 7.
As shown in Fig. 7, upconversion luminescence from A549 cells lysate of CSLNs should be obtained under the 980 nm excitation wavelength. All the spectra featured three distinct Er3+ emission bands. The green emissions between 514 and 534 nm and between 534 and 560 nm are the result of transitions from 2H11/2 and 4S3/2 to 4I15/2 of Er3+, respectively. A red emission is observed between 635 and 680 nm due to the transition from 4F9/2 to 4I15/2.38 Furthermore, the fluorescence intensity increased with increasing CSLNs ranging from 1 μg μL−1 to 50 μg μL−1. The results of upconversion luminescence further confirmed that the UCNPs coated with the lipid were effectively uptaken by A549 cells.28 At the same time, no upconversion luminescence was observed with UCNPs, which suggested that the UCNPs were difficult to enter into the cells in the absence of lipid. Fig. 4b showed obvious lipid membrane on the surface of CSLNs, therefore, the cellular uptake of CSLNs could be enhanced through the membrane fusion and endocytosis after coating with lipid.39
RNA interference (RNAi) is a promising therapeutic approach for combating human diseases through targeted post-transcriptional gene silencing of undesired gene expressions.40 Naked siRNA is susceptible to degradation by nucleases in blood serum and unable to cross the cell membrane due to its anionic charge.41 Lipids have been widely used for gene delivery, since the first cationic lipid N-[1-(2,3,-dioleyloxy) propyl]-N,N,N-trimethylammonium chloride (DOTMA) was introduced by Felgner et al. in 1987. Cationic lipids are gaining increasing popularity as the non-viral transfection vectors in RNAi therapeutics. In the present work, the CSLNs coated with lipid were confirmed for the ability to deliver siRNA into A549 cells which can stably express firefly luciferase. As shown in Fig. 8, outstanding gene silencing efficiency of firefly luciferase was achieved using CSLNs/siRNA at the mass ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The gene silencing efficiency of CSLNs was 70%, which was similar to that of CDO14. Therefore, the peptide-based cationic lipid might facilitate the cellular entry and subsequent endosomal escape of the CSLNs, thus contributing to the observed efficient gene inhibition. Cellular internalization of the CSLNs/siRNA complexes was already evidenced by upconversion luminescence imaging. Overall, the CSLNs showed the great potential for bioimaging and gene delivery, which attributed to the lipid nature as it can easily deliver the cargos into cells and release them once in the cells. The use of lipid will be rewarding in the design of the CSLNs as bifunctional vectors for the further in vivo study and even in clinical trials.
1. The gene silencing efficiency of CSLNs was 70%, which was similar to that of CDO14. Therefore, the peptide-based cationic lipid might facilitate the cellular entry and subsequent endosomal escape of the CSLNs, thus contributing to the observed efficient gene inhibition. Cellular internalization of the CSLNs/siRNA complexes was already evidenced by upconversion luminescence imaging. Overall, the CSLNs showed the great potential for bioimaging and gene delivery, which attributed to the lipid nature as it can easily deliver the cargos into cells and release them once in the cells. The use of lipid will be rewarding in the design of the CSLNs as bifunctional vectors for the further in vivo study and even in clinical trials.
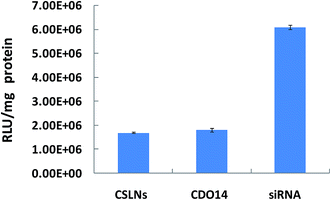 | ||
| Fig. 8 Gene silencing mediated by CSLNs. (CSLNs were formulated with siRNA at a mass ratio of 3/1, the amount of siRNA was 0.5 μg per well in 96-well plates). | ||
As an important factor in biocompatibility, in vitro cytotoxicity of gene delivery materials is usually measured by the MTT method.42–44 Cell viabilities were evaluated in A549 cells after exposure to CSLNs for 48 h across the entire range of mass ratios used in the transfection experiments. The lipid and UCNPs were at a mass ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 against A549 cells, which were formulated with siRNA at a mass ratio of 3/1. The cell viability of negative control cells was designated as 100%.
1 against A549 cells, which were formulated with siRNA at a mass ratio of 3/1. The cell viability of negative control cells was designated as 100%.
