Open Access Article
Open Access ArticleDesorption electrospray ionization mass spectrometry imaging reveals chemical defense of Burkholderia seminalis against cacao pathogens†
F. D. S. Araújo *a,
R. L. Vieiraa,
E. P. L. Molanob,
H. J. Máximoc,
R. J. D. Dalioc,
P. H. Vendraminia,
W. L. Araújod and
M. N. Eberlina
*a,
R. L. Vieiraa,
E. P. L. Molanob,
H. J. Máximoc,
R. J. D. Dalioc,
P. H. Vendraminia,
W. L. Araújod and
M. N. Eberlina
aThoMSon Mass Spectrometry Laboratory, Institute of Chemistry, University of Campinas, POB 6154, 13084-970, Campinas, SP, Brazil. E-mail: fdsaraujo@gmail.com
bGenomic and Expression Laboratory, Department of Genetics, Evolution and Bio-agents, Institute of Biology, University of Campinas, 13083-970, Campinas, SP, Brazil
cBiotechnology Laboratory, Centro de Citricultura Sylvio Moreira/Agronomic Institute, 13490-970, Cordeirópolis, SP, Brazil
dDepartment of Microbiology, Institute of Biomedical Sciences, University of São Paulo, 05508-900, São Paulo, SP, Brazil
First published on 9th June 2017
Abstract
Fungal and oomycete diseases are responsible for significant economic losses of cacao crops worldwide. Such diseases are caused mainly by the fungus Moniliophthora perniciosa, and the oomycetes Phytophthora citrophtora, P. capsici, and P. palmivora. The endophytic bacterium Burkholderia seminalis has showed antagonistic action against several plant pathogens. Herein, desorption electrospray ionization mass spectrometry imaging (DESI-MSI) was applied to map spatial metabolic distributions from B. seminalis and cacao pathogen monocultures, as well as to monitor interkingdom interactions of the endophytic bacterium with the fungus and oomycetes. DESI-MSI was successful in the direct analysis of fungus and oomycete pathogens on dehydrated agar plate, detecting several phospholipids and other unidentified metabolites. Chemical defense of B. seminalis against cacao pathogens was evidenced by the detection of partially diffuse rhamnolipid, which probably act in synergism with other unidentified diffuse metabolites.
1. Introduction
The main threats to cacao (Theobroma cacao L.) production are the fungal and oomycete diseases. On a global scale, the most important disease is the brown rot caused by Phytophthora spp., followed the witches' broom caused by Moniliophthora perniciosa and the frost rot caused by Moniliophthora roreri. It is estimated that these diseases are responsible for losses around 20% of cacao beans worldwide.1–3M. perniciosa, previously known as Crinipellis perniciosa, has reduced the cacao production in South American and Caribbean countries. The witches' broom attacks the young developing leaves and the meristematic tissue of embedded shoots. The flowers and young green beans are also susceptible to infections, directly reducing the crop yields.4,5 Fungicides alone are ineffective against witches' broom and biological control has been used to control the disease propagation.6,7
The black pod of cacao, caused by several species of genus Phytophthora, occurs in all producing countries. Of the three species that cause the black pod, P. citrophthora is the most virulent followed by P. palmivora and P. capsici. In Brazil, the P. hevea species has also been reported, but its pathogenicity to cacao has been considered moderate. Black pod infects the whole plant, but the greatest damages are observed in the fruits.8–11
The traditional methods of chemical control of fungal cacao diseases are often too expensive for small farmers, as well as hazardous to human health and the environment. Several studies involving endophytic microorganisms as biocontrol agents have therefore been developed trying to alter the microbial ecology of the cacao tree in the hope to suppress the diseases.4,8,12–19 Cladobotryum amazonense, a fungus found parasitizing M. perniciosa basidiocarps, produced an antibiotic substance capable of inhibiting the mycelial growth and in vitro germination of fungus basidiospores.20 The fungus Trichoderma stromaticum was promisor in the biocontrol of M. perniciosa at saprophytic phase, demonstrating the reduction of growth, sporulation and propagation of the pathogen through direct mycoparasitism or production of bioactive metabolites.4 The co-inoculation of cacao leaves with a pool of seven endophytic fungi showed higher resistance to Phytophthora sp.17 Endophytic fungi isolated from cacao trees resistant to the M. perniciosa were identified and evaluated in vitro and in vivo against this pathogen, verifying that Gliocladium catenulatum (currently named Clonostachys rosea) reduced the incidence of the disease around 70%.7 Cacao endophytic fungi demonstrated in vitro antagonism against M. roreri, P. palmivora and M. perniciosa, and field tests against M. roreri and P. palmivora in four farms in Panama significantly reduced pod losses caused by P. palmivora in the treatment with Colletotrichum gloeosporioides, and reduction of lesions caused by M. roreri when treated with C. rosea.18
The endophytic bacterium Burkholderia seminalis strain TC3.4.2R3, isolated from the surface of disinfected sugarcane roots, has recently been characterized by secondary metabolite productions in monoculture and coculture with Fusarium oxysporum using matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI).21 This bacterium showed antagonism against plant pathogens Fusarium verticillioides and Xanthomonas albilineans,22 and to suppress orchid necrosis, caused by Burkholderia gladioli.41
Desorption electrospray ionization mass spectrometry imaging (DESI-MSI) has emerged as an efficient technique to monitor the spatial distributions of microbial metabolites,23–25 including cross talking processes.26–28 In the present study, we report an investigation of the potential of B. seminalis TC3.4.2R3 in the inhibition of the main cacao pathogens (M. perniciosa, P. capsisi, P. palmivora, and P. citrophtora) by DESI-MSI.
