Open Access Article
Open Access ArticleRetracted Article: Long non-coding RNA KCNQ1OT1 promotes osteosarcoma progression by increasing β-catenin activity
Changsheng Zhang†
 *a,
Shengyang Du†b and
Lei Caob
*a,
Shengyang Du†b and
Lei Caob
aDepartment of Minimally Invasive Spine Surgery, Henan Luoyang Orthopedic Hospital (Henan Provincial Orthopedic Hospital), No. 100 Yongping road, Zhengzhou, 450000, Henan, China. E-mail: hlohchanger@sina.com; Tel: +86-0371-85965160
bDepartment of Orthopaedics, The First People's Hospital of Xuzhou, Quanshan, 221000, Xuzhou, Jiangsu, China
First published on 8th November 2018
Abstract
Objective: Long non-coding RNA KCNQ1OT1 has been associated with the development of different types of cancers. The present research investigated the role of KCNQ1OT1 in osteosarcoma. Methods: Expression level of KCNQ1OT1 in osteosarcoma and paired non-cancerous tissue specimens from 56 osteosarcoma patients and its association with patients' clinicopathological features was investigated. KCNQ1OT1 overexpression and knockdown in primary-cultured osteosarcoma cells was constructed by lentiviral transduction. Influence of KCNQ1OT1 overexpression or knockdown on osteosarcoma cell growth, apoptosis, migration, invasion, epithelial-to-mesenchymal transition and beta-catenin activation was investigated. Results: Expression of KCNQ1OT1 in osteosarcoma tissue specimens was significantly increased in comparison to that in adjacent counterparts. High expression of KCNQ1OT1 significantly associated with osteosarcoma progression and patients' decreased survival. Overexpression of KCNQ1OT1 significantly increased osteosarcoma cell growth, proliferation, migration, invasion, epithelial-to-mesenchymal transition and beta-catenin activation while reducing cell apoptosis in vitro, and KCNQ1OT1 knockdown showed opposite effects. Inhibition of beta-catenin/TCF activity by ICG-001 treatment significantly attenuated the promoting effect of KCNQ1OT1 overexpression on osteosarcoma cell malignancy described above. Conclusion: KCNQ1OT1 might be a potential prognostic factor in osteosarcoma. High expression of KCNQ1OT1 might promote osteosarcoma development by increasing the activation of WNT/beta-catenin signaling pathway.
Introduction
Osteosarcoma is currently the most frequent primary bone tumor.1,2 Long non-coding RNAs (lncRNAs) have been found to regulate cell proliferation and metastasis of osteosarcoma cells through different mechanisms.3,4 LncRNAs are non-coding transcripts that regulate gene expression or protein function by interacting with DNA, RNA or proteins.5 Particularly, lncRNAs might function as competitive endogenous RNA than bind to and “sponge” microRNAs, preventing these small RNAs from binding to their target mRNAs that induces mRNA degradation or represses protein translation.6 LncRNA KCNQ1OT1 (previously named LIT1) is an antisense lncRNA encoded in human chromosome 11p15 that have been implicated in Beckwith–Wiedemann syndrome, a congenital overgrowth disorder characterized by imprinting disruption in chromosome 11p15 and increased risk of childhood cancer.7 Knockdown of KCNQ1OT1 has been reported to decrease the malignancy of glioma,8 hepatocellular carcinoma9 and tongue cancer cells10 while reducing chemoresistance of other cancer types,10–12 suggesting the cancer-promoting role of this lncRNA. Notably, Li et al. have reported that KCNQ1OT1 could increase beta-catenin protein level in hepatocellular carcinoma cells, presumably by reducing beta-catenin degradation via miR-504 sponging,9 while Sunamura et al. have demonstrated that beta-catenin can bind to KCNQ1OT1 promotor and activate its transcription in colon cancer cells, thus contributing to colorectal cancer development.13The role of KCNQ1OT1 in osteosarcoma hasn't been addressed before, but previous studies have confirmed that Beckwith–Wiedemann syndrome increases the risk of rhabdomyosarcoma in association with abnormal increase in KCNQ1OT1 expression due to loss-of-imprinting.14,15 A previous research by Gao et al. demonstrated that overexpressing KCNQ1OT1 in osteoblast cells might increase beta-catenin expression and its activity, presumably by binding to beta-catenin.16 Beta-catenin is a transcription co-activator, complexing with TCF/LEF family transcription factors to activate the transcription of target genes such as MYC, JUN, WISP1 and CCND1 that are involved in cell proliferation, survival and mobility.17,18 Activation of WNT/beta-catenin signaling pathway has been demonstrated to promote osteosarcoma cell proliferation, metastasis and chemoresistance, and inhibition of this osteosarcoma-promoting pathway might synergize with current therapeutic approaches and improve patients' outcome.19–21
In the present research we investigated whether KCNQ1OT1 was involved in osteosarcoma development. Our data demonstrated that KCNQ1OT1 expression was significantly increased in osteosarcoma tissue specimens comparing to non-cancerous counterparts. High expression of KCNQ1OT1 significantly correlated with osteosarcoma progression. Overexpression of KCNQ1OT1 significantly increased malignancy of primary osteosarcoma cells in vitro, while KCNQ1OT1 knockdown showed opposite effect. We further demonstrated that KCNQ1OT1 increased beta-catenin activity, which was involved in the osteosarcoma-promoting role of KCNQ1OT1.
