Open Access Article
Open Access ArticleExchanging the order of carotenogenic genes linked by porcine teschovirus-1 2A peptide enable to optimize carotenoid metabolic pathway in Saccharomyces cerevisiae†
Xiang Jiaoab,
Wenyi Suna,
Yue Zhanga,
Xiangjian Liua,
Qi Zhanga,
Qian Wanga,
Sufang Zhang *ac and
Zongbao Kent Zhao
*ac and
Zongbao Kent Zhao ac
ac
aDivision of Biotechnology, Dalian Institute of Chemical Physics, CAS, Dalian 116023, PR China
bUniversity of Chinese Academy of Sciences, Beijing, 100049, PR China
cDalian National Laboratory for Clean Energy, Dalian Institute of Chemical Physics, CAS, Dalian 116023, PR China. E-mail: zsfjxy@dicp.ac.cn
First published on 12th October 2018
Abstract
The yeast Saccharomyces cerevisiae serves as a promising host for the production of a wide range of chemical compounds and fuels. Currently, simultaneous expression of several genes could be achieved via the use of 2A viral peptides, yet detailed characterizations to assess the discrepancy of different orders of genes linked by 2A peptides are rarely sufficient. In this study, we investigated the effects of the order of genes linked by porcine teschovirus-1 2A (P2A) peptide on the metabolic pathway in S. cerevisiae. A heterologous carotenoid biosynthetic system involving nine kinds of polycistronic expression of codon-optimized carotenogenic genes GGPPS, CARB and CARRP from Blakeslea trispora was introduced into S. cerevisiae. The order of genes in the polycistronic segment was exchanged; β-carotene production by engineered yeasts was significantly different. The highest β-carotene yield was achieved in transformants carrying the plasmid, with CARB as the first gene in the polycistronic construct regardless of the location of GGPPS, CARRP. In addition, we found that β-carotene production was coupled with the growth in engineered strain with the highest β-carotene content during the shake flask fermentation and fed-batch fermentation. A novel microbial heterologous carotenoid production system was established by optimizing the order of genes linked by P2A peptide sequences in a polycistronic expression construct. The observation of the importance of the order in a polycistronic construct may be used to increase yields in other P2A peptide-containing expression systems.
Introduction
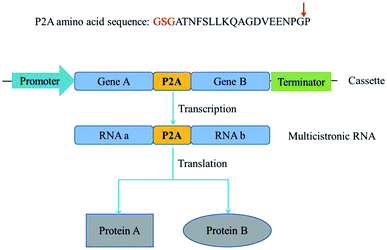
Metabolic engineering usually involves introduction of multiple genes on a single plasmid into a host. However, eukaryotic proteins are usually encoded by monocistronic genes, and gene expression requires individual and specific promoter and terminator sequences, which requires a large set of gene sequences and brings metabolic burden. Self-cleaving 2A peptides are among the best-known polycistronic systems for gene co-expression in eukaryotes.1–3 2A peptides drive a translational recoding event, with a cleavage of the polypeptide backbone between the last two amino acids in the C-terminus of the 2A peptide, glycine and proline (Fig. 1).4–6 These peptides could be used as a multigene manipulation tool, to create polycistronic transcripts from a series of naturally monocistronic genes.There are several advantages to the use of 2A peptides. First, the choice of constitutive promoters and terminators for driving gene expression is restricted in some not well-characterized species. If the same promoter and terminator were employed to drive the expression of multiple genes, the probability of homologous recombination between promoter and terminator would be increased. Second, the larger the size of the vector, the more difficult it is to construct. Expression of various genes from a polycistronic segment, with