2. Experimental
2.1 Strains and materials
M. perniciosa, P. palmivora, P. citrophthora, and P. capsici strains were isolated from cacao. The sugarcane root endophyte B. seminalis TC3.4.2R3 was obtained from Saccharum officinarum. All strains were maintained in culture medium PDA (potato dextrose agar, Difco) or TSA (tryptic soy agar, Merck). Methanol HPLC grade (≥99.9%) was purchased from J. T. Baker (Center Valley, PA, USA), and acetonitrile (≥99.9%) from Sigma-Aldrich (Steinheim, Germany). n-Butylamine (99.5%), taurocholic acid sodium hydrate (≥95%), and sodium dodecyl sulfate (≥98.5%) were purchased from Sigma-Aldrich (Steinheim, Germany), caffeine from Sigma Chemical Company (Saint Louis, MO, USA), MRFA from Thermo Scientific (Germany), Ultramark 1621 from Abcr (Karlsruhe, Germany), glacial acetic acid (99.7%) from Synth (Diadema, SP, Brazil).2.2 Preparation of microbial monocultures and cocultures
B. seminalis and cacao pathogens were cultivated on PDA medium in biochemical oxygen demand (B.O.D.) incubator at 28 °C. For monocultures, the bacterium cells grown for 48 h were suspended in sterile water and 1 μL of this suspension was applied on a glass slide placed in a sterile Petri dish and previously recovered with a thin film of PDA medium. The pathogen mycelia, with 24 to 48 h of incubation, were cut and applied on glass slides also containing the PDA medium. All glass slides with microorganisms were inoculated in B.O.D. at 28 °C. After incubation, the slides were removed from the Petri dishes, dehydrated in a desiccator under vacuum, and individually analyzed by DESI-MSI.21,23 For cocultures, B. seminalis cells suspended in sterile water (1 μL) were inoculated on glass slides recovered with PDA medium, and pathogen mycelia was applied 5 mm away. All samples were incubated in B.O.D., at 28 °C, for 30 (M. perniciosa coculture), 12 (P. capsici coculture), and 18 h (P. citrophthora and P. palmivora cocultures). After, the microbial glass slides were dehydrated under vacuum and analyzed by DESI-MSI. Prior the instrumental analyses, all dehydrated agar plates containing the fungi and oomycetes were sprayed with methanol to ensure a uniform surface.2.3 DESI-MSI analysis
An ionization source from Prosolia (Model OS-3201) coupled with mass spectrometer Thermo Scientific Q Exactive Hybrid Quadrupole-Orbitrap was used for imaging experiments, which were optimized by signal intensity and image quality. The analyses were performed in positive and negative ion modes using methanol flow of 3.0 μL min−1. The spray solution was electrically charged to 5 kV, the ion transfer capillary was held at 320 °C, the nebulizer gas pressure was set at 150 psi, and S-lens of 100 V. The scan range was acquired of m/z 150–1500, with spatial resolution of 200 μm, and scan speed of 740 μm s−1. An incidence angle was of 55° to the surface plane was used for DESI spray. The instrument was previously calibrated using an Ion Max API source with HESI probe attached. For positive ion mode was used a blend containing caffeine, MRFA, ultramark 1621, and n-butylamine H2O:acetonitrile:methanol with 0.1% of acetic acid, and for negative ion mode a blend containing taurocholic acid, sodium hydrate, sodium dodecyl sulfate, and ultramark 1621 in the same solvent. The spectrometer was controlled by Xcalibur 2.0 software (Thermo Fisher Scientific).2.4 Imaging dataset analysis
The Firefly 2.1.05 software was used to convert the DESI data to the imaging datasets. The BioMAP 3.8.04 software was used to visualize the images, in mass range of m/z 150–1500, and using a hypothetical color assigned to a fixed value, in order to enable a relative comparison among each experiment replicates.2.5 Identification of interest metabolites