Experimental
Patients and tissue specimens
Osteosarcoma tissue and paired non-cancerous tissue specimens were obtained by biopsy via fine needle aspiration from 56 osteosarcoma patients before treatment in Henan Luoyang Orthopedic Hospital during 2005–2016. Tumor tissue specimens were stored in liquid nitrogen for further analysis. Patients' basic information and clinicopathological features related to this research were presented in Table 1. All experiments were performed in accordance with the Guidelines of “Experimental Animals Ethics Committee for Animal Research of Henan Luoyang Orthopedic Hospital (Henan Provincial Orthopedic Hospital)”, and Experiments were approved by the ethics committee at “Henan Luoyang Orthopedic Hospital (Henan Provincial Orthopedic Hospital)”. Informed consents were obtained from human participants of this study.| Categories | n | KCNQ1OT1 expression | p value | ||
|---|---|---|---|---|---|
| Low | High | ||||
| a LN, lymph node.b p < 0.05 by Chi-square test.c p < 0.05 by Chi-square test for trend. | |||||
| Age (year) | |||||
| ≤30 | 35 | 18 | 17 | 0.7825 | |
| >30 | 21 | 10 | 11 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Gender | |||||
| Male | 29 | 13 | 16 | 0.4224 | |
| Female | 27 | 15 | 12 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Tumor location | |||||
| Tibia/femur | 39 | 18 | 21 | 0.3833 | |
| Other | 17 | 10 | 7 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Tumor stage | |||||
| I and II | 29 | 19 | 10 | 0.0161b | |
| III and IV | 27 | 9 | 18 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Tumor size | |||||
| T1 | 23 | 15 | 8 | 0.0445c | |
| T2 | 23 | 10 | 13 | ||
| T3 | 10 | 3 | 7 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| LNa metastasis | |||||
| No | 43 | 25 | 18 | 0.0267b | |
| Yes | 13 | 3 | 10 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Distal metastasis | |||||
| No | 39 | 23 | 16 | 0.0419b | |
| Yes | 17 | 5 | 12 | ||
Establishment of osteosarcoma primary cell culture
A 27 year-old male Chinese osteosarcoma patient received biopsy for diagnosis purpose before chemotherapy and surgery. The biopsy sample was washed with warm PBS and minced under sterile condition, followed by digestion with 150 U mL−1 (in DMEM complete culture medium, described later) of Type I collagenase for 3 hours under 37 °C. Single cells suspension was obtained by passing the biopsy sample after digestion through a 70 μm cell strainer (Sangon Biotech, Shanghai, China) and centrifugation. The cell pellet was resuspended in DMEM high glucose medium (Zhong Qiao Xin Zhou Biotechnology, Shanghai, China) supplemented with 10% FBS (Invitrogen) and 1% penicillin/streptomycin stock solution (Sciencell), and the cells were cultured in a 35 mm Petri dish in a humidified atmosphere at 37 °C with 5% CO2 until confluent without changing the medium. Cells were then sub-cultured at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 split ratio, and osteosarcoma cell colonies were picked out with a cell scraper (Costar). The osteosarcoma cells were maintained in vitro as described above, and their tumorigenicity was confirmed by tumor formation assay using 6 week-old male BALB/c nude mice (Vital river, Beijing, China).
5 split ratio, and osteosarcoma cell colonies were picked out with a cell scraper (Costar). The osteosarcoma cells were maintained in vitro as described above, and their tumorigenicity was confirmed by tumor formation assay using 6 week-old male BALB/c nude mice (Vital river, Beijing, China).