the genes linked by 2A peptide sequences, would considerably reduce the size of the expression vector.7 Third, to validate protein's function in vivo, its translation is first confirmed by western blotting. The complexity of the target protein itself, or the lack of specific antibodies, might lead to false negative results or no results. Linkage of genes encoding fluorescent proteins or antibiotic resistance determinants with 2A peptide sequences upstream of the gene(s) encoding target protein(s) constitute an alternative detection approach.8 Because of the simultaneous and efficient expression of multiple genes, 2A-mediated cleavage was universally exploited in biomedicine and biotechnology. E.g. an entire penicillin biosynthesis pathway was expressed using a 2A peptide-based system in Aspergillus nidulans strains.9 Similarly, porcine teschovirus-1 2A with glycine–serine–glycine spacer exhibits high cleavage efficiency in insect cell lines and was used to successfully cleave vitellogenin receptor fragment linked with EGFP in the silkworm glands.10
β-Carotene is used in functional foods, nutraceuticals, pharmaceuticals and cosmetics because of its anti-oxidative and anti-cancer activities. Microorganisms, such as Escherichia coli and Saccharomyces cerevisiae, have been manipulated for heterologous β-carotene production.11,12 Recently, Xanthophyllomyces dendrorhous crtI, crtE and crtYB genes were expressed in S. cerevisiae from a single polycistronic construct, with the individual Crt proteins separated by Thosea asigna virus 2A (T2A) peptide.13 The authors explored the utility of T2A-based polycistronic gene expression for pathway engineering of β-ionone production in S. cerevisiae, but the effect of the order of genes in the T2A construct on β-ionone production was unclear. Nevertheless, the order of genes in a 2A construct might be important because the differential expression of several proteins was resulted from the processing of the 2A peptide.14 Thus, it is greatly meaningful to optimize a heterologous expression pathway for nature product via exchanging the order of pathway genes in a polycistronic 2A peptide construct.
In this study, S. cerevisiae strain BY4741 was used as a host of three codon-optimized carotenoid genes from B. trispora linked via P2A peptide sequences to explore the effect of the gene order in such a polycistronic construct on the carotenoid metabolic pathway.
Materials and methods
Strains, medium, and reagents
E. coli DH5α was cultured at 37 °C in Luria–Bertani (LB) medium (10 g L−1 NaCl, 10 g L−1 tryptone, and 5 g L−1 yeast extract). The yeast carrying plasmids with the URA selectable marker was selected and cultivated at 30 °C by synthetic dropout (SD) plate, which consists of 20 g L−1 glucose, 6.7 g L−1 yeast nitrogen bases (YNB) with (NH4)2SO4, and 20 g L−1 agar supplemented with 20 mg L−1 histidine, 20 mg L−1 methionine, and 100 mg L−1 leucine.PrimeStar MAX, restriction enzymes, and other enzymes were purchased from Takara (Dalian, China). DNA gel purification and plasmid extraction kits were purchased from Sangon Biotech (Shanghai, China). Yeast nitrogen base with (NH4)2SO4 and without amino acids was purchased from Difco (Detroit, USA). Yeast extract and tryptone were purchased from Oxiod (Basingstoke, UK). Amino acids and agar powder were supplied by Dingguo Biotech (Beijing, China). Oligonucleotides were synthesized by Sangon Biotech or Dingguo Biotech.
Plasmid construction
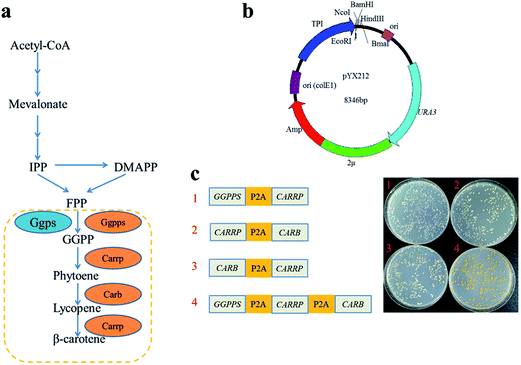