The metabolites of each microorganism, previously inoculated in adequate conditions, were extracted with methanol (1 mL), sonicated (30 min), centrifuged (15 min), and the supernatants were separated of the cells. The extracts were analyzed by ESI-MS to acquire high accuracy mass and fragmentation ion patterns obtained by collision-induced dissociation with collision energy of 10–45 eV and isolation widths of 0.4 Da. The ESI experiments were performed using the Ion Max API source with HESI probe, operating in positive and negative ion modes. The instrumental parameters were as follows: flow rate, 5 μL min−1; nitrogen sheath gas, 100 psi; spray voltage, 3.5 kV; source temperature, 320 °C. The mass range was of m/z 150–1500, with resolution of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. The data was acquired and processed using Xcalibur 2.0 software (Thermo Fisher Scientific). The identifications were performed by searches in METLIN and LipidMaps databases, combined to fragmentation studies based in previous reports.
000. The data was acquired and processed using Xcalibur 2.0 software (Thermo Fisher Scientific). The identifications were performed by searches in METLIN and LipidMaps databases, combined to fragmentation studies based in previous reports.
3. Results and discussion
3.1 DESI-MSI of B. seminalis monocultures
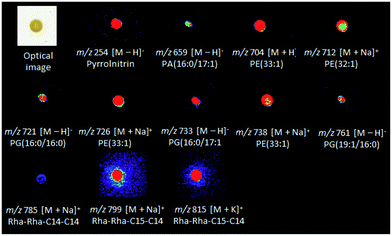
Diffuse and colony-associated metabolites were detected in B. seminalis by DESI(±)-MSI (Fig. S1 and S2†). Tandem mass spectrometry (ESI-MS/MS) of these ions was performed using methanolic extracts of B. seminalis. The high accurate masses and fragmentation patterns allowed seaches for their structures in METLIN, and LIPID MAPS databases, as well as previous reports as detailed below. The metabolites of m/z 704, 712, 726 and 738 were identified as protonated molecules of the glycerophosphoethanolamine (PE) class, respectively [PE(33:1) + H]+, [PE(32:1) + Na]+, [PE(33:1) + Na]+, and [PE(34:2) + Na]+ (Table S1†). The MS/MS of these ions were characterized for showing neutral losses of ethanolamine phosphate (141 Da) (Fig. S3 and S4†). The sodiated molecules (Fig. S4†) showed a neutral loss of ethyleneamine (43 Da) as well as the product ion of m/z 164 corresponding to sodiated phosphoethanolamine, and the neutral loss of phosphoric acid (98 Da) from the fragment [M + H − 43]+.29,30 Phospholipids were also detected as their deprotonated molecules in the negative ion mode, that is, glycerophosphatidic acid [PA(16:0/17:1) − H]− of m/z 659 (Fig. S5†), and the following glycerophosphoglycerols: [PG(16:0/16:0) − H]−, [PG(16:0/17:1) − H]− and [PG(19:1/16:0) − H]− of m/z 721, 733, and 761, respectively (Fig. S6†). These phospholipids are the major components of bacterial cell membranes, playing central roles as permeability barrier for controlling the diffusion of ions and molecules in and out of bacterial cells.31 The metabolite of m/z 785 was identified as the sodiated molecule of the rhamnolipid Rha–Rha–C14–C14 (Fig. S7†), a cytotoxic exolipid previously detected in Burkholderia pseudomallei.32 The rhamnolipid Rha–Rha–C15–C14 detected by the ion of m/z 799 [M + Na]+ (Fig. S7†) was also detected by DESI-MSI, together with its corresponding potasiated molecule [M + K]+ of m/z 815. The gene cluster for biosynthesis and efflux of rhamnolipids (Bsem_06536-06543) was already annotated in the B. seminalis TC3.4.2R3 genome,42 and the roles in motility, cell signaling, biofilm formation and anti-bacterial activity have been proposed.43 The ion of m/z 254 was identified as the pyrrolnitrin metabolite with an accurate m/z 254.9746 (Table S1 and Fig. S5†), a classic antifungal found in Pseudomonas spp., Myxococcus fulvus, and Enterobacter agglomerans.33–35 Fig. 1 shows the DESI-MSI of these metabolites identified in B. seminalis colonies. This result confirmed the genome information, which described the presence of a cluster (Bsem 0612-06815) encoding the complete pathway for this antifungal metabolite.3.2 DESI-MSI of pathogen monocultures