Lentiviral transduction and ICG-001 treatment
KCNQ1OT1 overexpression or knockdown on osteosarcoma cells was achieved by lentiviral transduction. Customized viral particles for KCNQ1OT1 overexpression or knockdown by shRNA expression were purchased from Cyagen (Suzhou, China) and were used following manufacturer's instructions. Briefly, 2 × 105 cells were incubated with 10 μL of lentiviral particle suspension (at a titer of about 1 × 108 TU ml−1) with the presence of 5 μg mL−1 polybrene (EMD Millipore) for 72 hours. Cells transduced with empty vector (WT) was used as control for lentiviral transduction. Cells with positive transduction were selected with puromycin (Sigma). To inhibit the transcriptional activity of beta-catenin/TCF complex, cells were treated in vitro with 100 μM of ICG-001 (Selleck, Shanghai, China) for 24 hours before further analysis. ICG-001 was dissolved in DMSO (Sigma-Aldrich) as a 100 mM stock before use.RT-qPCR and western blot
For RT-qPCR, total RNA in cell culture or tissue specimens were extracted using Trizol reagent (Invitrogen). RT-qPCR detecting KCNQ1OT1 abundance in total RNA samples was performed using cDNA synthesis kit and SYBR green qPCR master mix (QP009 and QP005, respectively, Genecopoeia, Guangzhou, China) following manufacturer's instructions. GAPDH mRNA was used as internal reference for comparing KCNQ1OT1 expression level using 2−ΔΔCt method. Primer sequences used for RT-qPCR were as follows: KCNQ1OT1 FW: TTGGTAGGATTTTGTTGAGG; KCNQ1OT1 RV: CAACCTTCCCCTACTACC; GAPDH FW: TTTGGTCGTATTGGGCGCCTGGTCA; GAPDH RV: TTGTGCTCTTGCTGGGGCTGGTGGT. Western blot was performed using reducing SDS-PAGE gel to minimized background noise. Primary antibodies for western blot detecting protein level of GAPDH (NB100-56875), PCNA (NB500-106), MCM-2 (H00004171-M01), and cleaved PARP (NBP2-67454) were purchased from Novus Biologicals. Primary antibodies corresponding to protein of cleaved Caspase-3 (ab32042) N-cadherin (ab18203), E-cadherin (ab1416), GSK3B (ab131356), beta-catenin (ab16051) and beta-catenin Serine37 phosphorylation (ab75777) were purchased from Abcam. Secondary antibodies (ab6789, ab97057 and ab6721) were purchased from Abcam. Primary antibodies were used at a working concentration of 1 μg ml−1, and secondary antibodies were used at a dilution factor of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Western blot was detected with Pierce ECL substrate and X-ray films (32209 and 34090, respectively, Thermo Fisher Scientific). Analysis on relative protein expression level was performed using ImageJ software, and GAPDH protein was used as internal reference.
000. Western blot was detected with Pierce ECL substrate and X-ray films (32209 and 34090, respectively, Thermo Fisher Scientific). Analysis on relative protein expression level was performed using ImageJ software, and GAPDH protein was used as internal reference.
Cell functional assays in vitro
Cell growth was assayed by evaluating cell viability at different timepoint. About 4000 cells per well of each group were seeded on 96-well-plate, and cell viability at 24, 48 and 72 hours post seeding was monitored using CCK-8 kit (C0040, Beyotime, Shanghai, China) following manufacturer's instructions. Apoptosis in cell culture was evaluated by FITC-Annexin V/PI double staining followed by flow cytometry. Cells were harvested by trypsin digestion and labeled with Annexin A5 Apoptosis Detection Kit (640932, Biolegend) following