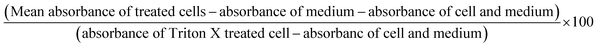Open Access Article
Open Access ArticleQuantum curcumin mediated inhibition of gingipains and mixed-biofilm of Porphyromonas gingivalis causing chronic periodontitis†
Ashish Kumar Singhab,
Shivangi Yadava,
Kavanjali Sharmac,
Zeba Firdausd,
Prerana Aditid,
Kaushik Neogie,
Monika Bansalf,
Munesh Kumar Guptaa,
Asheesh Shankerg,
Rakesh Kumar Singhb and
Pradyot Prakash *a
*a
aBacterial Biofilm and Drug Resistance Research Laboratory, Department of Microbiology, Institute of Medical Sciences, Banaras Hindu University, Varanasi 221005, India. E-mail: pradyot_micro@bhu.ac.in
bMolecular Immunology Laboratory, Department of Biochemistry, Institute of Science, Banaras Hindu University, Varanasi 221005, India
cDepartment of Pathology, Institute of Medical Sciences, Banaras Hindu University, Varanasi 221005, India
dDepartment of Medicinal Chemistry, Institute of Medical Sciences, Banaras Hindu University, Varanasi 221005, India
eDepartment of Pharmaceutical Engineering and Technology, Indian Institute of Technology, Banaras Hindu University, Varanasi 221005, India
fFaculty of Dental Sciences, Institute of Medical Science, Banaras Hindu University, Varanasi 221005, India
gDepartment of Bioinformatics, Central University of South Bihar, Gaya 824236, Bihar, India
First published on 5th December 2018
Abstract
Periodontitis is a biofilm-associated irreversible inflammation of the periodontal tissues. Reports suggest the role of Porphyromonas gingivalis specific Arg- and Lys-specific proteinases in the orchestration of the initiation and progression of periodontal diseases. These proteinases are precisely termed as gingipains R and K. Curcumin is an active polyphenol that is extracted from the rhizomes of Curcuma longa. However, the molecule curcumin owing to its high hydropathy index and poor stability has not been able to justify its role as frontline drug modality in the treatment of infectious and non-infectious diseases as claimed by several investigators. In the present study, at first, we synthesized and characterized quantum curcumin, and investigated its biocompatibility. This was subsequently followed by the evaluation of the role of quantum curcumin as an antimicrobial, anti-gingipains and antibiofilm agent against Porphyromonas gingivalis and select reference strains. We have successfully synthesized the quantum curcumin utilizing a top-down approach with the average size of 3.5 nm. Apart from its potent antimicrobial as well as antibiofilm properties, it also significantly inhibited the gingipains in a dose-dependent manner. At the minimal concentration of 17.826 μM, inhibition up to 98.7% and 89.4% was noted for gingipain R and K respectively. The data was also supported by the in silico docking experiments which revealed high exothermic enthalpies (−7.01 and −7.02 cal mol−1). Besides, the inhibition constant was found to be 7.24 μM and 7.1 μM against gingipains R and K respectively. The results suggest that quantum curcumin is a potential drug candidate which needs further clinical validation.
Introduction
Periodontitis is a biofilm-associated inflammatory disease of the gingiva and the supporting structures of the periodontium (gingival, alveolar bone, periodontal ligament, and cementum) which has multiple etiologies; however, microbial and immunological factors are reported to be pivotal.1,2 More precisely, periodontitis insinuates the plaque-induced irreversible inflammation of the periodontal tissues which ultimately obliterates the periodontal ligament and alveolar bone.3 An estimate from World Health Organization reveals 10–15% global load of this disease among total adult population.4 Plaque is an established biofilm which exemplifies the socialization wherein mixed consortia of bacteria co-exist which in turn determines the biofilm matrix composition and mutual cooperativity.1,5 After microbial inhabitation, the immune system comes into play and the disease is aggravated further by the destructive host inflammatory responses.6–9 However, reports suggest the involvement of only a few sorts of bacteria inhabiting the subgingival niche which orchestrate the initiation and progression of periodontal disease.10 In this context, Porphyromonas gingivalis, a Gram-negative anaerobe becomes especially important because of its diverse armamentarium both structural and functional, needed for its success (pathogenesis and progression of the inflammatory events) in the said micro-environment. It seems to be the keystone species in the development of chronic periodontitis.3,11 Datta et al. reported this bacterium in 85.75% of subgingival plaque samples from patients with chronic periodontitis.12 Surprisingly, for its survival, this bacterium depends on the fermentation of amino acids for energy in deep periodontal pockets where sugar availability is scarce.13 A plethora of literature document the essentiality of exceedingly high concentrations of cysteine proteinases with trypsin-like activities in the adult-onset periodontitis.6,11,13–16 Besides, to initiate and prolong the inflammatory response during periodontitis, a number of biochemical cascades are needed which require proteolytic activities. As per the report of de Diego et al., gingipain R and K collectively account for 85% of the extracellular proteolytic activity of P. gingivalis at the site of infection.17 Gingipains orchestrate diverse functions, which aid in bacterial survival. It activates kallikrein/kinin cascade, inactivates host proteinase inhibitors, foster dysregulation of coagulation and complement cascade, degrades immunoglobulins, bactericidal proteins, Fe-transporting proteins etc.3,14–16The routine therapeutic modality for periodontitis management is scaling and root planing (SRP) wherein supra and subgingival plaque and calculus are removed.18 However, the recalcitrance of biofilm dwellers and recolonization at the treated sites cannot be overruled.19 Possibly, owing to this, now a day's application of nonspecific broad-spectrum antibiotics locally at sites are in vogue as an addendum to the mechanical debridement. Drugs like chlorhexidine, delmopinol, tetracycline, minocycline, doxycycline, metronidazole etc. in the form of mouthwash etc. are routinely used to curb the re-proliferation of periodontopathic bacteria following SRP.20 However, the poor effectiveness of these drugs against the plaque biofilms and bacterial stochastic variations demands new effective measures for its management. The lack of selectivity of these drugs further complicates the situation by affecting the beneficial commensals as well as impart undesired side effects such as vomiting, diarrhea, addiction, or teeth discoloration.18,20–22
The therapeutic inadequacies of the drugs are due to the lack of selectivity and upsurge of antimicrobial resistance within the plaque biofilms.20 The situation becomes more complex when microbial shifting occurs during the course of biofilm development, especially during plaque formation and maturation.5 A report by Marsh et al. has indicated the role of dysbiosis in the oral cavity that leads to periodontitis.23 A recent report by Dewhirst et al. suggested the presence of over 700 bacterial species in the oral cavity.21 Interestingly, early bacterial colonizers in the oral cavity are reported to actively recruit other bacteria as well. For instance, Streptococcus gordonii was found to promote and help establish the biofilm of Porphyromonas gingivalis via several genetic mechanisms.13 As periodontitis develops, the oral microbiota shifts from the one consisting largely of Gram-positive aerobes to one consisting predominantly of Gram-negative anaerobes. This cooperative recruitment of metabolically and phylogenetically diverse bacteria leads to the overall functional heterogeneity which confers several advantages like metabolic cooperation, wherein the waste of one bacterial species serves as the food for the other.5 One noteworthy illustration is the fermentation of sugars to lactic acid by Streptococci which is then disintegrated to propionate and acetate by Veillonella spp.5 Furthermore, bacterial cross-talk in the form of quorum sensing help them to regulate their social behaviour. Reports suggest that the oral bacteria produce and respond to the autoinducer-2 signaling.24 These all conducive underpinnings further tunes the matrix chemistry for their survival, ultimately influencing the therapeutic outcomes. Therefore, it becomes prudent for the scientific community to look for easily available, cost-effective therapeutic agent(s) having both antimicrobial and anti-biofilm activities with no or minimal side-effects or toxicity.
Etiology suggests that the periodontitis can only be successfully tackled after simultaneously addressing the bacterial and inflammatory components responsible for the condition. Curcumin fits in our requirement window. A plethora of literature documents the anti-inflammatory, antioxidant, anticancer, antibacterial and anti-fungal properties of curcumin, a hydrophobic curcuminoid, extracted from the rhizomes of Curcuma longa.25,26 Curcumin was found to be highly effective against a wide array of microbial pathogens ranging from Trichomonas vaginalis, Helicobacter pylori to Candida albicans and Paracoccidioides brasiliensis.26–29 Besides, it was also reported to inhibit the biofilm formation by Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumonia, Staphylococcus aureus, and Staphylococcus epidermidis.25 Among the oral bacteria, curcumin was reported to inhibit sortase A of Streptococcus mutans, a major cariogenic bacteria.30
The scientific literature is inundated with the reports pertaining to the determination of minimum inhibitory concentration (MIC) of curcumin against various pathogens.31–34 However, we faced difficulties in determining and interpreting the results due to its insolubility and instability. These factors resulted in the colored precipitation which obscured the precise and accurate MIC determination as per CLSI guidelines. Therefore, as a drug, the use of curcumin is limited due to several such constraints. Nevertheless, attempts have been made to increase its aqueous solubility and bioavailability by the nanoparticle-based approach.33 For this, liposomes, polymeric nanoparticles, and recently lipid-drug hybrid nanoparticles have been extensively studied, where they have shown improved efficacies.35–38 Nonetheless, the innate toxicity issues cannot be overlooked. Recently, the newer quantum approach has also been utilized for its stability, solubility and therapeutic potential by making quantum curcumin against aerobic pathogens. However, its detailed toxicity evaluation is yet to be done.25
In the present work, we synthesized, characterized and then evaluated the antimicrobial and antibiofilm potential of CurQDs against representative periodontic bacterium Porphyromonas gingivalis along with other facilitating bacteria like Actinomycetemcomitans viscosus and Streptococcus mutans. Moreover, degradative effects of CurQDs on P. gingivalis and on mixed P. gingivalis–S. mutans–A. viscosus biofilm formation were also examined. Furthermore, we tried to elucidate the action of quantum curcumin against the most significant virulence factor of anaerobe Porphyromonas gingivalis: gingipain R and K which was further validated by docking experiments. These studies were made after the detailed investigations pertaining to the biocompatibility of quantum curcumin which included in vitro cell viability assay by methylthiazoltetrazolium salt, haemo-compatibility assay, membrane integrity assay using lactate dehydrogenase as marker and investigation of the production of reactive oxygen species (ROS) using flow cytometry.