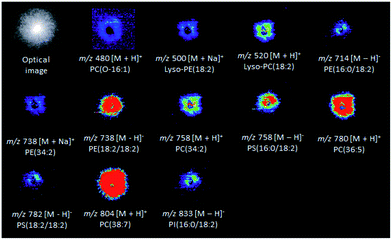
Several colony-associated metabolites were detected in M. perniciosa, P. citrophthora, P. capsici, and P. palmivora colonies by DESI(±)-MSI (Fig. S8–S15†). The analysis in the positive ion mode of M. perniciosa (Fig. 2) identified phospholipids of the glycerophosphoethanolamine and glycerophosphocholine classes. The ions of m/z 500 and 738 were characterized by the formation in MS/MS experiments of the product ion of m/z 164 attributed to sodiated phosphoethanolamine, as well as by a neutral loss of phosphoethanolamine (141 Da), and were therefore identified as sodiated Lyso-PE(18:2), and PE(34:2), respectively (Fig. S16 and Table S2†). The ions of m/z 480, 520, 758, 780, and 804 were respectively identified as [LysoPC(O-16:1) + H]+, [LysoPC(18:2) + H]+, [PC(34:2) + H]+, [PC(36:5) + H]+, and [PC(38:7) + H]+. Their MS/MS showed the product ion of m/z 184, which is typical of phospholipids containing phosphocholines, as well as by a neutral loss of the (CH3)3N molecule (59 Da) (Fig. S17–S19†).29,36 In the negative ion mode (Fig. 2), the ion of m/z 758 was identified as the deprotonated molecule of glycerophosphoserine PS[16:0/18:2] metabolite (Fig. S20 and Table S2†). This ion showed by MS/MS a neutral loss of 87 Da, which is characteristic of the serine polar head group. Its product ion of m/z 391 [(M − H)− − 87 − R2COOH], which is more abundant that of m/z 415, results from the loss of a fatty acid (FA) at the sn-2 position, whereas the ion of m/z 415 [M − H − 87 − R1COOH]− results from the loss of another FA at the sn-1 position. The product ions of m/z 409 [M − H − 87 − R′2COOH]− and 433 [M − H − 87 − R′1COOH]− are formed due to ketene losses from the sn-2 and sn-1 positions, respectively.29,37 The metabolite of m/z 782 also showed the same characteristic product ions and was, therefore, identified as deprotonated PS[18:2/18:2] (Fig. S20 and Table S2†). The ions of m/z 714 and 738 (Fig. S21 and Table S2†) were respectively identified via MS/MS as the deprotonated forms of PE(16:0/18:2) and PE(18:2/18:2), by the characteristic fragment ions of m/z 196 [M − H − R1COOH − R2CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O]− corresponding to neutral loss of the sn-2 fatty acid as a ketene.29,38 The ion of m/z 833 was identified as deprotonated glycerophosphoinositol PI(16:0/18:2) showing in its MS/MS the product ion of m/z 241, which is characteristic of the inositol head group. The most abundant fragment ion of m/z 279 corresponded to the deprotonated FA (R2COO−) from the sn-2 position, whereas the less abundant product ion of m/z 255 corresponded to the FA (R1COO−) at the sn-1 position (Fig. S21 and Table S2†).
O]− corresponding to neutral loss of the sn-2 fatty acid as a ketene.29,38 The ion of m/z 833 was identified as deprotonated glycerophosphoinositol PI(16:0/18:2) showing in its MS/MS the product ion of m/z 241, which is characteristic of the inositol head group. The most abundant fragment ion of m/z 279 corresponded to the deprotonated FA (R2COO−) from the sn-2 position, whereas the less abundant product ion of m/z 255 corresponded to the FA (R1COO−) at the sn-1 position (Fig. S21 and Table S2†).