manufacturer's instructions. Cells with positive FITC-Annexin V labeling were considered apoptotic cells. Cell invasion or migration was assayed in vitro using cell culture inserts (8 mm pore size in 6-well-plate format, Thermo Fisher Scientific) with or without collagen precoat. About 2 × 105 cells suspended in FBS-deprived culture medium were added into the cell culture insert, which was loaded on 6-well-plate in complete culture medium. The cells were incubated under cell culture conditions for 24 hours, and cells migrated to the bottom of the insert were stained with crystal violet (Beyotime) and counted under microscope.Wnt reporter activity assay
Wnt reporter activity assay was performed using TCF Reporter Plasmid Kit (17-285, Sigma-Aldrich). 5000 osteosarcoma cells were seeded into each well on a 24-well-plate and pre-cultured for 24 hours. Osteosarcoma cells in each well were then transfected with 250 ng of TOPflash reporter plasmids and 50 ng of FOPflash reporter plasmids using 1.5 μL of Lipofectamin 3000 (Thermo Fisher Scientific). 48 hours after transfection, osteosarcoma cells were serum deprived for 24 hours to stimulate the activation of WNT/beta-catenin signaling, after which the cells were lysed, and firefly/Renilla luciferase activity was evaluated using the dual-luciferase reporter assay system (E1910, Promega) with a microplate reader following manufacturer's instructions. Activity of Renilla luciferase expressed by FOPflash plasmid was used as internal reference for that of firefly luciferase expressed by TOPflash in response to beta-catenin/TCF activity.Statistical analysis
Statistical analysis was performed using IBM SPSS (Ver.19) software. Data group in each graph represents 6 independent replicates and was presented as fold change in mean ± SD form when applicable, unless otherwise indicated. Data in Table 1 were analyzed by Chi-square test or Chi-square test for trend; data in each graph were analyzed by Students' t test or Tuckey's test. A p < 0.05 was considered significance.Results and discussion
High KCNQ1OT1 expression associated with osteosarcoma development
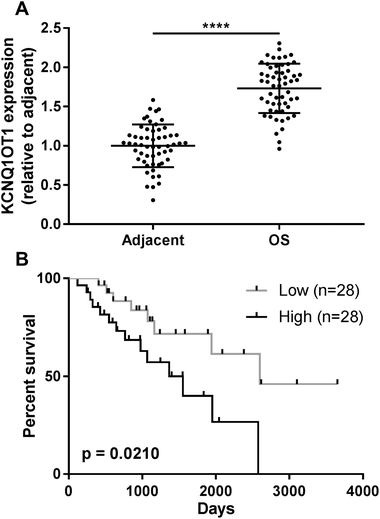
The present research aimed to investigated the role of KCNQ1OT1 in osteosarcoma development. To do this, we first analyzed KCNQ1OT1 expression level in paired osteosarcoma and non-cancerous tissue specimens obtained from 56 patients. As shown in Fig. 1A, KCNQ1OT1 expression level in osteosarcoma tissue specimens was significantly higher than that in non-cancerous ones. We stratified the patients to KCNQ1OT1 high expression and low expression groups according to the KCNQ1OT1 median expression level, and investigated the association between KCNQ1OT1 expression level and osteosarcoma patients' clinicopathological features. As demonstrated in Table 1 and Fig. 1B, high expression of KCNQ1OT1 significantly associated with increased tumor stage, tumor size, lymph node and distal metastasis as well as decreased overall survival. These data suggested that increase in KCNQ1OT1 expression might participate in osteosarcoma development.KCNQ1OT1 promote osteosarcoma cell growth, migration, invasion and epithelial-to-mesenchymal transition while reducing cell apoptosis in vitro
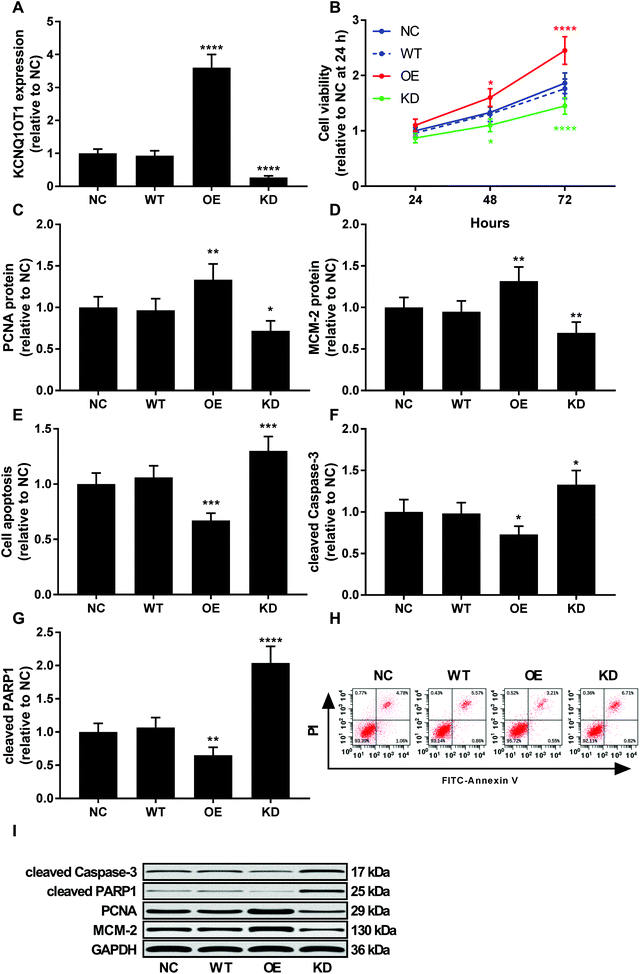
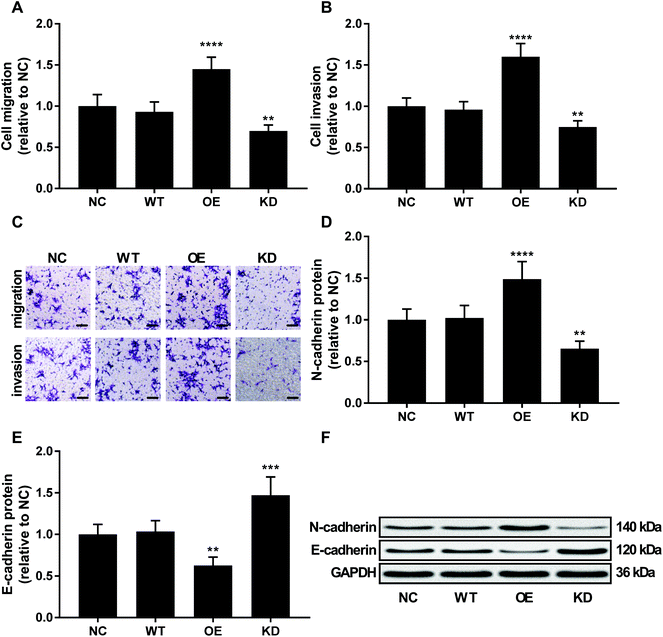