Materials and methods
Chemicals
Curcumin (>95% pure) was procured from TCI chemicals, Japan. Phosphate Buffered Saline (PBS, pH 7.2), Tris–HCl buffer (pH 7.6) were prepared in-house while Dulbecco's modified Eagle's medium, DABCO and propidium iodide from Life technologies, Invitrogen were used. Flat-bottom polystyrene 96-well tissue culture plates and 8-well chambered slides were procured from SPL biosciences, Korea. Penicillin, streptomycin and gentamicin were from SRL laboratories, India while iodonitrotetrazolium chloride, Hanks balanced salt solution (HBSS), heparin, sodium bicarbonate, dimethylsulphoxide (DMSO), benzoyl-L-arginine-4-methylcoumaryl-7-amide, t-butyl-oxycarbonyl-L-valyl-L-leucyl-L-lysine-4-methylcoumaryl-7-amide, Nα-p-tosyl-L-lysine chloromethyl ketone hydrochloride, and methylthiazoltetrazolium salt were from Sigma-Aldrich, USA. Trisodium citrate, calcium chloride (CaCl2), sodium chloride (NaCl), crystal violet (CV), dimethyl sulfoxide (DMSO) and acetone were procured from Merck, USA while paraformaldehyde, brain heart infusion (BHI) broth, Colombia blood agar, yeast extract, hemin, menadione, cysteine, and fetal bovine serum were procured from HiMedia laboratories, Mumbai.Synthesis of quantum curcumin (CurQDs)
The synthesis of CurQDs was hierarchical and was done by the two-step top-down method as described earlier with modifications.25 Initially, a stock solution of 20 mM curcumin was prepared in DMSO. A stock solution of 150 mM trisodium citrate was also prepared in deionized water. CurQDs formation was investigated using different concentrations curcumin 0.125 mM to 2.5 mM at different pH ranging from 4 to 7. In a typical experiment, 0.1 mM of curcumin was taken in deionized water and its pH adjusted using trisodium citrate. Further, 2.5 g of zirconia beads (0.1 mm in diameter) were put in the 50 ml oak ridge tube and spun at 2500 rpm for 15 min with the progressive increase in the drug loading up to 2.5 mM. The resultant solution was siphoned out and dried using rotary evaporator at 70 °C. From this, a stock solution of 1 mM was prepared in acetone which was then added dropwise to hot water (25 ml) at 75 ± 5 °C at the rate of 0.2 ml min−1 with continuous ultra-sonication for about an hour with power input of 750 W, frequency 20 kHz, and intensity 30 W cm−2.The final solution was kept at room temperature for 48 h and centrifuged at a speed of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min at 4 °C to remove any residual native curcumin if present. The suspension was finally dried and re-suspended in deionized water and used for further characterizations. For XRD and Raman spectroscopic measurements, the isolated quantum curcumin were freeze-dried to obtain a dry powder.
000 rpm for 15 min at 4 °C to remove any residual native curcumin if present. The suspension was finally dried and re-suspended in deionized water and used for further characterizations. For XRD and Raman spectroscopic measurements, the isolated quantum curcumin were freeze-dried to obtain a dry powder.
Physical characterization
The prepared quantum curcumin was monitored spectroscopically at a wavelength range of 300–700 nm. All the absorbance and fluorescence readings were made in Lambda 25 UV-visible spectrometer, Perkin Elmer and LS fluorescence spectrophotometer Perkin Elmer. Morphological features of quantum curcumin were analyzed by electron microscopy. A drop of the sample was allowed to dry on carbon-coated copper grids in the air. For TEM analysis, the images were acquired through FEI Tecnai 200 TEM operated at 80 kV and high-resolution images in Tecnai transmission electron microscope at 300 kV. The phase compositions and crystalline nature of CurQDs were further studied by XRD in Rigaku MINIFLEX 600, Japan using Cu radiation at k = 0.1546 nm at a voltage of 40 kV and a current of 30 mA. Additionally, surface composition of the synthesized quantum curcumin was investigated by high-resolution X-ray photoelectron spectroscopy. Besides, Raman spectroscopy was also used to assess the phase variability and structure. The Raman spectrum was obtained on Renishaw In-Via Raman spectrometer using 50× objective from Lica. The sample was excited by 532 nm solid state diode laser and acquisition time for each window was selected as 50 s. The zeta potential of CurQDs was measured on Delsa Nano (Beckman-coulter, United Kingdom) utilizing zeta dip cells. For the measurement, the sample was prepared by mixing CurQDs in 10 mM NaCl in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 proportion and then 1 ml of the diluted sample was used.
10 proportion and then 1 ml of the diluted sample was used.
Chemical characterization
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 330.578 Hz, and the digital resolution was 0.157 Hz, with an acquisition time of 3.17 s. A total of 16 scans were taken. 13C NMR measurements were done at 300 K at 500.13 MHz, and the chemical shifts (ppm) were assigned. The spectral width was 26
330.578 Hz, and the digital resolution was 0.157 Hz, with an acquisition time of 3.17 s. A total of 16 scans were taken. 13C NMR measurements were done at 300 K at 500.13 MHz, and the chemical shifts (ppm) were assigned. The spectral width was 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 455.02 Hz with the digital resolution of 1.61 Hz and acquisition time of 0.30 s. The spectrum was obtained with 1024 scans.
455.02 Hz with the digital resolution of 1.61 Hz and acquisition time of 0.30 s. The spectrum was obtained with 1024 scans.Toxicity profiling of quantum curcumin
MTT assay for in vitro cell viability assay
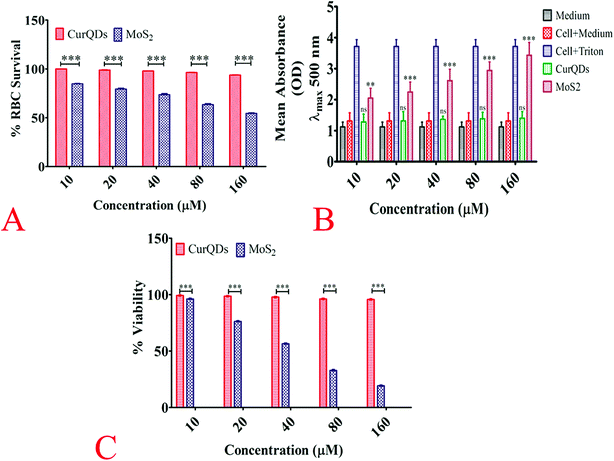
Methylthiazoltetrazolium bromide (MTT) assay was carried out to evaluate the cell viability. Briefly, Vero cells were seeded in 96-well plate at a density of 1 × 104 cells per well in a final volume of 150 μl Dulbecco's modified Eagle's medium and incubated with various concentrations of quantum curcumin and MoS2 QDs (10 μM, 20 μM, 30 μM, 40 μM, 50 μM, 60 μM, and 70 μM) at 37 °C for 24 h. After incubation, the used medium was replaced with 180 μl fresh medium and 20 μl MTT solution (10 mM) with subsequent incubation for 3 h. After that, the medium was replenished with 150 μl of DMSO (to dissolve formazan crystals) per well and incubated for 60 min in dark. The absorbance was measured using a microplate reader at 590 nm.Haemo-compatibility analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 with PBS before subjecting to hemolysis assay. Briefly, to the pellet, RPMI 1640 medium containing 10% autologous plasma, sodium bicarbonate (2 mg ml−1), penicillin (100 IU ml−1), streptomycin (100 mg ml−1), and gentamicin (30 mg ml−1), were added. The cells were maintained at 37 °C temperature, 5% CO2 and 95% air in the humidified atmosphere in a CO2 incubator. The RBCs were treated with quantum curcumin at (10 μM, 20 μM, 30 μM, 40 μM, 50 μM, 60 μM, and 70 μM) concentrations and the toxicity was analyzed through the haemolytic studies. Briefly, to 100 μl of the RBC suspension, 400 μl of the desired concentration of quantum curcumin suspended in PBS was added. 400 μl of H2O and PBS served as the positive and negative controls respectively. After incubation for 3 h, the samples were vortexed, centrifuged and the supernatant was collected. The absorbance of the supernatant at 577 nm was recorded and the percentage hemolysis calculated by the formula:
10 with PBS before subjecting to hemolysis assay. Briefly, to the pellet, RPMI 1640 medium containing 10% autologous plasma, sodium bicarbonate (2 mg ml−1), penicillin (100 IU ml−1), streptomycin (100 mg ml−1), and gentamicin (30 mg ml−1), were added. The cells were maintained at 37 °C temperature, 5% CO2 and 95% air in the humidified atmosphere in a CO2 incubator. The RBCs were treated with quantum curcumin at (10 μM, 20 μM, 30 μM, 40 μM, 50 μM, 60 μM, and 70 μM) concentrations and the toxicity was analyzed through the haemolytic studies. Briefly, to 100 μl of the RBC suspension, 400 μl of the desired concentration of quantum curcumin suspended in PBS was added. 400 μl of H2O and PBS served as the positive and negative controls respectively. After incubation for 3 h, the samples were vortexed, centrifuged and the supernatant was collected. The absorbance of the supernatant at 577 nm was recorded and the percentage hemolysis calculated by the formula:| (Sample absorbance − absorbance of negative control) ÷ (absorbance of positive control − absorbance of negative control) × 100 |
Membrane integrity analysis by lactate dehydrogenase (LDH) assay
The cytotoxicity of quantum curcumin was assessed using Vero cells by measuring lactate dehydrogenase (LDH) activity (LDH-Cytotoxicity Assay Kit, BioVision, Inc., Milpitas, CA) in the culture media wherein the known cytotoxic MoS2 QDs were used as a reference for comparisons (in concentration range 10 μM, 20 μM, 40 μM, 80 μM, 160 μM). Briefly, the 12 h treated cells were spun at 400 × g for 5 min and the growth medium was collected. The broth and the LDH reagent was incubated in the ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for 30 min followed by the absorbance reading at 500 nm.
1 for 30 min followed by the absorbance reading at 500 nm.
Reactive oxygen species production analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events was acquired for each sample.