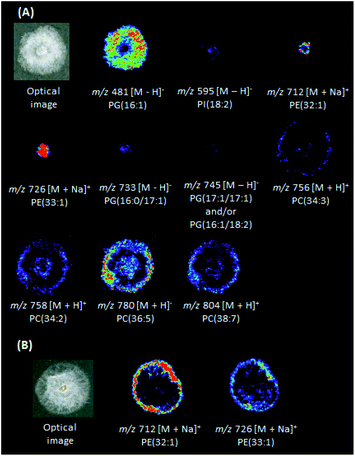
DESI(+)-MSI (Fig. 3A) was also able to identify in the P. citrophthora colonies glycerophosphoethanolamines [PE(32:1) + Na]+ (m/z 712) and [PE(33:1) + Na]+ (m/z 726), and the glycerophosphocholines [PC(34:3) + H]+ (m/z 756, Fig. S22†), [PC(34:2) + H]+ (m/z 758), [PC(36:5) + H]+ (m/z 780), and [PC(38:7) + H]+ (m/z 804, Table S2†). In the negative ion mode, the glycerophosphoglycerols PG(16:1) of m/z 481, PG(16:0/17:1) of m/z 733, and PG(17:1/17:1) and/or PG(16:1/18:2) of m/z 745, glycerophosphoinositol PI(18:2) of m/z 435, and the glycerophosphatidic acid PA(17:1) of m/z 595 (Fig. S23, S24 and Table S2†) were also identified. For P. capsici, a few phospholipids were also identified: [PE(32:1) + Na]+ (m/z 712) and [PE(33:1) + Na]+ (m/z 726) (Fig. 3B and Table S2†). For P. palmivora several ions were detected (Fig. S14 and S15†), but remained unidentified.
3.3 Interactions between B. seminalis and pathogens by DESI-MSI
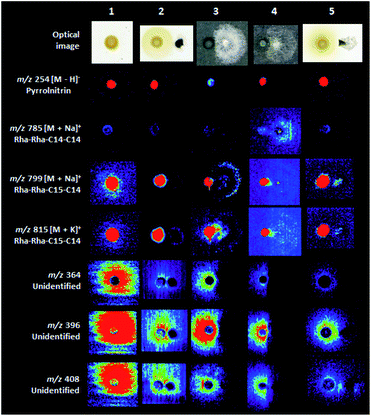
The coculture experiments between B. seminalis and M. perniciosa pointed the considerable inhibition of the fungus growth when evaluated for 48 h (Fig. 4 and S25†). Also, the interaction of B. seminalis with the oomycetes showed significant inhibition of P. palmivora for 48 h, followed of P. citrophthora and P. capsisi when evaluated for 24 and 18 h, respectively (Fig. 4 and S26–S28†). The defense mechanism of B. seminalis is based on the production of antimicrobial metabolites. The partially diffuse rhamnolipid (m/z 799 and 815, Fig. 4) identified in B. seminalis may have an important role in the antifungal properties. Rhamnolipids from Pseudomonas aeruginosa showed antifungal activity against oomycete and fungi such as Phytophthora capsici, and were able to suppress the development of C. orbiculare infection on leaves of cucumber plants, as well as to control red rot disease of sugarcane caused by the fungus Colletotrichum falcatum.39,40 The antifungal pyrrolnitrin plays a central role in the inhibition of pathogens,33–35 however their DESI images were localized within the B. seminalis colony (m/z 254, Fig. 4) suggesting a low involvement in the inhibitory activity in the equipament conditions and incubation times used in our experiments. Many diffuse metabolites of unknown identify (Fig. S1 and S2†) were detected in B. seminalis by DESI-MSI (for instance, m/z 364 and 396 in Fig. 4), and more ions were found in comparison with previous MALDI-MSI analysis.21 The siderophore pyochelin detected by MALDI-MSI21 was not found by DESI-MSI in the conditions analyzed. Probably the defense mechanism of B. seminalis is resulted of the synergistic action of partially diffuse rhamnolipids, pyochelin and other unidentified diffuse metabolites.4. Conclusions
DESI-MSI was successful in the analysis of fungus and oomycete pathogens directly from dehydrated agar plates, being able to detect and characterize many phospholipids. The mapping of B. seminalis showed many diffuse and colony-associated metabolites potentially involved in the antimicrobial activity. The interaction experiments pointed to the potential of B. seminalis in the growth inhibitions of cacao pathogens.Acknowledgements
The authors acknowledge the financial support from the FAPESP, Brazil (Grant 2013/24860-9 and 2015/18454-3), which enabled the present contribution.References
- R. Ploetz, in Cacao Diseases, ed. B. A. Bailey and L. W. Meinhardt, Springer International Publishing, 2016, pp. 33–59 Search PubMed.