To investigate the role of KCNQ1OT1 in osteosarcoma cells, we constructed KCNQ1OT1 overexpression or knockdown in primary osteosarcoma cells (Fig. 2A). Results of cell viability assay and western blot evaluating the protein level of cell proliferation marker PCNA and MCN-2 suggested that KCNQ1OT1 overexpression significantly increased primary osteosarcoma cell growth and proliferation in vitro, while KCNQ1OT1 knockdown showed the opposite effects (Fig. 2B–D). Osteosarcoma cell apoptosis in vitro was significantly decreased by KCNQ1OT1 overexpression and increased by its knockdown, as suggested by the results of flow cytometry assay and western blot evaluating caspase-3 activation and PARP cleavage as indicators of cell apoptosis (Fig. 2E–I). Transwell migration and invasion assay further indicated that KCNQ1OT1 overexpression significantly enhanced osteosarcoma cell migration and invasion in vitro, which was significantly impaired by KCNQ1OT1 knockdown (Fig. 3A–C). Epithelial-to-mesenchymal transition is a critical process involved in osteosarcoma cell survival, proliferation and metastasis.22 Western blot evaluating the protein level of mesenchymal marker protein N-cadherin and epithelial marker protein E-cadherin revealed that KCNQ1OT1 overexpression significantly increased mesenchymal phenotype and decreased epithelial phenotype in osteosarcoma cells in vitro, while KCNQ1OT1 knockdown showed opposite effects (Fig. 3D–F), suggesting that KCNQ1OT1 could promote epithelial-to-mesenchymal transition in osteosarcoma cells.KCNQ1OT1 increased beta-catenin activity in osteosarcoma cells in vitro
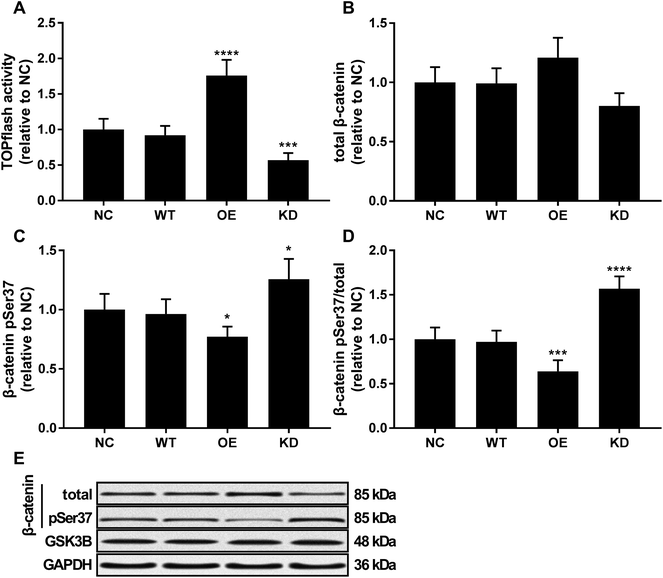
Beta-catenin activated by the canonical WNT/beta-catenin signaling pathway has been demonstrated to promote osteosarcoma development by interacting with TCF/LEF complex in the nucleus, activating the transcription of other tumor promoting genes such as MYC and CCND1.20 Using a TOPflash/FOPflash system monitoring beta-catenin activity, we found that KCNQ1OT1 overexpression significantly increased beta-catenin activity in osteosarcoma cells in vitro, which was significantly reduced by KCNQ1OT1 knockdown (Fig. 4A). No significant impact by KCNQ1OT1 overexpression or knockdown on beta-catenin protein level in osteosarcoma was found (Fig. 4B); we also performed RT-qPCR evaluating CTNNB1 mRNA expression but found no significant change after KCNQ1OT1 overexpression or knockdown (data not shown). However, KCNQ1OT1 overexpression significantly reduced beta-catenin Ser37 phosphorylation, a marker of beta-catenin inactivation and ubiquitin-mediated degradation, which is significantly increased by KCNQ1OT1 knockdown (Fig. 4C and D). GSK3B is the serine/threonine kinase responsible for beta-catenin Ser37 phosphorylation, and we found that GSK3B protein level in osteosarcoma cells was not altered by KCNQ1OT1 overexpression or knockdown (Fig. 4E). These data seemed to suggest that KCNQ1OT1 could increase beta-catenin activity not by regulating its expression but by reducing its inactivation.KCNQ1OT1 increased the malignancy of osteosarcoma cells through beta-catenin