000 events was acquired for each sample.Growth inhibition assays of quantum curcumin (CurQDs)
For native curcumin, the stock solution was prepared by dissolving 5.434 mM of CurQDs in 1 ml of deionized water and was diluted in a series of two-fold dilutions ranging from 1.114 μM to 17.826 μM in sterile BHI broth (HiMedia laboratories, India) in a microtiter plate. The aforementioned bacteria were grown anaerobically in 10 ml BHI broth for 48 h. Bacterial cultures (500 μl) were then diluted to 1.5 ml fresh BHI broth. Each well of 96-well tissue culture plate was then inoculated with 170 μl of standardized cell suspension (105 CFU ml−1) and incubated at 37 °C for next 48 h along with the 30 μl diluted drug such that effective concentration of the drug per well ranges from 0.1114 to 8.913 μM in two fold dilutions. The MIC was defined as the lowest concentration of CurQDs at which sharp colour change (from light pink to orange/reddish-orange) was observed after addition of 50 μl of iodonitrotetrazolium chloride (1 mg ml−1). Positive controls did not contain any drug moiety while the sterile broth was used as negative control. For comparison, we also used metronidazole dissolved in the nutrient liquid medium as per CLSI guidelines.
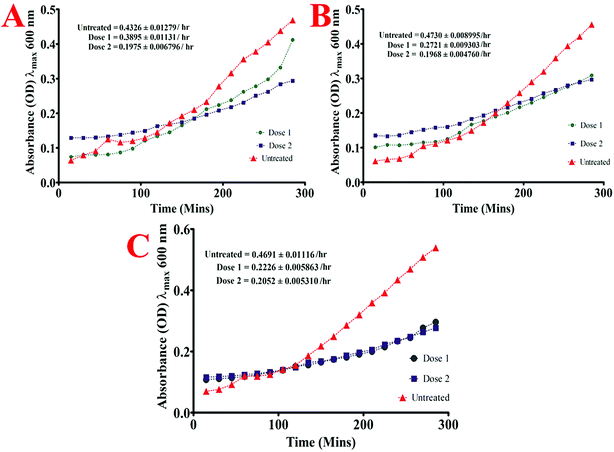
Briefly, the standard bacterial test inoculum (106 CFU ml−1) was prepared from the late-exponential phase of bacterial growth and were added (10 μl) to the wells of 96 well microtiter plate already containing 2 fold lower and 2 fold higher dose than the MIC dose of quantum curcumin in 190 μl of BHI broth such that its OD at λmax 600 nm was approximately 0.01. The optical density of each well was then monitored at λmax 600 nm by periodic measurements after every 15 min for 4.5 h. To determine the maximum growth rate of each isolate both in presence and absence of CurQDs, the slope of the linear part of the growth curve (R2, ≥0.98) was determined for at least 5 data points of the semi-logarithmic plot of absorbance (ln[OD600]) vs. incubation time (in hours). For exact determination of absorbance of bacterial growth, absorbances of drugs were subtracted from those of the cumulative absorbance readings.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in pre-reduced BHI broth containing 1 mg ml−1 yeast extract, 5 mg ml−1 hemin, and 1 mg ml−1 menadione and allowed to grow anaerobically in an atmosphere of 80% N2, 10% CO2, and 10% H2 at 37 °C. A volume of 90 μl of each diluted bacterial suspension was then dispensed into flat-bottom lysine coated polystyrene 96-well tissue culture plate and 10 μl of drug solution was added to attain final concentrations ranging from 0.557 to 17.826 μM. Wells without drugs were set up as positive controls. Plates were incubated at 37 °C with shaking at 110 rpm for 24 h in the case where biofilm inhibitory action of the drugs was investigated. After incubation, biofilm was quantitated by crystal violet (CV) assay (Merck, Germany) as described earlier.25
100 in pre-reduced BHI broth containing 1 mg ml−1 yeast extract, 5 mg ml−1 hemin, and 1 mg ml−1 menadione and allowed to grow anaerobically in an atmosphere of 80% N2, 10% CO2, and 10% H2 at 37 °C. A volume of 90 μl of each diluted bacterial suspension was then dispensed into flat-bottom lysine coated polystyrene 96-well tissue culture plate and 10 μl of drug solution was added to attain final concentrations ranging from 0.557 to 17.826 μM. Wells without drugs were set up as positive controls. Plates were incubated at 37 °C with shaking at 110 rpm for 24 h in the case where biofilm inhibitory action of the drugs was investigated. After incubation, biofilm was quantitated by crystal violet (CV) assay (Merck, Germany) as described earlier.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (105 cells per ml) in BHI broth containing 1% (w/v) sucrose to a 6-well plate. The bacterial suspensions were then incubated statically at 37 °C in 5% CO2 for 72 h, with intermittent replacement with fresh broth every 24 h.
100 (105 cells per ml) in BHI broth containing 1% (w/v) sucrose to a 6-well plate. The bacterial suspensions were then incubated statically at 37 °C in 5% CO2 for 72 h, with intermittent replacement with fresh broth every 24 h.The effect of native and quantum curcumin on mixed biofilm was investigated in two different growth conditions. In the first condition, P. gingivalis, S. mutans, and A. viscosus were grown together for 48 h. After the biofilm was established, it was initially treated with said drugs for 60 min followed by thorough washing with PBS (pH 7.2) to remove the residual drug. Finally, the biofilm was quantitated by crystal violet (CV) assay. In the second set of experiment, S. mutans and A. viscosus were first grown anaerobically for 24 h followed by the addition of P. gingivalis which was grown further for next 24 h. The treatment regime with the said drugs was for 60 min. The residual drug was then removed by thorough washing with PBS and the biofilm was estimated by CV-assay as described earlier.42
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 with the fresh broth to OD at λmax 600 nm equivalent to 0.2. Fifty microliters of a diluted suspension of representative bacterial isolates (S. mutans and A. viscosus) were then dispensed into chambered slide containing 450 μl of BHI broth. The biofilm was grown anaerobically with shaking 110 rpm for 36 h. This was followed by addition of 50 μl P. gingivalis (ATCC 33277) and was regrown anaerobically for the next 72 h. These established biofilms were then exposed to 4 different concentrations of quantum curcumin (1.114–8.913 μM) for 45 min. Prior to staining, the residual broth was aspirated out followed by three washes with phosphate buffer (pH 7.2). Biofilm was fixed using 4% (v/v) paraformaldehyde for 60 min and washed thrice with PBS (pH 7.2). After fixation, concanavalin A, labelled with tetramethyl-rhodamine isothiocyanate (TRITC) (Life Technologies, USA) was used to visualize the biofilm matrix as it binds with the sugars of the biofilm. The stock (1 mg ml−1) of the conjugated lectin was prepared in 10 mM phosphate buffer (pH 7.5). For use, stock solutions were diluted again with phosphate buffer to a lectin concentration of 10 μg ml−1. Ten-microliters was the aliquot were added to the wells and incubated at room temperature for 20 min in the dark. After incubation, the residual dye was removed by washing with PBS thrice. The sections were then analyzed and images acquired using the Zeiss LSM 510 inverted confocal laser scanning microscope (Carl Zeiss, Jena, Germany). Optical sections of fluorescent specimens were obtained via a Plan-Neofluar 40×/1.3 oil objective with a z-step of 2.0 μm using a HeNe laser (λ = 543 nm) and an argon laser (λ = 458 nm) using a pinhole of 250. Each image was acquired within 2 min, to minimize photo-degradation.
100 with the fresh broth to OD at λmax 600 nm equivalent to 0.2. Fifty microliters of a diluted suspension of representative bacterial isolates (S. mutans and A. viscosus) were then dispensed into chambered slide containing 450 μl of BHI broth. The biofilm was grown anaerobically with shaking 110 rpm for 36 h. This was followed by addition of 50 μl P. gingivalis (ATCC 33277) and was regrown anaerobically for the next 72 h. These established biofilms were then exposed to 4 different concentrations of quantum curcumin (1.114–8.913 μM) for 45 min. Prior to staining, the residual broth was aspirated out followed by three washes with phosphate buffer (pH 7.2). Biofilm was fixed using 4% (v/v) paraformaldehyde for 60 min and washed thrice with PBS (pH 7.2). After fixation, concanavalin A, labelled with tetramethyl-rhodamine isothiocyanate (TRITC) (Life Technologies, USA) was used to visualize the biofilm matrix as it binds with the sugars of the biofilm. The stock (1 mg ml−1) of the conjugated lectin was prepared in 10 mM phosphate buffer (pH 7.5). For use, stock solutions were diluted again with phosphate buffer to a lectin concentration of 10 μg ml−1. Ten-microliters was the aliquot were added to the wells and incubated at room temperature for 20 min in the dark. After incubation, the residual dye was removed by washing with PBS thrice. The sections were then analyzed and images acquired using the Zeiss LSM 510 inverted confocal laser scanning microscope (Carl Zeiss, Jena, Germany). Optical sections of fluorescent specimens were obtained via a Plan-Neofluar 40×/1.3 oil objective with a z-step of 2.0 μm using a HeNe laser (λ = 543 nm) and an argon laser (λ = 458 nm) using a pinhole of 250. Each image was acquired within 2 min, to minimize photo-degradation.Results
TEM, HR-TEM, UV-visible absorption and photo-luminescence (PL) spectra of CurQDs
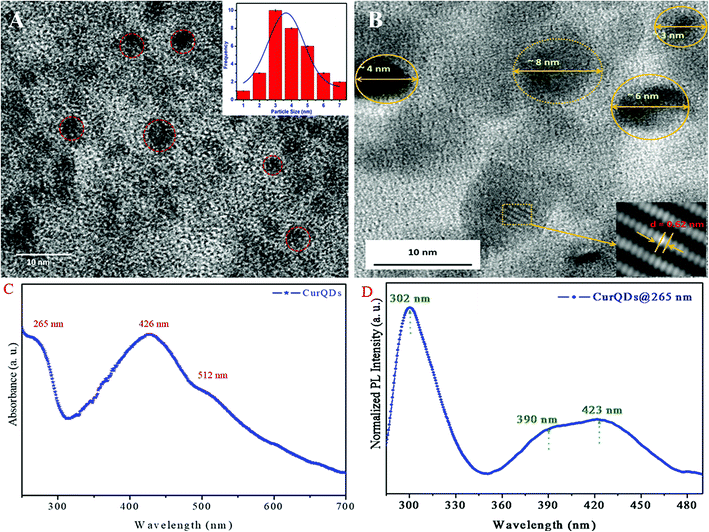
The TEM micrograph of CurQDs is depicted in Fig. 1A CurQDs are encircled with red color. Inset of the Fig. 1A shows the particle distribution estimated over 30–35 particles. The particle distribution bar graph has been fitted for Gaussian function and the average particle size is found to be 3.5 nm. The HR-TEM has been shown in the Fig. 1B. Inset therein shows an interlayer spacing of ∼0.63 nm corresponding to the (002) plane of curcumin.UV-visible absorption spectra of CurQDs were recorded in water as depicted in Fig. 1C. One milliliter CurQDs suspension was used for photoluminescence studies using excitation wavelength 265 nm as represented in Fig. 1D. Three discrete absorption bands were obtained for CurQDs: one in UV region (263 nm) and two in the visible region (425 and 512 nm).