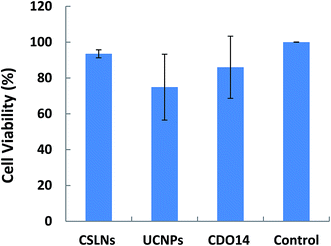
The results in Fig. 9 revealed that the cell viabilities were still greater than 90% even after the incubation with the CSLNs for 48 h. Compared with UCNPs, they displayed lower cytotoxicity at the same experiment conditions. Generally, the toxicity of cationic lipids is due to the positive charge of the head group. The peptide cationic lipids have drawn great advantage, as peptides are more biocompatible. We selected the lipid with headgroups of tri-ornithine to coat the UCNPs, because ornithine was found in a number of structurally characterized siRNA binding domains of proteins. After UCNPs coated with the lipid, the results confirmed that the CSLNs were biocompatible with A549 cells and they could be applied to the theranostics applications including multimodal bioimaging, bioseparation and photodynamic therapy. To the best of our knowledge, this is the first study that gives a significant evidence for the nontoxic nature of UCNPs coated with the lipid used in A549 cells.
3. Conclusion
A simple and pragmatic method was used for the preparation of bifunctional CSLNs by coating UCNPs with the peptide cationic lipid. We demonstrated the possibility of bioimaging and gene delivery of the bifunctional CSLNs. The cellular uptake studies clearly demonstrated that CSLNs could effectively enter into A549 tumor cells; therefore, the CSLNs exhibited good capability for bioimaging due to the coating with the lipid. The results of siRNA delivery in vitro by the CSLNs against A549 cells showed high transfection efficiency. Moreover, it also showed much lower cytotoxicity against the tumor cells than UCNPs due to the introduction of the lipid. In combination with the cellular uptake and RNAi results, the lipid appeared particularly suited for applications in coating UCNPs for both of bioimaging and siRNA delivery. This article provides a novel technology to construct hydrophilic gene delivery vector based on upconversion nanoparticles. In general, our studies open up new perspectives for gene delivery of cancer therapeutics and diagnosis of bioimaging with great biocompatibility.4. Experimental
4.1 Materials
Yttrium chloride (YCl3·6H2O, 99.9%), ytterbium chloride (YbCl3·6H2O, 99.9%), erbium chloride (ErCl3·6H2O, 99.9%), ammonium fluoride (NH4F, 98%) and ethanol were purchased from Aladdin. Anti-luciferase siRNA was purchased from Sigma-Aldrich (USA). Human lung carcinoma cells (A549 cells) were from Leaf's lab of the University of North Carolina at Chapel Hill. DMEM, RPMI1640, fetal bovine serum (FBS) and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Gibco (USA). All water used in this study was purified using a Milli-Q Plus 185 water purification system (Millipore, USA).4.2 Synthesis of β-NaYF4:Yb,Er nanoparticles
β-NaYF4:Yb,Er nanoparticles were synthesized by a solvothermal approach. In a typical procedure, YCl3·6H2O, YbCl·6H2O and ErCl3·6H2O solutions were added to a round bottom flask and stirred well. Oleic acid (OA) and 1-octadecene (ODE) were added to the former solutions of chloride and stirred for 30 min until the homogenous solution appeared at 150 °C. Under stirring, appropriate amount of NH4F and sodium hydroxide (NaOH) were added into the solution to obtain the final reaction mixture. The mixture was then kept stirring for 1 h at 280 °C. After the reaction, the solution was cooled to room temperature naturally. The nanoparticles were collected by centrifugation and washed with ethanol and deionized water three times.4.3 Preparation of CSLNs
To prepare CSLNs, UCNPs were formulated in combination with the lipid CDO14 at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mass ratio. 0.5 mg UCNPs and 0.5 mg CDO14 were dissolved in 1 mL of chloroform in the glass vial at room temperature. The solvent was removed under a stream of nitrogen gas, followed by high vacuum desiccation. Ten the dry lipid film was resuspended in 1 mL distilled water. After being subject to vortex and ultrasonication, CSLNs with a concentration of approximately 1 mg mL−1 was given.
1 mass ratio. 0.5 mg UCNPs and 0.5 mg CDO14 were dissolved in 1 mL of chloroform in the glass vial at room temperature. The solvent was removed under a stream of nitrogen gas, followed by high vacuum desiccation. Ten the dry lipid film was resuspended in 1 mL distilled water. After being subject to vortex and ultrasonication, CSLNs with a concentration of approximately 1 mg mL−1 was given.