ICG-001 is a small molecule that binds to CREB-binding protein (CBP) and block the interaction between beta-catenin and CBP, thus reducing the transcriptional activity of beta-catenin.23,24 To investigate whether beta-catenin activity was involved in KCNQ1OT1 promoting osteosarcoma development, we used ICG-001 treatment to inhibit the transcriptional activity of beta-catenin. As shown in Fig. 5, treatment with ICG-001 significantly reduced cell proliferation, survival, migration, invasion and epithelial-to-mesenchymal transition of osteosarcoma cells with different KCNQ1OT1 gene manipulation. Notably, osteosarcoma cells with KCNQ1OT1 showed increased sensitivity to ICG-001 treatment comparing to wild type one, while those with KCNQ1OT1 knockdown were less prone to ICG-001 treatment induced beta-catenin inhibition. These data indicated that KCNQ1OT1 might promote osteosarcoma development in part through beta-catenin.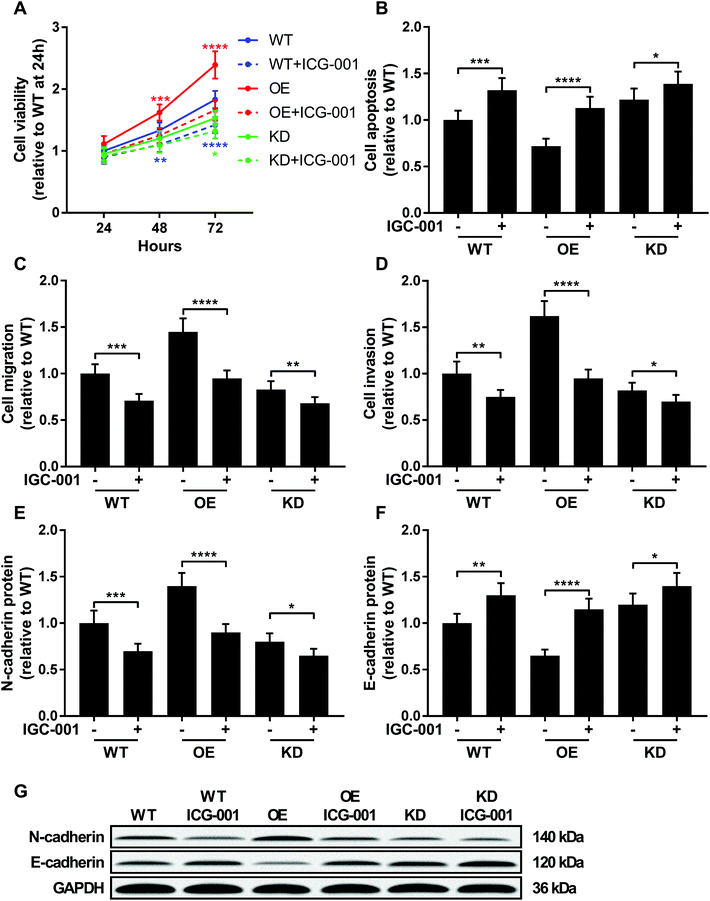 | ||
| Fig. 5 KCNQ1OT1 increased osteosarcoma cells malignancy through beta-catenin. Wild type (WT), KCNQ1OT1 overexpressing (OE) or KCNQ1OT1 knockdown (KD) osteosarcoma cells were treated with or without 100 μM of ICG-001 for 24 hours before assay. (A to D) Cell viability, apoptosis, migration, invasion and (E to G) epithelial-to-mesenchymal transition were assayed as described in Fig. 2 and 3. (+) treatment with IGC-001; (−) treatment with isometric vehicle (DMSO). * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001. | ||
Discussions
WNT/beta-catenin signaling pathway participates in osteosarcoma cell proliferation, metastasis, stemness gain and chemoresistance, and inhibition of this pathway might inhibit osteosarcoma progression.25–28 Previous research demonstrated that KCNQ1OT1 increases beta-catenin protein level and its transcriptional activity in osteoblast cells.16 We investigated in the present research whether KCNQ1OT1 was involved in osteosarcoma development. Our data for the first time demonstrated a significant increase in KCNQ1OT1 expression level in osteosarcoma and its association with osteosarcoma progression. Cell functional assays results demonstrated that KCNQ1OT1 promoted osteosarcoma cell malignancy in vitro, possibly by increasing the transcriptional activity of beta-catenin.Beta-catenin activity in osteosarcoma cells has been found regulated by other lncRNAs. For incidence, Li et al. demonstrated that lncRNA HOTTIP promoted osteosarcoma cell proliferation and cisplatin resistance in vitro possibly by regulating beta-catenin protein level,29 while Zhao et al.'s report showed similar effect of lncRNA HNF1A-AS1 on osteosarcoma cell proliferation and metastasis.30 Although the present research have implicated KCNQ1OT1 with osteosarcoma development, presumably by increasing beta-catenin signaling, the mechanism of action of KCNQ1OT1 in osteosarcoma cells remains largely unknown. KCNQ1OT1 has been found to promote cancer progression mostly via miRNA sponging. Gong et al. previously reported that KCNQ1OT1 might promote glioma development by sponging miR-370,8 the overexpression of which has been found to reduce beta-catenin protein level and transcriptional activity in osteosarcoma cells.31
LncRNAs interacting with beta-catenin has been demonstrated to prevent beta-catenin degradation,32,33 and research by Gao et al. have shown the KCNQ1OT1-beta-catenin association.16 In the present research we didn't observe any significant influence on beta-catenin protein level in osteosarcoma by KCNQ1OT1 overexpression or knockdown; however, KCNQ1OT1 overexpression significantly increased beta-catenin's transcriptional activity and reduced beta-catenin Ser37 phosphorylation level, which is one of the initiative steps leading to beta-catenin degradation, while KCNQ1OT1 knockdown showed opposite effects. Intuitively, association of beta-catenin with APC and