X-ray photoelectron spectroscopy (XPS) analysis
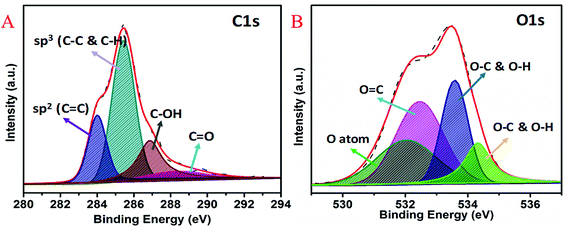
XPS was used to study the chemical states and electronic structure of quantum curcumin (Fig. 2A and B). HR-XPS spectrum of C1s (Fig. 2A) was found deconvoluted into four peaks of binding energies ∼284.0 eV, 285.5 eV, 286.8 eV, and 288.8 eV respectively. These four deconvoluted XPS peaks correspond to sp2 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), sp3 (C–C & C–H), C–OH and C
C), sp3 (C–C & C–H), C–OH and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O binding energies respectively. Presence of C–OH and C
O binding energies respectively. Presence of C–OH and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O at binding energies ∼286.6 eV and 288.8 eV confirm the presence of alcoholic and carbonyl (keto) functional groups of the CurQDs. HR-XPS spectrum of O1s is shown in the Fig. 2B where four deconvolutions appeared at positions ∼532 eV, 532.4 eV, 533.6 eV, and 534.3 eV respectively. These peaks correspond to oxygen atoms, O
O at binding energies ∼286.6 eV and 288.8 eV confirm the presence of alcoholic and carbonyl (keto) functional groups of the CurQDs. HR-XPS spectrum of O1s is shown in the Fig. 2B where four deconvolutions appeared at positions ∼532 eV, 532.4 eV, 533.6 eV, and 534.3 eV respectively. These peaks correspond to oxygen atoms, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and O–C & O–H respectively which again confirm the no change in the chemistry of curcumin while being converted into quantum form.
C and O–C & O–H respectively which again confirm the no change in the chemistry of curcumin while being converted into quantum form.
Raman spectroscopy
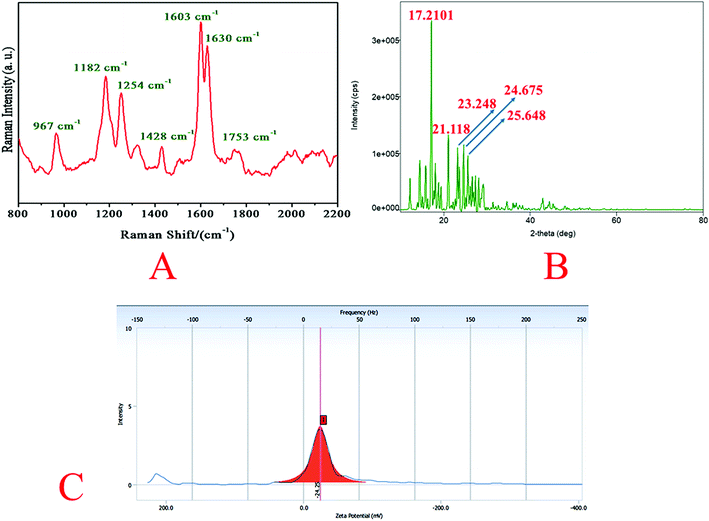
Raman spectrum of quantum curcumin is presented in Fig. 3A. The spectrum was similar to that of the native curcumin indicating no chemical and structural change during synthesis. The absence of other additional peaks indicated the purity of the synthesis. In 1600–1800 cm−1 region, the peak at 1630 cm−1 is assigned to the vibration of ϑ (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). The distal benzene rings of the curcumin molecule remain intact as expected after quantum dot formation as indicated by C–C stretching of the aromatic ring at 1603 cm−1. At 1428 cm−1 stretching of phenolic C–O is also observed. These are the signature peaks of curcumin which remained unaltered. At ∼1753 cm−1, a new broad-band appears in CurQDs possibly owing to keto–enol tautomerism (due to ϑ (C
O). The distal benzene rings of the curcumin molecule remain intact as expected after quantum dot formation as indicated by C–C stretching of the aromatic ring at 1603 cm−1. At 1428 cm−1 stretching of phenolic C–O is also observed. These are the signature peaks of curcumin which remained unaltered. At ∼1753 cm−1, a new broad-band appears in CurQDs possibly owing to keto–enol tautomerism (due to ϑ (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)). However, peaks at 1254 cm−1 and 1182 cm−1 were due to the stretchings of enolic C–OH and C–O–C stretchings. Interestingly, a peak at 967 cm−1 is also noted that has appeared due to the in-plane bending of C–H.
O)). However, peaks at 1254 cm−1 and 1182 cm−1 were due to the stretchings of enolic C–OH and C–O–C stretchings. Interestingly, a peak at 967 cm−1 is also noted that has appeared due to the in-plane bending of C–H.
X-ray diffraction analysis
The core size of CurQDs was estimated by Scherrer formula:D = 0.9λ/β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ θ |
Zeta potential
The zeta potential of CurQDs was found to be −24.25 mV (Fig. 3C).Nuclear magnetic resonance spectroscopy
Biocompatibility assays
Cell leakage study
| Medium absorbance (λmax 500) | Cell + medium absorbance (λmax 500) | Cell + 2% Triton X 100 absorbance (λmax 500) | Concentration (μM) | CurQDs treated cells absorbance (λmax 500) | MoS2 treated cells absorbance (λmax 500) | % cytotoxicity by MoS2 |
|---|---|---|---|---|---|---|
| 1.12 ± 0.153 | 1.31 ± 0.264 | 3.712 ± 0.223 | 10 | 1.35 ± 0.251 | 2.451 ± 0.314 | 0.8742 |
| 20 | 1.47 ± 0.302 | 2.646 ± 0.32 | 8.992 | |||
| 40 | 1.59 ± 0.105 | 2.913 ± 0.367 | 20.168 | |||
| 80 | 1.63 ± 0.213 | 3.42 ± 0.275 | 41.215 | |||
| 160 | 1.78 ± 0.211 | 3.913 ± 0.413 | 61.74 |
The formula used for % cytotoxicity calculation is as follows:
Flow cytometry
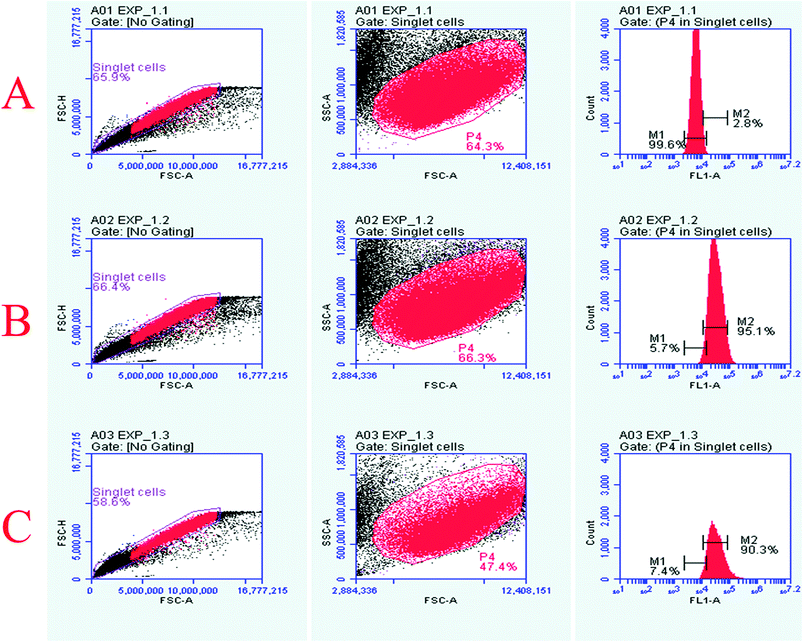
The Vero cells were exposed to 160 μM concentrations of quantum curcumin and MoS2 QDs, followed by evaluation of ROS generation by probing with 2′,7′-dichlorfluorescein-diacetate (DCFH-DA) using flow cytometry. We observed significant differences in the ROS generation profile of the two discrete QDs as evident from the increased intensity/count in the DCF fluorescence (Fig. 6) and increase in the cell count (those took up the dye) when exposed to MoS2 quantum dots. MoS2 QD treated cells exhibited a significant increase in the DCF fluorescence compared to the control, whereas no significant increase in the fluorescence was perceived when the cells were treated with quantum curcumin. This showed that CurQDs yielded significantly low ROS in comparison to the MoS2 QDs. Interestingly; the administration of quantum curcumin has improved the cellular health as evidenced with the restoration and increase of cell population in the gated region (more granularity) under investigation. The results of flow cytometry are in agreement with the results obtained from preceding assays.Antimicrobial assay of quantum curcumin against clinical isolates of Porphyromonas gingivalis and select reference strains
We tried to observe antibacterial activity of CurQDs over eleven clinical isolates of P. gingivalis isolated from the patients of chronic periodontitis as well as antibiofilm activity of the drug over the individual as well as mixed biofilm of P. gingivalis (ATCC 33277), A. viscosus (ATCC 29522), and S. mutans (ATCC 25175).| Bacterial isolate | MIC (μM) | MBIC50 (μM) | MBIC90 (μM) | MIC (mM) for metronidazole | MBIC90 (mM) for metronidazole |
|---|---|---|---|---|---|
| Porphyromonas gingivalis (ATCC 33277) | 1.114 | 0.557 | 17.826 | 1.46 | 93.48 |
| Actinomycetemcomitans viscosus (ATCC 29522) | 2.228 | 0.557 | 17.826 | 2.921 | 186.97 |
| Streptococcus mutans (ATCC 25175) | 8.913 | 1.14 | >17.826 | 11.68 | 373.94 |
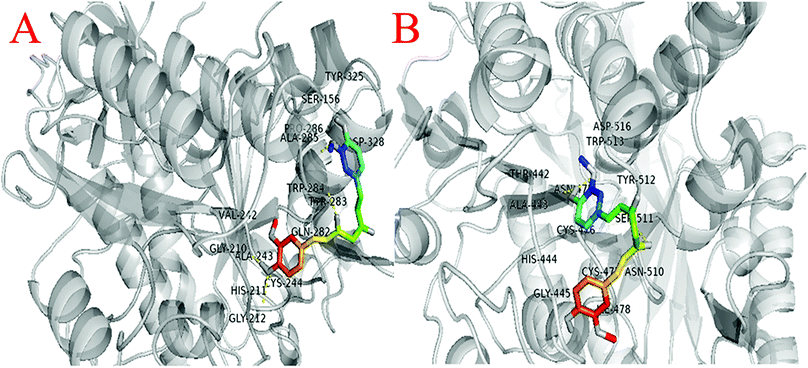
Similarly, molecular docking were also performed between gingipain K (PDB ID: 4RBM) with curcumin to analyze their interaction. The interaction of THR-442, ALA-443, HIS-444, GLY-445, ASN-475, CYS-476, CYS-477, ILE-478, ASN-510, SER-511, TYR-512, TRP-513, and ASP-516 amino acids were observed around 4 Å of curcumin (Fig. 10B).