- P. K. Hebbar, Phytopathology, 2007, 97, 1658 CrossRef PubMed.
- G. J. Samuels, C. Suare, K. Solis, K. A. Holmes, S. E. Thomas, A. Ismaiel and H. C. Evans, Mycol. Res., 2006, 110, 381 CrossRef PubMed.
- F. H. V. Medeiros, A. W. V. Pomella, J. T. Souza, G. R. Niella, R. Valle, R. P. Bateman, D. Fravel, B. Vinyard and P. K. Hebbar, Crop Prot., 2010, 29, 704 CrossRef CAS.
- V. T. Queiroz, C. T. Guimarães, D. Anhert, I. Schuster, R. T. Daher, M. G. Pereira, V. R. M. Miranda, L. L. Loguercio, E. G. Barros and M. A. Moreira, Plant Breed., 2003, 122, 268 CrossRef CAS.
- R. Bateman, Gro-Cocoa Newsletter, 2002, 1, 4 Search PubMed.
- M. R. Rubini, R. T. Silva-Ribeiro, A. W. V. Pomella, C. S. Maki, W. Araújo, D. R. Santos and J. L. Azevedo, Int. J. Biol. Sci., 2005, 1, 24 CrossRef CAS PubMed.
- R. J. Hanada, A. W. V. Pomella, H. S. Costa, J. L. Bezerra, L. L. J. Loguercio and O. Pereira, Fungal Biol., 2010, 114, 901 CrossRef PubMed.
- H. C. Evans, Phytopathology, 2007, 97, 1640 CrossRef PubMed.
- F. G. Faleiro, E. D. M. N. Luz, A. O. Cerqueira, C. S. S. Rocha, A. Dantas Neto, A. B. Flores, R. C. S. Bahia and A. S. G. Faleiro, Fitopatol. Bras., 2004, 29, 303 CrossRef.
- E. D. M. N. Luz and S. D. V. M. Silva, in Doenças causadas por Phytophthora no Brasil, ed. E. D. M. N. Luz, A. F. D. Santos, K. Matsuoka and J. L. Bezerra, Campinas: Rural, 2001, pp. 175–265 Search PubMed.
- U. Krauss, E. Hidalgo, R. Bateman, V. Adonijah, C. Arroyo, J. García, J. Crozier, N. A. Brown, G. M. ten Hoopen and K. A. Holmes, Biol. Control, 2010, 54, 230 CrossRef CAS.
- B. A. Bailey, H. Bae, M. D. Strem, D. P. Roberts, S. E. Thomas, J. Crozier, G. J. Samuels, I. K. Choi and K. A. Holmes, Planta, 2006, 224, 1449 CrossRef CAS PubMed.
- J. Crozier, S. E. Thomas, M. C. Aime, H. C. Evans and K. A. Holmes, Plant Pathol., 2006, 55, 783 CrossRef CAS.
- M. Aneja, T. J. Gianfagna and P. K. Hebbar, Physiol. Mol. Plant Pathol., 2005, 67, 304 CrossRef CAS.
- K. A. Holmes, H. J. Schroers, S. E. Thomas, H. C. Evans and G. J. Samuels, Mycol. Progr., 2004, 3, 199 CrossRef.
- A. E. Arnold, L. C. Mejía, D. Kyllo, E. I. Rojas, Z. Maynard, N. Robbins and E. A. Herre, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 15649 CrossRef CAS PubMed.
- L. C. Mejía, E. I. Rojas, Z. Maynard, S. Van Bael, A. E. Arnold, P. Hebbar, G. J. Samuels, N. Robbins and E. A. Herre, Biol. Control, 2008, 46, 4 CrossRef.
- G. J. Samuels, S. L. Dodd, W. Gams, L. A. Castlebury and O. Petrini, Mycologia, 2002, 94, 146 CrossRef PubMed.
- C. N. Bastos, H. C. Evans and R. A. Samson, Trans. Br. Mycol. Soc., 1981, 77, 273 CrossRef.