Genes GGPPS (UniProtKB accession number Q9P885), CARB (UniProtKB accession number Q67GI0) and CARRP (UniProtKB accession number Q9UUQ6) of the β-carotene pathway were codon-optimized for expression in S. cerevisiae and synthesized by Genewiz (Suzhou, China). P2A peptide sequence was synthesized in Synbio Tech (Suzhou China). To assemble plasmid pJX11, GGPPS-P2A, CarB-P2A, and CarRP-P2A were PCR amplified with primer pairs ADH2-EcoRI-GGPPS-F/GGPPS-P2A-BamHI-R, BamHI-CarB-F/CarB-P2A-KpnI-R, and KpnI-CarRP-F/CarRP-P2A-HindIII-R, then digested with EcoRI/BamHI, BamHI/KpnI, and KpnI/HindIII respectively. The resulting fragments were inserted into plasmid pUC18. The EcoRI-GGPPS-F and CarRP-NcoI-R primer set were used to amplify a 4.7 kb GGPPS-P2A-CarB-P2A-CarRP fragment with plasmid pJX11 as the template. The 4.7 kb fragment and plasmid pYX212 were treated with EcoRI/NcoI, and then linked each other to construct the plasmid pJX4. With the same methods, the plasmid pJX5 and pJX6 were successfully constructed.The second gene of the plasmid pJX4, pJX5 and pJX6 were deleted by the restriction-free (RF) cloning procedure to assemble the plasmid pJX1, pJX2 and pJX3, respectively.15 The second gene of the plasmid pJX4, pJX5 and pJX6 were amplified using primer pairs CarRP1373-F/RP-CarB-pYX-R, CarRP1373-F/RP-GGPPS-pYX-R, and CarB1279-F/RP-GGPPS-pYX-R, then were inserted into the plasmid pJX1, pJX2 and pJX3 to construct plasmid pJX7, pJX8 and pJX9, respectively. Strains and plasmids are listed in ESI Table 1.† Primes are listed in ESI Table 2.† All the recombinant plasmids were transformed into BY4741 according to previous standard protocols.16
Strains fermentation
Shake flask fermentations for production of β-carotene were conducted in minimal medium consisting of 20 g L−1 glucose, 14.4 g L−1 (NH4)2SO4, 3 g L−1 KH2PO4, 0.5 g L−1 MgSO4·7H2O, trace metal and vitamin solutions supplemented with 40 mg L−1 histidine, 40 mg L−1 methionine, and 100 mg L−1 leucine. While for testing β-carotene content during the process of fermentation, the minimal medium contains 50 g L−1 glucose. Inoculum were prepared at 30 °C for 24 h, then inoculated in 50 mL minimal medium with an initial OD600 of 0.1. The cultivation was carried out for 72 h at 200 rpm, 30 °C.The fed-batch fermentations for production of β-carotene were performed in 3 L bioreactors (Baoxing Shanghai China), with an initial working volume of 2 L. Fermentation medium contains 50 g L−1 glucose, 14.4 g L−1 (NH4)2SO4, 3 g L−1 KH2PO4, 0.5 g L−1 MgSO4·7H2O, trace metal and vitamin solutions supplemented with 0.5 g L−1 histidine, 0.5 g L−1 methionine, and 1.25 g L−1 leucine. The temperature was kept at 30 °C, agitation set to 400 rpm and the pH was kept at 6.0 by automatic addition of 2 M NaOH. A 750 g L−1 glucose solution was used as the feed medium.
Measurement of β-carotene production
Glucose was quantified by a glucose analyzer (SBA-50B; Shandong Academy of Sciences, Jinan, China). 30 mL culture were harvested by centrifugation at 8000g for 5 min, washed twice with distilled water, and dried at 105 °C to a constant dry cell weight (DCW). The extraction of carotenoid was performed based on a published method.17Carotenoid analysis was performed using a high-performance liquid chromatography (HPLC) system equipped with YMC carotenoid C30 column (250 × 4.6 mm, 3 μm) and UV230 II detector (Elite, Dalian, China). Gradient analysis method was used to resolve the compounds (ESI Table 3†); the mobile phase consisted of buffer A [methanol/H2O (92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8, v/v) and 10 mM ammonium acetate] and buffer B (methyl tert-butyl ether), with a flow rate of 1 mL min−1, at 33 °C.
8, v/v) and 10 mM ammonium acetate] and buffer B (methyl tert-butyl ether), with a flow rate of 1 mL min−1, at 33 °C.