The analysis suggests that the curcumin interacts with almost similar affinity with both gingipain R and gingipain K protein.
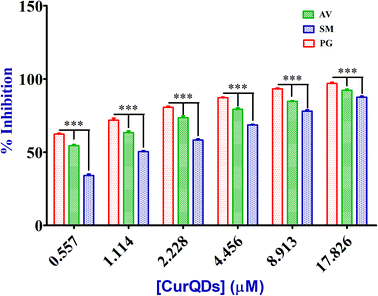
Analysis of the effect of CurQDs on biofilm formation by TCP assay
| Bacterial strains | Biofilm inhibition as % decrease in biomass/matrix | |||||
|---|---|---|---|---|---|---|
| Porphyromonas gingivalis, ATCC 33277 | 97.43 | 93.59 | 87.8 | 81.76 | 72.57 | 61.24 |
| Actinomycetemcomitans viscosus, ATCC 29522 | 91.29 | 84.67 | 79.43 | 73.67 | 65.42 | 53.13 |
| Streptococcus mutans, ATCC 25175 | 88.77 | 78.18 | 69.15 | 58.23 | 49.6 | 35.25 |
| Mixed biofilm, P. gingivalis, S. mutans and A. viscosus together | 93.28 | 88.54 | 79.67 | 71.41 | 60.87 | 49.32 |
| Mixed biofilm, S. mutans + A. viscosus then P. gingivalis | 81.65 | 72.88 | 60.37 | 51.25 | 39.76 | 28.26 |
| Effective conc. (μM) | 17.826 | 8.913 | 4.456 | 2.228 | 1.114 | 0.557 |
Confocal microscopic evaluation of the effect of CurQDs on biofilm architecture
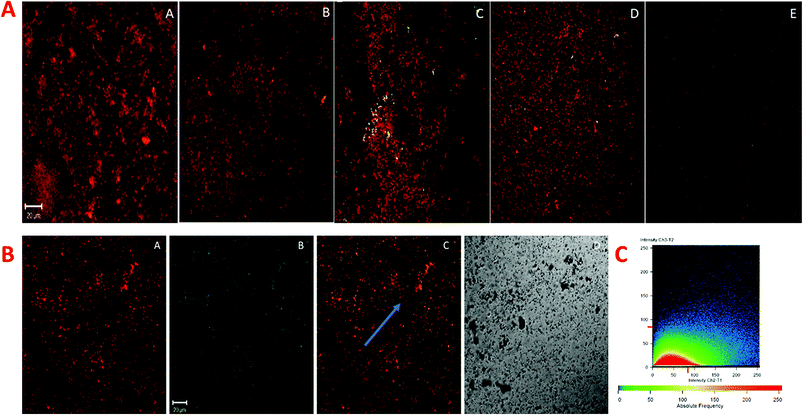
P. gingivalis, S. mutans, and A. viscosus isolates were selected and grown simultaneously for the biofilm study because of denser and relatively robust biofilm forming potential together, which is more closely associated with chronic periodontitis. The degradative effect of quantum curcumin on mixed biofilm was examined using CLSM. As shown in Fig. 12A and B, CurQDs corroded the mixed biofilm in a dose-dependent manner (P < 0.001). With initial exposure to 0.557 μM CurQDs, the biofilm matrix slackened off (became more porous which indicates the loosening of the matrix material) which can be seen unambiguously in Fig. 12A(B). This loosening intensified to its maxima when the exposed concentration was escalated to 8.913 μM (Fig. 12A(D)). Interestingly, upon exposure of 17.826 μM, one can note the scattered planktonic cells. These results are in consonance with the observations of TCP assay.For the assessment of the nature of the interaction of the quantum curcumin and the matrix, we selectively studied (Fig. 12B) the panel D of Fig. 12A. One can note the ubiquitous distribution of quantum curcumin (as green fluorescent signals, Fig. 12B(B)) in the biofilm matrix (red fluorescent signals, Fig. 12B(A). Panel 12B(C) shows the intermediary orange coloured signals which show the substantial interaction of quantum curcumin with the matrix materials. Panel 12C, a fluorogram, portrays the co-localization of CurQDs and the matrix for the region where a line is arbitrarily drawn for the stability assessment of the nature of co-localization (Fig. 12C). The unvarying linearity was observed for the signals in the fluorogram that revealed the stable association/interaction.
Discussion
The primary objective of the present study was the synthesis and the appraisal of antibacterial and antibiofilm potential of quantum curcumin (CurQDs) on pathogens especially involved in progression and aggravation of oral disease(s) such as chronic periodontitis (P. gingivalis ATCC 33277, A. viscosus ATCC 29522 and S. mutans ATCC 25175). We further attempted to observe the inhibitory effects of quantum curcumin over P gingivalis specific gingipains (Arg- and Lys-specific proteinases). Besides, we also evaluated the biocompatibility of quantum curcumin in detail.Quantum dots (QDs) are the discrete class of fluorescent particles with smaller physical dimensions compared to the exciton Bohr radius. These QDs have high quantum efficiency, long-term photo-stability, narrow emission, and continuous absorption spectra.43 Excitation–emission profile reveals that it always emits the same light irrespective of excitation wavelength. This simply implies that a single emitted colour can be seen throughout by using single laser excitation source. This emitted wavelength can further be used for imaging owing to excellent photo-stability.44 Our synthesized quantum curcumin was found to be green fluorescent and this property was exploited well in the confocal microscopy. No additional fluorescent tag was needed for the evaluation of its biological activity using CLSM or any other method that requires fluorescent probing/tagging. In our present study, after synthesizing CurQDs as per protocol from our previous study, we analysed the QDs particle on some additional parameters such as X-ray photoelectron spectroscopy, X-ray diffraction analysis and NMR apart from the previously described parameters.
Essentially, biomedical applications of QDs require their high-quality and high aqueous solubility. Quantum dots could be made directly in water but they are polydisperse. They have either narrow available size ranges or wide size distribution. This obscures its use as imaging material in biological systems.44–47 But bestows an additional advantage in trickling down across the layers of biofilm matrices with assorted pore sizes. For imaging pursuits, monodisperse quantum dots can be produced from high-temperature organic solvent synthetic strategies possessing wide emission colours which may range from ultraviolet to near infrared (300–2500 nm) by simply tuning its size, composition and/or structure. However, these QDs are insoluble in water.48
QDs have unique physico-electro-optical properties due to the combination of innate material characteristics & its dimensionality. Our PL and UV-vis profile is in consonance with the said characteristics. The rationale behind the stronger activity of quantum curcumin is related to the reduction in its particle size in the range of 0.5 to 7 nm (average size 3.5 nm), which is far less than the size of native curcumin particles (average size 2350 nm). This may be the reason for better penetration and interaction with the biofilm matrix and higher uptake by the bacterial cells.
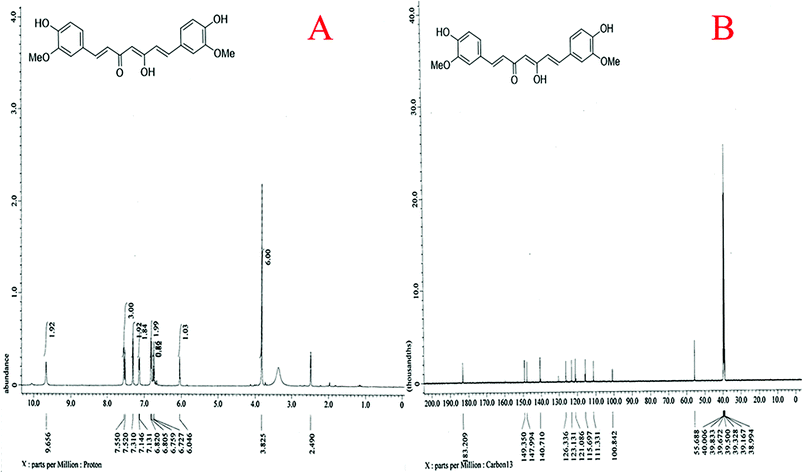
The basic challenge of quantum dot synthesis is to control its uniform growth using stabilizers to avoid agglomeration or aggregation that may culminate into the formation of microcrystals. However, during our synthesis, when acetone dissolved curcumin was injected slowly at the rate of 0.2 ml min−1 in the aqueous phase at 75 ± 5 °C with continuous ultra-sonication, the solubilized curcumin spread simultaneously with acetone with rapid volatilization. As acetone is miscible with water with density almost half of it, its rapid volatilization in presence of high energy sonication, resulted in sizing down of curcumin to quantum dot level, ruled out the need of addition of stabilizers to prevent conglomeration. The TEM profile revealed that the average size of the CurQDs was 3.5 nm. The HR-TEM shows an interlayer spacing of ∼0.63 nm corresponding to the (002) plane of curcumin. The XRD data of quantum curcumin reveals number of peaks in the range of 2θ (10–40°) implying its crystalline nature. XRD was used to verify the crystalline content of the drug present in the formulation. Surface characteristics and charge of CurQDs were investigated by zeta potential analysis and was found to be −25.6 mV. This value provides indirect evidence of the potential stability of the quantum curcumin. The UV-vis spectra showed three bands at ∼265, 426 and 512 nm. A broad absorption band was obtained at ∼426 nm along with a shoulder at ∼512 nm extending to the visible region. The absorption band at 426 nm in the visible region can be assigned to low energy π–π* transitions of curcumin in water arising possibly due to change in the tautomeric form of the keto-enol-enolate group. The chemistry of the synthesized quantum curcumin was further ascertained by using 1H and 13C NMR spectroscopy. Besides, Raman spectroscopy was also used to assess the phase variability and the structure of the quantum curcumin. The results clearly show the synthesis of curcumin quantum dots.