4.4 Characterization of UCNPs and CSLNs
The zeta-potential of CSLNs was measured on a Dynamic Laser Scattering NanoSizer (Nano ZS 90, Malvern, UK) at room temperature. 20 mL of the CSLNs was diluted to 1 mL with distilled water. FTIR (Shimadzu IR Prestige-21) spectra were recorded on KBr pellets in the range from 4000 to 1000 cm−1 at atmosphere. X-ray diffraction (XRD-6000 Shimadzu) data of UCNPs and CSLNs were collected at room temperature (25 °C) using a germanium monochromatic (Cu Kα). The data in the 2θ range of 10–80° were collected in a step of 2° with the remaining time 1 s per step under the tube conditions of 40 kV and 40 mA. The morphology and microstructure were examined using scanning electron microscopy (SEM-Hitachi S-4800) with an acceleration voltage of 20 kV. The particle size and morphology of UCNPs and CSLNs were performed on transmission electron microscopy (TEM JEM-2100 microscope, Japan), operated at an accelerating voltage of 200 kV. The TEM samples were prepared on a carbon-coated copper grid, followed with air drying for 10 min and then negatively stained in 2 wt% phosphotungstic acid solution for 10 s.4.5 SiRNA-binding assay
The performance of CSLNs condensing siRNA was investigated by electrophoresis on a 1% agarose gel. In order to determine the effect of the mass ratios of CSLNs to siRNA, siRNA complexes were prepared by adjusting the stoichiometry of CSLNs and siRNA (0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2
1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 4
1, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 6
1, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 8
1, 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 16
1 and 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). After incubation for 30 min, the polyplexes were loaded on a 1% agarose gel with tris-acetate (TAE) running buffer and subjected to electrophoresis at 90 V and 45 mA for 30 min.
1). After incubation for 30 min, the polyplexes were loaded on a 1% agarose gel with tris-acetate (TAE) running buffer and subjected to electrophoresis at 90 V and 45 mA for 30 min.
4.6 Upconversion luminescence study
A549 cells were cultured in 6-well plates with a density of 5 × 104 per mL in 2 mL per well overnight. Then cells were treated with various concentrations of CSLNs solutions (1, 5, 10, 20, 30, 40 and 50 μg μL−1) at 37 °C and 5% CO2 for 4 h. The medium RPMI1640 was removed and washed with phosphate buffer solution (PBS) three times to remove excess of CSLNs. Then cells were incubated with 1 mL per well lysis buffer (1% Triton X-100, 10% Glycerol, PBS) at room temperature for 20 min, and cell lysate solution was gathered. Luminescence was investigated by measuring the spectra of 1 mL A549 cell lysate at room temperature. The upconversion luminescence spectra in the range of 450–700 nm were recorded on a spectrophotometer (Hitachi F-4600) with a tunable 980 nm laser diode as the excitation source.4.7 Cellular imaging
A549 cells were seeded on 3.5 cm plates with a glass bottom and incubated at 37 °C in 5% CO2. At the confluency of about 80%, the culture medium was removed and washed by PBS. CSLNs were then added to the cells at different concentration. Following 4 h culture at 37 °C in 5% CO2, cells were washed with PBS extensively. Transfected cells with CSLNs were detected under a magnification of 60 × 10 in oil by microscopy (Olympus-BX53F Tokyo Japan).4.8 Cytotoxicity of the CSLNs
The cytotoxicity of the CSLNs was assessed by the MTT assay. The assay was performed in 96-well plates by maintaining the ratios as used in the transfection experiments. A549 cells were seeded in 96-well plates at 200 μL per well and incubated at 37 °C under 5% CO2 for 24 h to get a confluence of about 80%. After the replacement of the medium, the CSLNs/siRNA was added to the cells for further culture at 37 °C under 5% CO2 for 24 h and then MTT (5 mg mL−1 in PBS) was added and kept for 4 h. MTT-containing medium was removed and 150 μL DMSO was added to dissolve the formazan and the absorbance at 570 nm was monitored by using the enzyme mark instrument (Sunrise Tecan, Australia). Data were presented as the mean ± SD.4.9 In vitro gene silencing of the CSLNs
A549 cells were seeded in 24-well plates approximately 24 h before gene silencing. Cells were treated with anti-luciferase targeting complexes of CSLNs and siRNA at the mass ratio of 3/1 in Opti-MEM at 37 °C for 4 h. Cells were washed with PBS, followed by incubation with lysis buffer at room temperature for 20 min.Fluorescence intensity in cell lysate was determined using a Perkin-Elmer LS 50B luminescence spectrometer (Norwalk, CT) (λex, 494 nm; λem, 519 nm). Cell lysate (5 μg per well) was dissolved in BCA protein assay reagent, and total protein concentrations were determined at 570 nm. The absorbance at 570 nm was monitored by using the enzyme mark instrument (Sunrise Tecan, Australia). The silencing rate is expressed as relative light units (RLU) per milligram of total protein.