Western blotting
S. cerevisiae BY4741 transformants were cultured at 30 °C for 2 days in YPD medium, and 2 mL of culture were collected by centrifugation and washed with deionized water. Protein extraction was performed according to a previously published protocol.18 10 μL of protein samples were loaded on a 12% sodium dodecyl sulfate-polyacrylamide gel and electrophoresed at a constant current (10 mA). After electrophoresis, the proteins were transferred onto a nitrocellulose membrane (Salarbio, Beijing, China) in blotting buffer (48 mM Tris, 385 mM glycine, 0.1% (w/v) sodium dodecyl sulfate and 20% methanol; transfer conditions: 400 V, 100 mA, 1 h). The membrane was blocked in phosphate-buffered saline-Tween buffer (phosphate-buffered saline, 3% bovine serum albumin and 0.05% Tween-20) overnight at 4 °C. The nitrocellulose membrane was incubated for 2 h with a primary anti-2A peptide antibody (Cat # ABS31. Millipore, Temecula, USA) and then for another 2 h with the secondary antibody (horseradish peroxidase conjugate, goat anti-rabbit IgG (H+L), Beyotime, Shanghai, China). Protein signals were detected using Tanon High-sig ECL Western Blotting Substrate (Tanon, Shanghai, China). Quantification was based on relative band intensities. The band intensities on the membranes were determined using TanonImage software. β-Actin was selected as an internal reference gene.Statistical analysis
The amounts of β-carotene produced by engineered yeast strains were compared using t-test in GraphPad Prism (version 6.0). The data are presented as the averages ± standard deviations of three biological replicates. P-value <0.05 was considered to indicate statistically significant differences.Results
Design of biosynthetic pathway
P2A peptide sequence was used in the assembly of polycistronic constructs because of its highest cleavage efficiency among all 2A peptides analyzed to date.4,19 In such an expression system, the polycistronic construct is transcribed into polycistronic RNA, and several intact proteins are consequently generated after cleavage. In order to simply assess that carbon flux of metabolic pathway was related to the order of complete metabolic pathway genes in a polycistronic P2A peptide construct, the carotenogenic genes, GGPPS (encoding a geranylgeranyl pyrophosphate synthase), CARB (encoding a phytoene desaturase) and CARRP (encoding a bifunctional phytoene synthase and lycopene cyclase) from B. trispora were used to construct heterologous carotenoid pathway in S. cerevisiae (Fig. 2a). The different combinations of two genes linked by P2A peptide sequences were inserted into vector pYX212 (Fig. 2b and c). Due to the absence of CARB, an essential gene of β-carotene metabolic pathway, transformants carrying plasmid pJX1 displayed white. Although strains JX2 and JX3 harbored complete β-carotene pathway, the yields of β-carotene were below the detection limit of HPLC (data not shown). Thus, we were unable to predicate the relationship between carbon flux and the order of genes in such a polycistronic construct. Overexpression of geranylgeranyl diphosphate synthase contributed to increase the production of carotenoid precursors.12,20 Therefore, a polycistronic construct containing three genes, GGPPS, CARB, and CARRP was constructed. As expected, colonies harboring three-genes construct clearly showed a yellow phenotype (Fig. 2c).Carotenoid analysis in the engineered S. cerevisiae strains
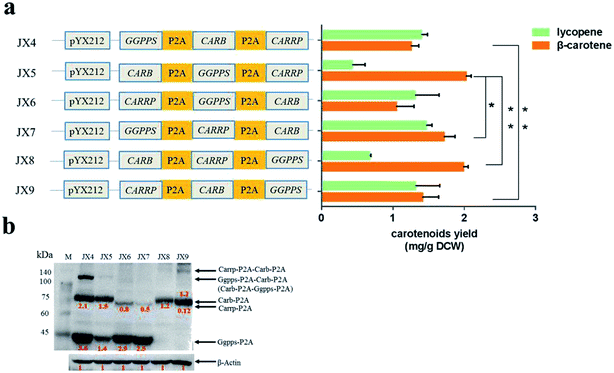
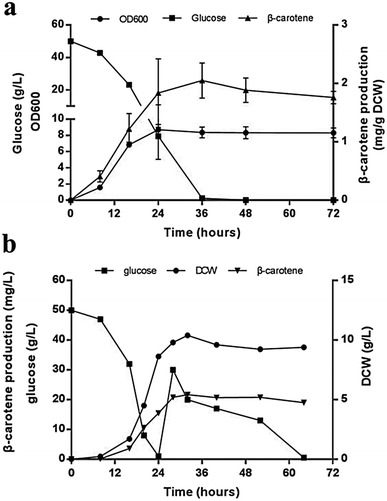
Compared with that in the strains carrying two carotenogenic genes, the yield of β-carotene in transformants harboring three genes was significantly increased. To optimize heterologous β-carotene production in transformants, we further designed another five orders of three carotenogenic genes in polycistronic construct. The carotenoids production were quantified by HPLC and strain JX5, which had CARB locating the first position in the polycistronic construct, produced the highest β-carotene specific titer (2.02 ± 0.08 mg g−1 DCW). Strain JX8, carrying plasmid pJX8 with CARB also as the first gene of the polycistronic segment, produced comparable β-carotene content (1.98 ± 0.07 mg g−1 DCW) (Fig. 