GSK-3beta proteins who plays central part in the beta-catenin destruction complexs should reduce beta-catenin's nuclear entry and its transcriptional activity. Considering that Sunamura et al.'s research showed that beta-catenin could trans-activate KCNQ1OT1 expression,13 we anticipated from our experimental results and previously published data cited above that KCNQ1OT1 might function as a positive feedback mechanism that augments WNT/beta-catenin signaling in osteosarcoma cells by reducing beta-catenin degradation, possibly by binding to it. We presume that binding of KCNQ1OT1 to beta-catenin might changes the conformation of the latter or blocks the binding site on the latter to APC and/or GSK-3beta, resulting the decrease in beta-catenin protein degradation. This possible mechanism should be better verified in osteosarcoma and other tumors.
Regulation of KCNQ1OT1 expression in osteosarcoma or other cancer cells remains uncharacterized. We previously utilized cBioportal website to explore the expression of KCNQ1OT1 in TCGA Soft Tissue Sarcoma dataset (http://www.cbioportal.org/study?id=sarc_tcga#summary),34,35 and we found that KCNQ1OT1 expression level inversely correlated to its gene methylation level (data not shown). KCNQ1OT1 expression in cancer cells might be regulated by epigenetic mechanism that deserves further investigation.
Conclusion
Overall, our data demonstrated that KCNQ1OT1 expression was significantly increased in osteosarcoma tissue comparing to non-cancerous counterparts. High expression of KCNQ1OT1 significantly associated with osteosarcoma progression and patients' decreased survival. Overexpressing KCNQ1OT1 significantly increased osteosarcoma cell proliferation, metastatic potential and epithelial-to-mesenchymal transition in vitro while increasing beta-catenin transcriptional activity. Based on our data and previous findings we suggest that KCNQ1OT1 might be a prognostic factor in osteosarcoma, and targeting this lncRNA might improve osteosarcoma treatment outcome and patients'prognosis.Funding
None.Conflicts of interest
The co-authors declare that they have no conflicts of interest.Acknowledgements
Not applicable.References
- M. S. Isakoff, S. S. Bielack, P. Meltzer and R. Gorlick, J. Clin. Oncol., 2015, 33, 3029–3035 CrossRef CAS PubMed.
- J. Ritter and S. S. Bielack, Ann. Oncol., 2010, 21(Suppl. 7), vii320–325 Search PubMed.
- Z. Yang, X. Li, Y. Yang, Z. He, X. Qu and Y. Zhang, Cell Death Dis., 2016, 7, e2389 CrossRef CAS PubMed.
- M. A. Smolle and M. Pichler, Noncoding RNAs, 2018, 4, 7 Search PubMed.
- A. M. Schmitt and H. Y. Chang, Cancer Cell, 2016, 29, 452–463 CrossRef CAS PubMed.
- C. Mayr, Annu. Rev. Genet., 2017, 51, 171–194 CrossRef CAS PubMed.
- K. Higashimoto, H. Soejima, T. Saito, K. Okumura and T. Mukai, Cytogenet. Genome Res., 2006, 113, 306–312 CrossRef CAS PubMed.
- W. Gong, J. Zheng, X. Liu, Y. Liu, J. Guo, Y. Gao, W. Tao, J. Chen, Z. Li, J. Ma and Y. Xue, Front. Cell. Neurosci., 2017, 11, 84 Search PubMed.
- C. Li, R. Miao, J. Zhang, K. Qu and C. Liu, Internet J. Oncol., 2018, 52, 1603–1612 Search PubMed.
- S. Zhang, H. Ma, D. Zhang, S. Xie, W. Wang, Q. Li, Z. Lin and Y. Wang, Cell Death Dis., 2018, 9, 742 CrossRef PubMed.
- H. Hu, L. Yang, L. Li and C. Zeng, Biochem. Biophys. Res. Commun., 2018, 503, 2400–2406 CrossRef CAS PubMed.
- K. Ren, R. Xu, J. Huang, J. Zhao and W. Shi, Cancer Chemother. Pharmacol., 2017, 80, 243–250 CrossRef CAS PubMed.