- F. D. S. Araújo, W. L. Araújo and M. N. Eberlin, J. Am. Soc. Mass Spectrom., 2017, XX, 1 Search PubMed.
- D. M. Luvizotto, J. Marcon, F. D. Andreote, F. Dini-Andreote, A. A. C. Neves, W. L. Araújo and A. A. Pizzirani-Kleiner, World J. Microbiol. Biotechnol., 2010, 26, 1829 CrossRef CAS.
- C. F. Angolini, P. H. Vendramini, F. D. S. Araújo, W. L. Araújo, M. N. Augusti, M. N. Eberlin and L. G. Oliveira, Anal. Chem., 2015, 87, 6925 CrossRef CAS PubMed.
- J. Watrous, P. Roach, T. Alexandrov, B. S. Heath, J. Y. Yang, R. D. Kersten, M. van der Voort, K. Pogliano, H. Gross, J. M. Raaijmakers, B. S. Moore, J. Laskin, N. Bandeira and P. C. Dorrestein, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E1743 CrossRef CAS PubMed.
- D. D. Nguyen, C. H. Wu, W. J. Moree, A. Lamsa, M. H. Medema and X. Zhao, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, E2611 CrossRef CAS PubMed.
- A. Tata, C. Perez, M. L. Campos, M. A. Bayfield, M. N. Eberlin and R. I. Demian, Anal. Chem., 2015, 87, 12298 CrossRef CAS PubMed.
- J. Watrous, N. Hendricks, M. Meehan and P. C. Dorrestein, Anal. Chem., 2010, 82, 1598 CrossRef CAS PubMed.
- M. F. Traxler, J. D. Watrous, T. Alexandrov, P. C. Dorrestein and R. Kolter, mBio, 2013, 4, e00459 CrossRef PubMed.
- M. Pulfer and R. C. Murphy, Mass Spectrom. Rev., 2003, 22, 332 CrossRef CAS PubMed.
- J. I. Zhang, N. Talaty, A. B. Costa, Y. Xia, A. Tao, R. Bell, J. H. Callahan and G. Cooks, Int. J. Mass Spectrom., 2011, 301, 37 CrossRef CAS.
- T. Y. Lin and D. B. Weibel, Appl. Microbiol. Biotechnol., 2016, 100, 4255 CrossRef CAS PubMed.
- S. Häußler, M. Nimtz, T. Domke, V. Wray and I. Steinmetz, Infect. Immun., 1998, 66, 1588 Search PubMed.
- K. Arima, H. Imanaka, M. Kousaka, A. Fukuta and G. Tamura, Agric. Biol. Chem., 1964, 28, 575 CrossRef CAS.
- L. Chernin, A. Brandis, Z. Ismailov and I. Chet, Curr. Microbiol., 1996, 32, 205 Search PubMed.
- K. Gerth, W. Trowitzsch, V. Wray, G. Höfle, H. Irschik and H. Reichenbach, J. Antibiot., 1982, 35, 1101 CrossRef CAS PubMed.
- S. Milne, P. Ivanova, J. Forrester and H. A. Brown, Methods, 2006, 39, 92 CrossRef CAS PubMed.
- F. F. Hsu and J. Turk, J. Am. Soc. Mass Spectrom., 2005, 16, 1510 CrossRef CAS PubMed.
- F. F. Hsu and J. Turk, J. Am. Soc. Mass Spectrom., 2000, 11, 892 CrossRef CAS PubMed.
- B. S. Kim, J. Y. Lee and B. K. Hwang, Pest Manage. Sci., 2000, 56, 1029 CrossRef CAS.
- D. Goswami, S. N. Borah, J. Lankar, P. J. Handique and S. Deka, J. Basic Microbiol., 2015, 55, 1265 CrossRef CAS PubMed.
- E. T. Mano, S. N. Minami, J. E. Loper and W. L. Araújo, Plant Dis., 2015, 99, 1642 CrossRef.
- W. L. Araújo, A. Creason, E. T. Mano, A. A. Camargo-Neves, S. N. Minami, J. Chang and J. E. Loper, Mol. Plant-Microbe Interact., 2016, 29, 435 CrossRef PubMed.
- A. M. Abdel-Mawgoud, F. Lépine and E. Déziel, Appl. Microbiol. Biotechnol., 2010, 86, 1323 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra03895j |
| This journal is © The Royal Society of Chemistry 2017 |