Acknowledgements
This research is financially supported by the National High-Tech Research and Development Program of China (863 Program, 2014AA020707), the National Natural Science Foundation of China (21503035, 21606041 and 21176046), the Fundamental Research Funds for the Central Universities (DC201502020205 and DC201501076).References
- A. Miyawaki, A. Sawano and T. Kogure, Nat. Cell Biol., 2003, 5, S1–S7 CrossRef.
- M. Bruchez Jr, M. Moronne, P. Gin, S. Weiss and A. P. Alivisatos, Science, 1998, 281, 2013–2016 CrossRef CAS.
- M. L. Becker, J. A. Fagan, N. D. Gallant, B. J. Bauer, V. Bajpai, E. K. Hobbie, S. H. Lacerda, K. B. Migler and J. P. Jakupciak, Adv. Mater., 2007, 19, 939–945 CrossRef CAS.
- I. I. Slowing, B. G. Trewyn and V. S. Lin, J. Am. Chem. Soc., 2007, 129, 8845–8849 CrossRef CAS PubMed.
- F. Wang, W. B. Tan, Y. Zhang, X. Fan and M. Wang, Nanotechnology, 2006, 17, R1–R13 CrossRef CAS.
- D. R. Larson, W. R. Zipfel, R. M. Williams, S. W. Clark, M. P. Bruchez, F. W. Wise and W. W. Webb, Science, 2003, 300, 1434–1437 CrossRef CAS PubMed.
- L. Y. Wang, R. X. Yan, Z. Y. Hao, L. Wang, J. H. Zeng, J. Bao, X. Wang, Q. Peng and Y. D. Li, Angew. Chem., Int. Ed., 2005, 44, 6054–6057 CrossRef CAS PubMed.
- D. K. Chatterjee and Z. Yong, Nanomedicine, 2008, 3, 73–82 CrossRef CAS PubMed.
- J. Pichaandi, J. C. Boyer, K. R. Delaney and F. C. J. M. vanVeggel, J. Phys. Chem. C, 2011, 115, 19054–19064 CAS.
- D. Q. Chen and P. Huang, Dalton Trans., 2014, 13, 11299–11304 RSC.
- L. Q. Xiong, Z. G. Chen, M. X. Yu, F. Y. Li, C. Liu and C. H. Huang, Biomaterials, 2009, 30, 5592–5600 CrossRef CAS PubMed.
- M. M. Lezhnina, T. Justel, H. Katker, D. U. Wiechert and U. H. Kynast, Adv. Funct. Mater., 2006, 16, 935–942 CrossRef CAS.
- C. X. Li and J. Lin, J. Mater. Chem., 2010, 20, 6831–6847 RSC.
- X. Wang, J. Zhuang, Q. Peng and Y. D. Li, Inorg. Chem., 2006, 45, 6661–6665 CrossRef CAS PubMed.
- F. Auzel, Chem. Rev., 2004, 104, 139–173 CrossRef CAS PubMed.
- L. Y. Wang and Y. D. Li, Chem. Mater., 2007, 19, 727–734 CrossRef CAS.
- H. X. Mai, Y. W. Zhang, R. Si, Z. G. Yan, L. D. Sun, L. P. You and C. H. Yan, J. Am. Chem. Soc., 2006, 128, 6426–6436 CrossRef CAS PubMed.
- S. Heer, K. Kompe, H. U. Gudel and M. Haase, Adv. Mater., 2004, 16, 2102–2105 CrossRef CAS.
- T. Y. Cao, Y. Yang, Y. Gao, J. Zhou, Z. Q. Li and F. Y. Li, Biomaterials, 2010, 32(11), 2959–2968 CrossRef PubMed.
- Z. G. Chen, H. L. Chen, H. Hu, M. X. Yu, F. Y. Li, Q. Zhang, Z. G. Zhou, T. Yi and C. H. Huang, J. Am. Chem. Soc., 2008, 130, 3023–3029 CrossRef CAS PubMed.
- V. Biju, Chem. Soc. Rev., 2013, 45(17), 744–764 Search PubMed.
- C. E. Ashley, E. C. Carnes, K. E. Epler, D. P. Padilla, G. K. Phillips, R. E. Castillo, D. C. Wilkinson, B. S. Wilkinson, C. A. Burgard, R. M. Kalinich, J. L. Townson, B. Chackerian, C. L. Willman, D. S. Peabody, W. Wharton and C. Jeffrey Brinker, ACS Nano, 2012, 3, 2174–2188 CrossRef PubMed.
- M. Rajesh, J. Sen, M. Srujan, K. Mukherjee, B. Sreedhar and A. Chaudhuri, J. Am. Chem. Soc., 2007, 129, 11408–11420 CrossRef CAS PubMed.