Various institutions across the globe have looked for its toxicity profile. Curcumin has been assessed by the Joint FAO/WHO Expert Committee on Food Additives (JECFA) and the EU Scientific Committee for Food (SCF).49 The JECFA set an acceptable daily intake (ADI) value of 0–3 mg kg−1 body weight per day for curcumin in 2004. The ADI value of 3 mg kg−1 body weight per day was based on the NOAEL (no observed adverse effect level) of 250–320 mg kg−1 body weight per day. The SCF, on the other hand, has not made any specification for the ADI value of curcumin.50 According to the directive 67/548/EWG as reported by Rimbach et al., curcumin neither poses a threat to man nor the environment.50 The JECFA evaluated four acute oral toxicity studies in mice and rats and found LD50 values of 2 to 10 g kg−1 body weight for mice and 5 to 10 g kg−1 body weight for rats. In the present study, the in vitro toxicity profiling was done in the range of 2–32 μg ml−1 and it qualified as non-toxic on all the concentrations.49 Therefore, the quantum curcumin (CurQDs) in the present study was found significantly effective as both antibacterial and antibiofilm agent that too in the concentration as low as 0.089 μg ml−1 while it was found non-toxic even at the concentration of 1024 μg ml−1 which is much below the maximum non-toxic threshold level as reported earlier.
To determine whether the synthesized CurQDs showed only anti-bacterial and antibiofilm efficacy without imparting any toxicity to healthy cells, its biocompatibility was evaluated. For this purpose, the morphology of Vero cells after treatment for 24 h of culture was monitored by phase contrast and fluorescence microscopy. Results clearly suggested that the Vero cells proliferated over the time forming a cell layer on the culture plate. Notably, the Vero cells displayed a more polygonal morphology upon CurQDs treatment which is the normal phenotype behavior of healthy Vero cells, suggesting that quantum curcumin restored/promoted Vero functionality.
The viability and proliferation rate of Vero cells were characterized by MTT assay, which was directly proportional to the optical density of the reaction product from the MTT working solution with live cells. As seen, clearly in Fig. S1,† after 3 days, the viability was significantly higher indicating that quantum curcumin has enhanced the cell viability.
Periodontitis continues to be a major health concern, particularly amid people with poor oral hygiene. Its development and progression involve multifactorial cascades and to a larger extent, it is dynamic in terms of its regulation and socialization.7,8 The primary microbiological factor which is significantly responsible for the disease progression is the shift in the oral microflora.5 Virulence factors of P. gingivalis are diverse and can be broadly classified on functionality. For instance, while fimbriae, hemagglutinins, outer membrane proteins, vesicles, and gingipains are mainly involved in colonization and attachment, proteinases (Arg- and Lys-gingipains), collagenase, fibrinolytic, keratinolytic, and other hydrolytic enzymes are involved in damaging host tissues.3,13
The study of the effect of either form of curcumin (native and quantum) against periodontopathic bacteria has not received much ponderance. However, a recent study by Izui et al. reports the MIC of curcumin against P. gingivalis ATCC 33277 to be 15 μg ml−1.41 However, the MIC in the present study was found to be 1.114 mM (0.41 μg ml−1), which is incongruous with the previous report. Interestingly, the MIC of quantum curcumin ranged from 0.557–8.913 μM (0.205–3.28 μg ml−1) in the present study. Socransky et al. suggested the strikingly significant role of P. gingivalis in periodontal disease.51 Therefore, the effect of CurQDs on the growth of P. gingivalis was investigated thoroughly wherein at significantly low concentrations (0.557 μM) quantum curcumin exhibited the strong inhibitory effects. The same efficacy was noted against other cariogenic pathogens in this study namely A. viscosus and S. mutans.
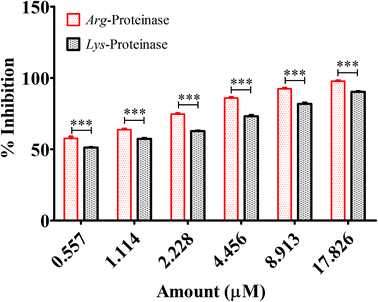
Fimbriae and proteinases are two major virulent factors that significantly weigh in to the P. gingivalis pathogenesis.3,13 To date, four P gingivalis proteinases are recognized serine, aspartate, thiol and metalloproteinase. Of these, the collagenases, aminopeptidases, and the trypsin-like proteinases are critical to P. gingivalis pathogenesis. The “thiol” proteases are “trypsin-like” enzymes those cleave polypeptides at the C-terminal after arginine or lysine residues and therefore named gingipain R and K.52 Both the Arg-gingipain and the Lys-gingipain do not share any structural/sequence similarity with any of the other known cysteine proteinase.3 These potent non-fimbrial adhesins fervently bind to several extracellular matrix proteins like fibrinogen, fibronectin, and collagen type V etc. Gingipains are belligerent endopeptidases, capable of degrading serum and tissue-derived proteins. The protein fragments obtained after degradation are ultimately subjected to further digestion by di- and tripeptidyl peptidases to release di- and tripeptides which are then taken up by P. gingivalis for its carbon and energy metabolism.14,15 Reports suggest that in densely populated biofilms, gingipains trim the cationic antimicrobial peptides to enable the bacterial survival. Degradation of cationic antimicrobial peptides obscures the neutralization of lipopolysaccharides, which may lead to intensified and unremitting production of pro inflammatory cytokines.7 Gingipains degrade different components of complement as well.3 In this study, we, therefore, investigated the effects of quantum curcumin on P. gingivalis Arg- and Lys-specific proteinases activities. Earlier polyphenol(s) and catechins(s) derivatives have been reported to inhibit Arg- and Lys-specific proteinases of P. gingivalis.53–55 In the present study, 58.2% reduction in Arg-specific proteinase activity was observed at 0.557 μM CurQDs while 98.7% of reduction was seen at the concentration of 17.826 μM in both ATCC 33277 and clinical isolates of P. gingivalis. The similar trend was seen over Lys-specific proteinase activities with 51.4% and 89.4% reduction upon administration with 0.557 μM and 17.826 μM CurQDs concentrations. The results obtained here was then investigated by in silico docking analysis by AutoDock. The thermodynamic profile of the interaction of curcumin with gingipains R and K indicated that the binding process was characterized by large exothermic enthalpies (−7.01 and −7.02 cal mol−1). Besides, the inhibition constant was found to be 7.24 μM and 7.1 μM against gingipains R and K respectively. In the present study, curcumin was found adopting conformations suitable for maximizing interactions with the gingipains. This may be due to its novel structural geometry and chemistry which include two hydrophobic phenyl domains connected by a flexible methylene linker along with keto–enol tautomerism which provides a strong and directed electrostatic interaction to increase favorable binding free energy.
Recently, the effect of native curcumin on mature, multispecies biofilm was also reported wherein the multispecies biofilm was allowed to mature for 7 days and then treated with curcumin for 24 h. After treatment, the outré-structural changes were observed using scanning electron microscopy.56,57 Curcumin-treated biofilms displayed significantly reduced metabolic activity and no visible extracellular matrix. Although the previous study investigated the effects of curcumin on matured biofilms, the current study investigates the effect of curcumin on the early as well as the late stages of biofilm formed by P. gingivalis with S. mutans and A. viscosus, mimicking the in vivo condition of biofilm associated lesion of periodontitis.
We observed when all the three bacteria were grown concurrently and then the drug was administered, the effect of the drug was maximum, and at 8.913 μM concentration, biofilm was completely suppressed within 90 min. The said observation was in consonance with the report which ascertains the involvement of P. gingivalis FimA and streptococcal cell surface glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in the early stages of biofilm formation.58 Therefore, it can be conjectured that quantum curcumin binds with high affinity to the P. gingivalis FimA which subsequently inhibits its binding to the streptococcal GAPDH, culminating in the suppression of the biofilm. Moreover, in accordance with other reports, it may also inhibit auto-aggregation and interaction of FimA with salivary proteins, resulting in suppression of P. gingivalis biofilm formation. However, we cannot confirm whether the observed inhibition is owing to the different mechanism(s) or simply it represents multifactorial selection of targets depending upon the site, ending up in biofilm inhibition. However, we hypothesized the killing action of quantum curcumin by membrane permeabilization and subsequent lysis.
The TCP assay results and our hypothesis pertaining to the mode of action was further validated by CLSM wherein P. gingivalis (late colonizer of dental biofilm) was added to antecedently formed biofilm of S. mutans and A. viscosus (facilitator). We found CurQDs significantly effective in degradation of the biofilm matrix at the dose as low as 8.913 μM (3.28 μg ml−1). The result revealed the interaction(s) between quantum curcumin and the biofilm matrix which eventually culminated in the loosening and ultimately degradation of the biofilm in the concentration dependent manner. Quantum curcumin is the green fluorescent moiety while concanavalin A (Con A), labelled with tetramethyl-rhodamine isothiocyanate (TRITC) exhibits red fluorescence. Con A-TRITC binds with matrix sugars and hence gives a gross approximation related to biofilm biomass. The colocalization map unfolds the juxtapositioning of Con A-TRITC and the quantum curcumin which is the indicative of its higher affinity for the biofilm matrix material. Accurate colocalization analysis requires distinct fluorescence emission spectra. Both Con A-TRITC and quantum curcumin colours are easily distinguishable and therefore yielded clear fluorogram with minimal bleed-through.