- N. Sunamura, T. Ohira, M. Kataoka, D. Inaoka, H. Tanabe, Y. Nakayama, M. Oshimura and H. Kugoh, Sci. Rep., 2016, 6, 20690 CrossRef CAS PubMed.
- F. Piersigilli, C. Auriti, V. Mondi, P. Francalanci, G. Salvatori and O. Danhaive, Indian J. Pediatr., 2016, 83, 1476–1478 CrossRef PubMed.
- M. M. Cohen Jr, Pediatr. Dev. Pathol., 2005, 8, 287–304 CrossRef PubMed.
- X. Gao, J. Ge, W. Li, W. Zhou and L. Xu, Cell Biosci., 2018, 8, 19 CrossRef PubMed.
- K. Saito-Diaz, T. W. Chen, X. Wang, C. A. Thorne, H. A. Wallace, A. Page-McCaw and E. Lee, Growth Factors, 2013, 31, 1–31 CrossRef CAS PubMed.
- R. Nusse and H. Clevers, Cell, 2017, 169, 985–999 CrossRef CAS PubMed.
- J. Tian, H. He and G. Lei, Tumor Biol., 2014, 35, 9439–9445 CrossRef CAS PubMed.
- C. H. Lin, T. Ji, C. F. Chen and B. H. Hoang, Adv. Exp. Med. Biol., 2014, 804, 33–45 CrossRef CAS PubMed.
- C. Li, X. Shi, G. Zhou, X. Liu, S. Wu and J. Zhao, Front. Biosci., Landmark Ed., 2013, 18, 1384–1391 CrossRef CAS.
- G. Yang, J. Yuan and K. Li, Med. Oncol., 2013, 30, 697 CrossRef PubMed.
- M. Eguchi, C. Nguyen, S. C. Lee and M. Kahn, Med. Chem., 2005, 1, 467–472 CrossRef CAS.
- W. R. Henderson Jr, E. Y. Chi, X. Ye, C. Nguyen, Y. T. Tien, B. Zhou, Z. Borok, D. A. Knight and M. Kahn, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 14309–14314 CrossRef PubMed.
- W. Liu, Z. Zhao, Y. Wang, W. Li, Q. Su, Q. Jia, J. Zhang, X. Zhang, J. Yin and J. Shen, Cell Death Dis., 2018, 9, 343 CrossRef PubMed.
- S. R. Martins-Neves, D. I. Paiva-Oliveira, C. Fontes-Ribeiro, J. Bovee, A. M. Cleton-Jansen and C. M. F. Gomes, Cancer Lett., 2018, 414, 1–15 CrossRef CAS PubMed.
- P. Chen, W. L. Gu, M. Z. Gong, J. Wang and D. Q. Li, Cell. Physiol. Biochem., 2017, 43, 1939–1949 CrossRef CAS PubMed.
- S. R. Martins-Neves, D. I. Paiva-Oliveira, P. M. Wijers-Koster, A. J. Abrunhosa, C. Fontes-Ribeiro, J. V. Bovee, A. M. Cleton-Jansen and C. M. Gomes, Cancer Lett., 2016, 370, 286–295 CrossRef CAS PubMed.
- Z. Li, L. Zhao and Q. Wang, Am. J. Transl. Res., 2016, 8, 2385–2393 CAS.
- H. Zhao, W. Hou, J. Tao, Y. Zhao, G. Wan, C. Ma and H. Xu, Am. J. Transl. Res., 2016, 8, 3503–3512 CAS.
- W. Zhang, N. Duan, Q. Zhang, T. Song, Z. Li, C. Zhang, X. Chen and K. Wang, Int. J. Biol. Sci., 2017, 13, 561–573 CrossRef CAS PubMed.
- H. Qian, L. Chen, J. Huang, X. Wang, S. Ma, F. Cui, L. Luo, L. Ling, K. Luo and G. Zheng, J. Mol. Med., 2018, 96, 1–12 CrossRef PubMed.
- S. Ouyang, X. Zheng, X. Zhou, Z. Chen, X. Yang and M. Xie, Oncotarget, 2017, 8, 92815–92826 Search PubMed.
- J. Gao, B. A. Aksoy, U. Dogrusoz, G. Dresdner, B. Gross, S. O. Sumer, Y. Sun, A. Jacobsen, R. Sinha, E. Larsson, E. Cerami, C. Sander and N. Schultz, Sci. Signaling, 2013, 6, pl1 CrossRef PubMed.
- E. Cerami, J. Gao, U. Dogrusoz, B. E. Gross, S. O. Sumer, B. A. Aksoy, A. Jacobsen, C. J. Byrne, M. L. Heuer, E. Larsson, Y. Antipin, B. Reva, A. P. Goldberg, C. Sander and N. Schultz, Cancer Discovery, 2012, 2, 401–404 CrossRef PubMed.
Footnote |
| † These authors contributed to this work equally. |
| This journal is © The Royal Society of Chemistry 2018 |