Open Access Article
Open Access ArticleInsight into the effect of quinic acid on biofilm formed by Staphylococcus aureus
Jin-Rong
Bai
 ,
Yan-Ping
Wu
,
Yan-Ping
Wu
 ,
Grosu
Elena
,
Kai
Zhong
* and
Hong
Gao
,
Grosu
Elena
,
Kai
Zhong
* and
Hong
Gao
College of Light Industry, Textile and Food Engineering and Healthy Food Evaluation Research Center, Sichuan University, Chengdu 610065, P. R. China. E-mail: eric211@163.com; Fax: +86 028 85405137; Tel: +86 028 85405236
First published on 29th January 2019
Abstract
The biofilm formation of Staphylococcus aureus on food contact surfaces is the main risk of food contamination. In the present study, we firstly investigated the inhibitory effect of quinic acid (QA) on biofilm formed by S. aureus. Crystal violet staining assay and microscopy analysis clearly showed that QA at sub-MIC concentrations was able to significantly reduce the biofilm biomass and cause a collapse on biofilm architecture. Meanwhile, fibrinogen binding assay showed that QA had obviously effect on the S. aureus bacteria adhesion. XTT reduction assay and confocal laser scanning microscopic images revealed that QA significantly decreased metabolic activity and viability of biofilm cells. In addition, qRT-PCR analysis explored the potential inhibitory mechanism of QA against biofilm formation, which indicated that QA significantly repressed the gene sarA and activated the gene agrA. Moreover, QA exhibited a highly ability to reduce the number of sessile S. aureus cells adhered on the stainless steel. So, it was suggested that QA could be used as a promising antibiofilm agent to control biofilm formation of S. aureus.
1. Introduction
Staphylococcus aureus is a leading Gram-positive pathogen associated with a series of diseases,1,2 including osteomyelitis, pneumonia, endocarditis and septicemia.3,4 It has been frequently found in many food materials, such as dairy products, egg, sea food and meat.5 Generally, food contaminated by S. aureus is able to cause food poisoning, which has been an extensive issue concerned by the international community and food industry.4In recent years, S. aureus with the excellent ability to form biofilm gives rise to a new concern in human health. Biofilms are surface associated bacterial aggregates in a self-produce extracellular polymeric matrix that enable bacteria to survive and persist on abiotic surfaces over long periods of time.6,7 Biofilms can easily adhere and form on fresh or processed foods, and food devices, such as dairy products, fishery products, stainless steel, glass, rubber, and plastic.8 Moreover, once biofilms adhere and form, they increase their resistance to commonly used antibiotics and disinfectants,8 which possesses a higher potential risk to the cross contamination and huge economic losses.9 Such issues have led to a great demand for potential antibiofilm compounds from plants with highly efficient activity against S. aureus biofilm, which will be beneficial to the development of safer and more productive food processing industries.
Organic acids, one kind of most common metabolites in plants, are routinely used in many countries to reduce foodborne pathogens contamination. In the United States, organic acids have been approved for use to prevent pathogen contamination in the meat decontamination area.10 Some of them have been reported to exhibit the excellent inhibitory activity against S. aureus biofilm formation, consisting of gallic acid,1 citric acid,11 ellagic acid,12 ginkgolic acid,13 caffeic acid and chlorogenic acid.14 Despite the excellent ability of these antibiofilm agents for inhibiting biofilm formation has been discovered, the molecular mechanism of the inhibitory action remains unclear and challenging. From this perspective, it is necessary to explore the inhibitory action mechanism of organic acids.
Quinic acid (QA), widely spread in many plants, is an important organic acid with many biological activities, including antioxidant, antimutagenic and anti-inflammatory activities.15,16 Although QA has been proved to have the excellent antibacterial activity against S. aureus through damaging the cell membrane and interfering with normal functions of cells in our previous study,17,18 its inhibitory effect against S. aureus biofilm formation is not clear. Therefore, in the present study, we firstly reported the capacity of QA to inhibit biofilm formation of S. aureus and explored its potential mechanism.