Reports pertaining to the maintenance of periodontal health using numerous natural products/phyto-extracts/plant secondary metabolites are in abundance. They were screened for their potential to inhibit periodontitis.53–57,59,60 For instance, extracts from E. globules leaf, exhibited the antibacterial activity with MIC values of 1–10 μg ml−1 against P. gingivalis and T. denticola, and was found to inhibit P. gingivalis Arg- and Lys-specific proteinase activities at 50–100 μg ml−1.55 Similarly, another controlled study showed that chewing gum containing eucalyptus extracts significantly inhibited the plaque formation and gingival inflammation.53 The MIC of CurQDs was some 244 times lower (1.114 μM, 0.41 μg ml−1) than that of all testified plant secondary metabolites against P. gingivalis. Besides, CurQDs also inhibited the growth of other anaerobes like A. viscosus as well and other cariogenic bacteria like S. mutans whereas other plant secondary metabolites had insignificant effects on the same. Recent reports suggest that green tea (Camellia sinensis) polyphenols, particularly epigallocatechin gallate, inhibited the growth of P. gingivalis at concentrations of 250–500 μg ml−1 and 50 mg ml−1.61,62 It is therefore, obvious that compared to the quantum curcumin the efficacy of the said phytochemicals are significantly compromised.
Recently, therapeutic efficacy of curcumin for the treatment of aggravated and chronic periodontitis was evaluated in a pilot study. In the study, it found that the patients who underwent scaling and root planing, followed by using curcumin mouthwash (20%) for 21 days, showed significant improvement in the gingival inflammation than those who underwent only scaling and root planing. However, they found insignificant difference in their plaque indices.63
Conclusion
The molecule curcumin, the most active polyphenol present in curcuma, owing to its high hydropathy index and instability has not been able to justify its role as frontline drug modality in the treatment of infectious and non-infectious diseases. The results of the current study indicate that quantum curcumin not only inhibited the growth of various periodontopathic bacteria but also exhibited potent antibiofilm activities against mixed biofilm formed by P. gingivalis, A. viscosus, and S. mutans. Being an asaccharolytic bacterium, P. gingivalis solely relies for its carbon and energy sources from proteins, which proves the inevitable role of these gingipains R and K for the growth and multiplication in the host that help in continuing the vicious life cycle leading to the proliferation and aggravation in the lesion of chronic periodontitis. Therefore, in this context, the activity of quantum curcumin against P. gingivalis specific Arg- and Lys-proteinase indicates its usefulness as first-line therapeutic modality. The bioassay utilized herein for evaluating inhibitors of P. gingivalis specific Arg- and Lys-specific proteinases may be used in future in screening of potential drugs candidates against such pathogens. However, for pharmacological investigations one may utilize appropriate animal models such as nonhuman primates (like rhesus monkeys (Macaca mulatta), cynomolgus monkeys (Macaca fascicularis), and baboons (Papio anubis)), miniature pigs (Minnesota miniature pig), dogs, ferrets (Mustela putorius) and rodents (rice rat (Oryzomys palustris), baker mouse) to substantiate the role of quantum curcumin in the management of chronic periodontitis. In all, these results add to the considerable body of evidence of antibacterial, antivirulence, antibiofilm activities of quantum curcumin over P. gingivalis.Author contributions
A. K. S. and P. P. conceived, designed and analyzed results of the study. A. K. S. and S. Y. designed the cultivation experiments, performed susceptibility experiments, bacterial growth rate analysis, gingipains inhibition assay along with biofilm degradation assay. A. K. S., Z. F., P. A. performed experiments on synthesis, physical characterizations and analyzed data. A. K. S., M. B., M. K. G. performed confocal microcopy experiments and analyzed the data. A. K. S., Z. F., P. A., K. N., and P. K. N. performed toxicity assay while A. K. S. and KS performed and analyzed results with flow cytometry. A. K. S. wrote the manuscript which was endorsed by all the co-authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We sincerely acknowledge the financial support from the Department of Science and Technology, Government of India [DST project (DST/TSG/NTS/2013/47) sanctioned to MB, and PP, contingency grant offered to AKS as JRF (2061430918 vide 22/06/2014(i) EU-V) by University Grants Commission, New Delhi and DST-PURSE grant sanctioned to Department of Microbiology, IMS, BHU.References
- R. C. Page and K. S. Kornman, The pathogenesis of human periodontitis: an introduction, Periodontol. 2000, 1997, 14, 9–11 CrossRef CAS.
- R. C. Page, S. Offenbacher, H. E. Schroeder, G. J. Seymour and K. S. Kornman, Advances in the pathogenesis of periodontitis: summary of developments, clinical implications and future directions, Periodontol. 2000, 1997, 14, 216–248 CrossRef CAS PubMed.
- K. Y. How, K. P. Song and K. G. Chan, Porphyromonas gingivalis: an overview of periodontopathic pathogen below the gum line, Front. Microbiol., 2016, 7, 53 Search PubMed.
- P. E. Petersen and H. Ogawa, The global burden of periodontal disease: towards integration with chronic disease prevention and control, Periodontol. 2000, 2012, 60, 15–39 CrossRef PubMed.
- A. B. Berezow and R. P. Darveau, Microbial shift and periodontitis, Periodontol. 2000, 2011, 55, 36–47 CrossRef PubMed.
- M. Nakayama and N. Ohara, Molecular mechanisms of Porphyromonas gingivalis-host cell interaction on periodontal diseases, Jpn. Dent. Sci. Rev., 2017, 134–140 CrossRef PubMed.
- G. Hajishengallis, Periodontitis: from microbial immune subversion to systemic inflammation, Nat. Rev. Immunol., 2015, 15, 30 CrossRef CAS PubMed.
- G. Hajishengallis, Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response, Trends Immunol., 2014, 35, 3–11 CrossRef CAS PubMed.
- P. M. Bartold and T. E. Van Dyke, Periodontitis: a host-mediated disruption of microbial homeostasis. Unlearning learned concepts, Periodontol. 2000, 2013, 62, 203–217 CrossRef PubMed.
- A. C. R. Tanner, C. Haffer, G. T. Bratthall, R. A. Visconti and S. S. Socransky, A study of the bacteria associated with advancing periodontitis in man, J. Clin. Periodontol., 1979, 6, 278–307 CrossRef CAS PubMed.
- J. Mysak, S. Podzimek, P. Sommerova, Y. Lyuya-Mi, J. Bartova and T. Janatova, et al., Porphyromonas gingivalis: major periodontopathic pathogen overview, J. Immunol. Res., 2014, 2014, 1–8 CrossRef PubMed.
- H. K. Datta, W. F. Ng, J. A. Walker, S. P. Tuck and S. S. Varanasi, The cell biology of bone metabolism, J. Clin. Pathol., 2008, 61, 577–587 CrossRef CAS PubMed.
- C. W. Cutler, J. R. Kalmar and C. A. Genco, Pathogenic strategies of the oral anaerobe, Porphyromonas gingivalis, Trends Microbiol., 1995, 3, 45–51 CrossRef CAS PubMed.
- J. Travis, R. Pike, T. Imamura and J. Potempa, Porphyromonas gingivalis proteinases as virulence factors in the development of periodontitis, J. Periodontal Res., 1997, 32, 120–125 CrossRef CAS.
- J. Potempa, A. Sroka, T. Imamura and J. Travis, Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes, Curr. Protein Pept. Sci., 2003, 4, 397–407 CrossRef CAS PubMed.
- T. Bengtsson, A. Khalaf and H. Khalaf, Secreted gingipains from Porphyromonas gingivalis colonies exert potent immunomodulatory effects on human gingival fibroblasts, Microbiol. Res., 2015, 178, 18–26 CrossRef CAS PubMed.
- I. de Diego, F. Veillard, M. Sztukowska, T. Guevara, B. Potempa and A. Pomowski, et al., Structure and mechanism of cysteine peptidase Kgp, a major virulence factor of Porphyromonas gingivalis in periodontitis, J. Biol. Chem., 2014 DOI:10.1074/jbc.M114.602052.
- A. D. Haffajee, M. A. Cugini, S. Dibart, C. Smith, R. L. Kent Jr and S. S. Socransky, Clinical and microbiological features of subjects with adult periodontitis who responded poorly to scaling and root planing, J. Clin. Periodontol., 1997, 24, 767–776 CrossRef CAS.
- L. Sbordone, L. Ramaglia, E. Gulletta and V. Iacono, Recolonization of the subgingival microflora after scaling and root planing in human periodontitis, J. Periodontol., 1990, 61, 579–584 CrossRef CAS PubMed.
- D. Herrera, M. Sanz, S. Jepsen, I. Needleman and S. Roldán, A systematic review on the effect of systemic antimicrobials as an adjunct to scaling and root planing in periodontitis patients, J. Clin. Periodontol., 2002, 29, 136–159 CrossRef PubMed.
- A. P. V. Colombo, S. K. Boches, S. L. Cotton, J. M. Goodson, R. Kent and A. D. Haffajee, et al., Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray, J. Periodontol., 2009, 80, 1421–1432 CrossRef CAS PubMed.
- J. G. Caton, S. G. Ciancio, T. M. Blieden, M. Bradshaw, R. J. Crout and A. F. Hefti, et al., Treatment with subantimicrobial dose doxycycline improves the efficacy of scaling and root planing in patients with adult periodontitis, J. Periodontol., 2000, 71, 521–532 CrossRef CAS PubMed.
- P. D. Marsh, D. A. Head and D. A. Devine, Prospects of oral disease control in the future–an opinion, J. Oral Microbiol., 2014, 6, 26176 CrossRef PubMed.
- J. Frias, E. Olle and M. Alsina, Periodontal pathogens produce quorum sensing signal molecules, Infect. Immun., 2001, 69, 3431–3434 CrossRef CAS PubMed.