3a). To determine whether polycistronic construct was translated into detached proteins, lysates of the six engineered strains were subjected to western blot analysis using anti-2A antibody. We found that comparing to other proteins, expression levels of Carb was probably highest in strain JX5 and JX8 (Fig. 3b). There was no big difference in β-carotene production in comparing the strain couples (JX4 vs. JX9, JX5 vs. JX8), suggesting that the order of GGPPS and CARRP was not essential for β-carotene production. Notably, carbon flux to β-carotene of metabolic pathway was to some extent related to the order of carotenogenic genes in a polycistronic P2A peptide construct.To analyze the accumulation of the β-carotene during the growth of the transformants, we conducted another shake flask fermentation using strain JX5 with 50 g L−1 glucose for 72 h. We found that the β-carotene content increased with the growth of the strains before 36 h. To further prolong the culture time, the carotenoids content kept stable. Furthermore, the strains had no longer grown despite some glucose remaining after 24 h, perhaps due to the exhaustion of certain amino acid. However, the β-carotene content slightly increased until glucose depleted at 36 h (Fig. 4a). This showed that as long as glucose was remaining in the medium, β-carotene production possibly continued to increase. Additionally, pH and dissolved oxygen (DO) are important to carotenoids fermentation.21 Thus, to further assess the relationship between β-carotene content and growth of strain JX5, fermentation was performed in 3 L bioreactor, in which pH was maintained at 6 by adding 2 M NaOH and DO was kept above 80% by adjusting stirring speed. After glucose was exhausted, we performed a supplement with a 750 g L−1 glucose solution. DCW reached its maximum (10.4 g L−1) at 32 h, and the production of β-carotene also reached its highest level (21.7 mg L−1, 2.1 mg g−1 DCW) (Fig. 4b). With the increase of culture time, the carotenoids content didn't further improve even if the carbon source was enough. Comparing the results with that of shake flask fermentation, β-carotene content didn't increase and maintained at a relatively stable level. We found that the accumulation of carotenoids mainly occurred during early fermentation without any overtly negative effects on biomass of transformants.
Discussion
Because of the simultaneous and efficient expression of multiple genes upon protein cleavage in 2A peptides construct, 2A self-cleavage peptides had been actively pursued in many species. Several studies investigated the cleavage activity of various 2A peptides, enabling the generation of multiple proteins from a single polycistronic mRNA.18,22 Others focused on the mechanism of ribosomal release of nascent proteins and the efficiency of cleavage by 2A peptides.2,4,23 In this study, we reconstituted a heterologous carotenoid biosynthetic pathway by linking three carotenogenic genes, GGPPS, CARB and CARRP, via P2A peptide sequences. The ultimate aim was to tune the expression of these genes to facilitate functional optimization of carotenogenic pathway.We asked if the alteration of the order of genes in the polycistronic P2A construct would impact carbon flux of carotenoid pathway. Consequently, we engineered different S. cerevisiae strains harboring variously ordered carotenogenic genes in a polycistronic segment, and evaluated their β-carotene production. Although a complete β-carotene pathway was assembled in strains JX2 and JX3, the yield of β-carotene was quite low due to the lack of adequate supply of precursor GGPP. Thus, to assess the relationship between the yield of β-carotene and the order of genes in the polycistronic construct, overexpression of gene GGPPS was highly necessary. Clearly, among the transformants containing polycistronic construct of different gene order, the yields of β-carotene were certainly different. The highest yield of β-carotene, the lowest yield of lycopene was obtained with strain JX5 and JX8, where CARB was the first gene in the polycistronic construct. To test the cleavage expression of proteins in polycistronic construct, western blot analysis of lysates of the six engineered strains was