- C. M. Lamanna, H. Lusic, M. Camplo, T. J. Mclntosh, P. Barthelemy and M. W. Grinstaff, Acc. Chem. Res., 2012, 7, 1026–1038 CrossRef PubMed.
- Y. N. Zhao, J. Zhu, H. J. Zhou, X. Guo, T. Tian, S. H. Cui, Y. H. Zhen, S. B. Zhang and Y. H. Xu, Colloids Surf., B, 2016, 145, 454–461 CrossRef CAS PubMed.
- X. Chen, W. Xu, H. Song, C. Chen, H. Xia, Y. Zhu, D. Zhou, S. Cui, Q. Dai and J. Zhang, ACS Appl. Mater. Interfaces, 2016, 8(14), 9071–9079 CAS.
- K. Ge, W. T. Sun, S. H. Zhang, S. X. Wang, G. Jia, C. M. Zhang and J. C. Zhang, RSC Adv., 2016, 6(26), 21725–21734 RSC.
- S. Gayathria, O. S. N. Ghosha, A. K. Viswanatha, P. Sudhakarab, M. J. K. Reddyc and A. M. Shanmugharaj, Int. J. Biol. Macromol., 2015, 72, 1308–1312 CrossRef PubMed.
- P. Sajan, R. S. Jayasree, S. Agouram and B. M. Junaid, Luminescence, 2015, 31(2), 544–550 CrossRef PubMed.
- I. Norrbo, P. Gluchowski, I. Hyppänen, T. Laihinen, P. Laukkanen, J. Mäkelä, F. Mamedov, H. S. Santos, J. Sinkkonen, M. Tuomisto, A. Viinikanoja and M. Lastusaari, ACS Appl. Mater. Interfaces, 2016, 8(18), 11592–11602 CAS.
- A. Li, D. Xu, Y. Zhang, H. Lin, S. Yan, Z. Chen and Y. Shao, J. Am. Ceram. Soc., 2016, 99(5), 1657–1663 CrossRef CAS.
- Y. Obata and S. N. Saito, Biochim. Biophys. Acta, 2009, 1788, 1148–1158 CrossRef CAS PubMed.
- D. Niculescu-Duvaz, J. Heyes and C. J. Springer, Curr. Med. Chem., 2003, 10, 1233–1261 CrossRef CAS PubMed.
- D. F. Zhi, S. B. Zhang, F. Qureshic, Y. N. Zhao, S. H. Cui, B. Wang, H. Y. Chen, Y. H. Wang and D. F. Zhao, Bioorg. Med. Chem. Lett., 2012, 22(11), 3837–3841 CrossRef CAS PubMed.
- H. Y. Chen, Y. N. Zhao, S. H. Cui, D. F. Zhi, S. B. Zhang and X. J. Peng, J. Appl. Polym. Sci., 2015, 132(35), 42469–42478 Search PubMed.
- C. Wang, L. Cheng, H. Xu and Z. Liu, Biomaterials, 2012, 33, 4872–4881 CrossRef CAS PubMed.
- S. K. Hong, I. H. Song, J. C. Kim, E. J. Kim, D. O. Jang and Y. S. Park, J. Controlled Release, 2006, 115(2), 234–241 CrossRef PubMed.
- Z. Chen, H. Chen, H. Hu, M. Yu, F. Li, Q. Zhang, Z. Zhou, T. Yi and C. Huang, J. Am. Chem. Soc., 2008, 130(10), 3023–3029 CrossRef CAS PubMed.
- Y. M. H. Charlie and U. Hasan, Biomaterials, 2012, 31, 7834–7848 Search PubMed.
- W. Qu, S. Y. Qin, Y. Kuang, R. X. Zhuo and X. Z. Zhang, J. Mater. Chem. B, 2013, 1, 2147–2154 RSC.
- H. X. Zeng, H. C. Little, T. N. Tiambeng, G. A. Williams and Z. B. Guan, J. Am. Chem. Soc., 2013, 135, 4962–4965 CrossRef CAS PubMed.
- X. Zhao and M. C. Tan, RSC Adv., 2016, 6(22), 18348–18356 RSC.
- Z. Z. Tu, Q. Zhang, M. Liu and Y. Qian, J. Mater. Sci., 2016, 51(6), 2972–2979 CrossRef CAS.
- Y. Chen, G. Jiang, Q. Zhou, Y. Zhang, K. Li, Y. Zheng, B. W. Zhang and X. S. Wang, RSC Adv., 2016, 6(28), 23804–23808 RSC.
| This journal is © The Royal Society of Chemistry 2017 |