- A. K. Singh, P. Prakash, R. Singh, N. Nandy, Z. Firdaus and M. Bansal, et al., Curcumin Quantum Dots Mediated Degradation of Bacterial Biofilms, Front. Microbiol., 2017, 8, 1517 CrossRef PubMed.
- R. De, P. Kundu, S. Swarnakar, T. Ramamurthy, A. Chowdhury and G. B. Nair, et al., Antimicrobial activity of curcumin against Helicobacter pylori isolates from India and during infections in mice, Antimicrob. Agents Chemother., 2009, 53, 1592–1597 CrossRef CAS PubMed.
- W. Jianhua and W. Hai, Antifungal susceptibility analysis of berberine, baicalin, eugenol and curcumin on Candida albicans, J. Med. Coll. PLA, 2009, 24, 142–147 CrossRef.
- E. Marini, M. Di Giulio, G. Magi, S. Di Lodovico, M. E. Cimarelli and A. Brenciani, et al., Curcumin, an antibiotic resistance breaker against a multiresistant clinical isolate of Mycobacterium abscessus, Phyther. Res., 2018, 32, 488–495 CrossRef CAS PubMed.
- B. Wachter, M. Syrowatka, A. Obwaller and J. Walochnik, In vitro efficacy of curcumin on Trichomonas vaginalis, Wien. Klin. Wochenschr., 2014, 126, 32–36 CrossRef CAS PubMed.
- P. Hu, P. Huang and M. W. Chen, Curcumin reduces Streptococcus mutans biofilm formation by inhibiting sortase A activity, Arch. Oral Biol., 2013, 58, 1343–1348 CrossRef CAS PubMed.
- J. Song, B. Choi, E. J. Jin, Y. Yoon and K. H. Choi, Curcumin suppresses Streptococcus mutans adherence to human tooth surfaces and extracellular matrix proteins, Eur. J. Clin. Microbiol. Infect. Dis., 2012, 31, 1347–1352, DOI:10.1007/s10096-011-1448-y.
- S.-H. Mun, S.-B. Kim, R. Kong, J.-G. Choi, Y.-C. Kim and D.-W. Shin, et al., Curcumin Reverse Methicillin Resistance in Staphylococcus aureus, Molecules, 2014, 19, 18283–18295, DOI:10.3390/molecules191118283.
- B. R. K. Bhawana, H. S. Buttar, V. K. Jain and N. Jain, Curcumin nanoparticles: preparation, characterization, and antimicrobial study, J. Agric. Food Chem., 2011, 59, 2056–2061, DOI:10.1021/jf104402t.
- P. Tyagi, M. Singh, H. Kumari, A. Kumari and K. Mukhopadhyay, Bactericidal activity of curcumin I is associated with damaging of bacterial membrane, PLoS One, 2015, 10, 1–15, DOI:10.1371/journal.pone.0121313.
- H. K. Syed, M. A. Iqbal, R. A. Haque and K.-K. Peh, Synthesis, characterization and antibacterial activity of a curcumin–silver(I) complex, J. Coord. Chem., 2015, 68, 1088–1100, DOI:10.1080/00958972.2014.1003051.
- O. Naksuriya, S. Okonogi, R. M. Schiffelers and W. E. Hennink, Curcumin nanoformulations: a review of pharmaceutical properties and preclinical studies and clinical data related to cancer treatment, Biomaterials, 2014, 35, 3365–3383, DOI:10.1016/j.biomaterials.2013.12.090.
- Y. Liu, Y. Cai, X. Jiang, J. Wu and X. Le, Molecular interactions, characterization and antimicrobial activity of curcumin–chitosan blend films, Food Hydrocolloids, 2015, 52, 564–572, DOI:10.1016/j.foodhyd.2015.08.005.
- R. Feng, Z. Song and G. Zhai, Preparation and in vivo pharmacokinetics of curcumin-loaded PCL-PEG-PCL triblock copolymeric nanoparticles, Int. J. Nanomed., 2012, 7, 4089–4098, DOI:10.2147/IJN.S33607.
- J. N. Eloff, A sensitive and quick microplate method to determine the minimal inhibitory concentration of plant extracts for bacteria, Planta Med., 1998, 64, 711–713 CrossRef CAS PubMed.
- R. Pike, W. McGraw, J. Potempa and J. Travis, Lysine-and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization, and evidence for the existence of complexes with hemagglutinins, J. Biol. Chem., 1994, 269, 406–411 CAS.
- S. Izui, S. Sekine, K. Maeda, M. Kuboniwa, A. Takada and A. Amano, et al., Antibacterial activity of curcumin against periodontopathic bacteria, J. Periodontol., 2016, 87, 83–90 CrossRef CAS PubMed.
- A. K. Singh, P. Prakash, A. Achra, G. P. Singh, A. Das and R. K. Singh, Standardization and classification of in vitro biofilm formation by clinical isolates of Staphylococcus aureus, J. Global Infect. Dis., 2017, 9, 93 CrossRef CAS PubMed.
- Y. Wang and L. Chen, Quantum dots, lighting up the research and development of nanomedicine, Nanomedicine, 2011, 7, 385–402, DOI:10.1016/j.nano.2010.12.006.
- X. Michalet, L. a Bentolila, J. M. Tsay and S. Doose, Quantum Dots for Live Cells in vivo Imaging, and Diagnostics, Science, 2005, 307, 538–544, DOI:10.1126/science.1104274.
- K. Sun, M. Vasudev, H.-S. Jung, J. Yang, A. Kar and Y. Li, et al., Applications of colloidal quantum dots, Microelectron. J., 2009, 40, 644–649, DOI:10.1016/j.mejo.2008.06.033.
- T. Jamieson, R. Bakhshi, D. Petrova, R. Pocock, M. Imani and A. M. Seifalian, Biological applications of quantum dots, Biomaterials, 2007, 28, 4717–4732, DOI:10.1016/j.biomaterials.2007.07.014.
- R. J. Byers and E. R. Hitchman, Quantum dots brighten biological imaging, Prog. Histochem. Cytochem., 2011, 45, 201–237, DOI:10.1016/j.proghi.2010.11.001.
- W. W. Yu, E. Chang, R. Drezek and V. L. Colvin, Water-soluble quantum dots for biomedical applications, Biochem. Biophys. Res. Commun., 2006, 348, 781–786, DOI:10.1016/j.bbrc.2006.07.160.
- E. F. S. Authority, Refined exposure assessment for curcumin (E 100), EFSA J., 2014, 12, 3876 CrossRef.
- T. Esatbeyoglu, P. Huebbe, I. M. A. Ernst, D. Chin, A. E. Wagner and G. Rimbach, Curcumin-from molecule to biological function, Angew. Chem., Int. Ed., 2012, 51, 5308–5332, DOI:10.1002/anie.201107724.
- S. S. Socransky and A. D. Haffajee, Evidence of bacterial etiology: a historical perspective, Periodontol. 2000, 1994, 5, 7–25 CrossRef CAS PubMed.
- S. C. Holt, L. Kesavalu, S. Walker and C. A. Genco, Virulence factors of Porphyromonas gingivalis, Periodontol. 2000, 1999, 20, 168–238 CrossRef CAS PubMed.
- H. Nagata, Y. Inagaki, M. Tanaka, M. Ojima, K. Kataoka and M. Kuboniwa, et al., Effect of eucalyptus extract chewing gum on periodontal health: a double-masked, randomized trial, J. Periodontol., 2008, 79, 1378–1385 CrossRef CAS PubMed.
- H. Nagata, Y. Inagaki, Y. Yamamoto, K. Maeda, K. Kataoka and K. Osawa, et al., Inhibitory effects of macrocarpals on the biological activity of Porphyromonas gingivalis and other periodontopathic bacteria, Oral Microbiol. Immunol., 2006, 21, 159–163 CrossRef CAS PubMed.
- K. Takahashi, M. Fukazawa, H. Motohira, K. Ochiai, H. Nishikawa and T. Miyata, A pilot study on antiplaque effects of mastic chewing gum in the oral cavity, J. Periodontol., 2003, 74, 501–505 CrossRef PubMed.
- C. C. C. Quishida, E. G. D. O. Mima, J. H. Jorge, C. E. Vergani, V. S. Bagnato and A. C. Pavarina, Photodynamic inactivation of a multispecies biofilm using curcumin and LED light, Lasers Med. Sci., 2016, 31, 997–1009 CrossRef PubMed.
- N. C. Araújo, C. R. Fontana, V. S. Bagnato and M. E. M. Gerbi, Photodynamic effects of curcumin against cariogenic pathogens, Photomed. Laser Surg., 2012, 30, 393–399 CrossRef PubMed.
- K. Maeda, H. Nagata, A. Nonaka, K. Kataoka, M. Tanaka and S. Shizukuishi, Oral streptococcal glyceraldehyde-3-phosphate dehydrogenase mediates interaction with Porphyromonas gingivalis fimbriae, Microbes Infect., 2004, 6, 1163–1170 CrossRef CAS PubMed.
- M. Shahzad, E. Millhouse, S. Culshaw, C. A. Edwards, G. Ramage and E. Combet, Selected dietary (poly) phenols inhibit periodontal pathogen growth and biofilm formation, Food Funct., 2015, 6, 719–729 RSC.
- N. C. Araújo, C. R. Fontana, M. E. M. Gerbi and V. S. Bagnato, Overall-mouth disinfection by photodynamic therapy using curcumin, Photomed. Laser Surg., 2012, 30, 96–101 CrossRef PubMed.
- A. Araghizadeh, J. Kohanteb and M. M. Fani, Inhibitory activity of green tea (Camellia sinensis) extract on some clinically isolated cariogenic and periodontopathic bacteria, Med. Princ. Pract., 2013, 22, 368–372 CrossRef PubMed.
- S. Sakanaka, M. Aizawa, M. Kim and T. Yamamoto, Inhibitory effects of green tea polyphenols on growth and cellular adherence of an oral bacterium, Porphyromonas gingivalis, Biosci., Biotechnol., Biochem., 1996, 60, 745–749 CrossRef CAS PubMed.
- S. Muglikar, K. C. Patil, S. Shivswami and R. Hegde, Efficacy of curcumin in the treatment of chronic gingivitis: a pilot study, Oral Health Prev. Dent., 2013, 11, 81–86 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra08435a |
| This journal is © The Royal Society of Chemistry 2018 |