carried out using anti-2A antibody. The last protein did not fuse with a 2A tag for western blot analysis, thus we could not detect it directly with anti-2A antibody. However, correct phenotype of those transformants indicated the presence of an expected functional protein. From western blot analysis, we found that the expression level of CARB gene was highest in strains JX5 and JX8 with highest β-carotene yield (Fig. 3a and b). This was coincidence with that the translation of the first gene in the polycistronic construct was highly efficient and the translation efficiency of genes located downstream from the P2A sequence gradually reduced.14 Moreover, probably due to the nature of gene CARRP itself, the western signal intensity of Carrp was less intense than the other two proteins even though the gene located in the first location of polycistronic construct. Clearly, incomplete processing of P2A peptide resulted in the formation of polyproteins, such as polyprotein Ggpps-P2A-Carb-P2A in strain JX4, Carb-P2A-Ggpps-P2A in JX5, and Carrp-P2A-Carb-P2A in JX9. To be honest, it was uncertain that the polyproteins were functional. Interestingly, the relative expression of proteins in strain JX4 was higher than that in other strains, but the yield of β-carotene in strain JX4 was relatively low. One possible explanation was that excessive precursor GGPP flowed to a bypass pathway such that the production of β-carotene was decreased. Comparing to Carb protein, Carrp (located at the second or third site of the polycistronic segment) was low in expression but may have a higher catalytic activity, more lycopene was further converted to β-carotene. Consequently, gene CARB could be a key gene of the carotenoid biosynthetic pathway; this was consistent with a previous report on the overexpression of phytoene desaturase gene, by increasing this gene's copy number, which greatly improved the production of β-carotene.12
From shake flask fermentation and fed-batch fermentation results of strain JX5, the yield of β-carotene was almost constant regardless of increased supply of necessary amino acids and glucose. This showed that the intracellular metabolic flux from glucose to β-carotene was severely insufficient. Thus, to further improve the performance of producing β-carotene in strain JX5, extensive strategies could be carried out, such as the increased supply of isoprenoid synthesis precursors in mevalonate pathway, ATP and NADPH in central metabolic pathway, overexpression of carotenogenic genes in β-carotene pathway, and improvement of storage of hydrophobic terpene products.24–26
An additional advantage of optimizing the performance of a biosynthetic pathway in a heterologous host by changing the order of 2A peptide sequence-linked genes in a polycistronic segment is that this approach enables the prediction and regulation of a key gene in a metabolic pathway. If necessary, overexpression of the key gene could further improve the yield of the end product. To the best of our knowledge, this is the first-ever report demonstrating that the optimization of a heterologous biosynthetic pathway may be achieved by exchanging the order in a gene series linked by 2A peptide sequences in a polycistronic segment.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Science Foundation of China (31370128 and 21325627).References
- A. Ahier and S. Jarriault, Genetics, 2014, 196, 605–613 CrossRef CAS PubMed.
- M. L. L. Donnelly, L. E. Hughes, G. Luke, H. Mendoza, E. T. Dam, D. Gani and M. D. Ryan, J. Gen. Virol., 2001a, 82, 1027–1041 Search PubMed.
- E. Minskaia, J. Nicholson and M. D. Ryan, BMC Biotechnol., 2013, 13, 67 CrossRef CAS PubMed.
- J. F. Atkins, N. M. Wills, G. Loughran, C. Y. Wu, K. Parsawar, M. D. Ryan, C. H. Wang and C. C. Nelson, RNA, 2007, 13, 803–810 CrossRef CAS PubMed.
- M. L. Donnelly, D. Gani, M. Flint, S. Monaghan and M. D. Ryan, J. Gen. Virol., 1997, 78, 13–21 CrossRef CAS PubMed.
- M. L. Donnelly, G. Luke, A. Mehrotra, X. J. Li, L. E. Hughes, D. Gani and M. D. Ryan, J. Gen. Virol., 2001b, 82, 1013–1025 Search PubMed.
- Z. Q. Liu, O. Chen, J. B. J. Wall, M. Zheng, Y. Zhou, L. Wang, H. R. Vaseghi, L. Qian and J. D. Liu, Sci. Rep., 2017, 7, 2193 CrossRef PubMed.
- J. M. Fang, J. J. Qian, S. L. Yi, T. C. Harding, G. H. Tu, M. Vanroey and K. Jooss, Nat. Biotechnol., 2005, 23, 584–590 CrossRef CAS PubMed.
- S. E. Unkles, V. Valiante, D. J. Mattern and A. A Brakhage, Chem. Biol., 2014, 21, 502–508 CrossRef CAS PubMed.
- Y. C. Wang, F. Wang, R. Y. Wang, P. Zhao and Q. Y. Xia, Sci. Rep., 2015, 5, 16273 CrossRef CAS PubMed.
- S. W. Kim, J. B. Kim, W. H. Jung, J. H. Kim and J. K. Jung, Biotechnol. Lett., 2006, 28, 897–904 CrossRef CAS PubMed.
- R. Verwaal, J. Wang, J. P. Meijnen, H. Visser, G. Sandmann, J. A. van den Berg and A. J. J. van Ooyen, Appl. Environ. Microbiol., 2007, 73, 4342–4350 CrossRef CAS PubMed.
- J. Beekwilder, H. M. van Rossum, F. Koopman, F. Sonntag, M. Buchhaupt, J. Schrader, R. D. Hall, D. Bosch, J. T. Pronk, A. J. A. van Maris and J. M. Daran, J. Biotechnol., 2014, 192, 383–392 CrossRef CAS PubMed.
- P. Felipe, G. A. Luke, L. E. Hughes, D. Gani, C. Halpin and M. D. Ryan, Trends Biotechnol., 2006, 24, 68–75 CrossRef PubMed.
- F. van den Ent and J. Lowe, J. Biochem. Biophys. Methods, 2006, 67, 67–74 CrossRef CAS PubMed.
- R. D. Gietz and R. H. Schiestl, Nat. Protoc., 2007, 2, 1–4 CrossRef CAS PubMed.
- C. L. Zhang, H. W. Shen, X. B. Zhang, X. Yue, H. Wang, S. Xiao, J. H. Wang and Z. B. Zhao, Biotechnol. Lett., 2016, 38, 1733–1738 CrossRef CAS PubMed.
- H. W. Liu, X. Zhao, F. J. Wang, Y. H. Li, X. N. Jiang, M. L. Ye, Z. B. K. Zhao and H. F. Zou, Yeast, 2009, 26, 553–566 CrossRef CAS PubMed.
- J. H. Kim, S. R. Lee, L. H. Li, H. J. Park, J. H. Park, K. Y. Lee, M. K. Kim, B. A. Shin and S. Y. Choi, PLoS One, 2011, 6, e18556 CrossRef CAS PubMed.
- F. Matthäus, M. Ketelhot, M. Gatter and G. Barth, Appl. Environ. Microbiol., 2014, 80, 1660–1669 CrossRef PubMed.
- S. L. Gao, Y. Y. Tong, L. Zhu, M. Ge, Y. A. Zhang, D. J. Chen, Y. Jiang and S. Yang, Metab. Eng., 2017, 41, 192–201 CrossRef CAS PubMed.
- C. Halpin, S. E. Cooke, A. Barakate, A. E. I. Amrani and M. D. Ryan, Plant J., 1999, 17, 453–459 CrossRef CAS PubMed.
- T. M. Souza-Moreira, C. Navarrete, X. Chen, C. F. Zanelli, S. R. Valentini, M. Furlan, J. Nielsen and A. Krivoruchko, FEMS Yeast Res., 2018, 18, foy036 CrossRef PubMed.
- V. J. J. Martin, D. J. Pitera, S. T. Withers, J. D. Newman and J. D. Keasling, Nat. Biotechnol., 2003, 21, 796–802 CrossRef CAS PubMed.
- J. Zhao, Q. Y. Li, T. Sun, X. N. Zhu, H. T. Xu, J. L. Tang, X. L. Zhang and Y. H. Ma, Metab. Eng., 2013, 17, 42–50 CrossRef CAS PubMed.
- T. Wu, L. J. Ye, D. D. Zhao, S. W. Li, Q. Y. Li, B. L. Zhang, C. H. Bi and X. L. Zhang, Metab. Eng., 2017, 43, 85–91 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra06510a |
| This journal is © The Royal Society of Chemistry 2018 |