2. Materials and methods
2.1. Chemicals, bacteria strain, and culture conditions
QA (≥98%) was purchased from the Aladdin Industrial Corporation (Shanghai, China). QA was dissolved in distilled water and diluted with the corresponding medium for the assays. The standard bacterial strain S. aureus ATCC 29213 (ATCC, Arlington, Virginia, USA) was used in the present study for its ability to form biofilms. The bacterial cells were shaken at 37 °C for 10 h in tryptone soy broth (TSB) supplemented with 1% glucose to obtain logarithmic phase cells for biofilm assay. The minimum inhibitory concentration (MIC) of QA against S. aureus ATCC 29213 was detected to be 5.0 mg mL−1.172.2. Biofilm formation
The biofilm of S. aureus ATCC 29213 was prepared in a 96-well microtitre plate by the previous method with a slight modification.19 In brief, logarithmic phase S. aureus cells were diluted with TSB with 1% glucose to obtain a bacterial suspension of 1 × 106 CFU mL−1. Then, 100 μL of the bacterial suspension was dispensed into each well of a microtitre plate in the presence of sub-MIC concentrations of QA. The negative control was the TSB with 1% glucose. All of wells were seeded in triplicate. The plate was incubated at 37 °C for 24 h without shaking to form the one-day-old biofilm.2.3. Assessment of growth inhibition and biofilm biomass
After one-day-old biofilm formation, we measured absorbance values at 600 nm using a microplate reader (Spectra MAX-190, Molecular Devices Co., Sunnyvale, CA, USA) to evaluate the effect of QA on the planktonic bacterial growth according the previous report.20 The planktonic cells were discarded from the wells, and then the wells were gently washed twice with PBS (0.01 M, pH 7.2). Subsequently, 200 μL of 0.4% crystal violet solution was added to the wells to stain the adherent sessile cells for 5 min. The stained biofilm cells were washed twice with distilled water and dissolved in 200 μL of 20% glacial acetic acid. After 30 min, the biofilm biomass was quantified by measuring the optical density at 570 nm. The reduction percentage of growth or biofilm was shown by using the following formula (Sivaranjani et al., 2016).21| Reduction percentage = [(Control OD600/570 nm − treated OD600/570 nm)/control OD600/570 nm] × 100 |
2.4. Scanning electron microscopic (SEM)
In order to visualize the inhibition of QA on the biofilm formation of S. aureus, SEM images were performed using a scanning electron microscope (SU3500, Hitachi, Japan) according to the previously reported method with minor modifications.14 In brief, 500 μL of QA (0.3125, 0.625, 1.25 mg mL−1) was added into the wells of a 24-well microplate containing sterile round glass slides (∅14 mm). Then, 500 μL of cell suspension at a density of 1 × 106 CFU mL−1 was added into the wells. After incubation at 37 °C for 24 h, the glass slides were gently taken out, fixed in 2.5% glutaraldehyde at 4 °C for 3 h, and then gently washed once with PBS (0.01 M, pH 7.2). Subsequently, biofilm samples were dehydrated in a sequence of ethanol, dried with critical point drier, and coated with gold prior to observation.2.5. Fibrinogen binding assay
Fibrinogen binding assay was performed according to the previously reported method with little modifications.20 In brief, 1 mL of logarithmic phase S. aureus cells (1 × 106 CFU mL−1) with different concentrations of QA were co-cultured at 37 °C with shaking at 120 rpm for 5 h. The bacterial cells were collected by centrifugation at 3000 rpm for 5 min, and suspended in PBS (0.01 M, pH 7.2). Then, 100 μL of the bacterial suspension was dispensed into each well of a 96-well microtitre plate coated with fibrinogen (incubated with 20 μg mL−1 of bovine fibrinogen at 4 °C for 12 h) and further incubated at 37 °C. After 1 h, the wells were washed once with PBS (0.01 M, pH 7.2) and fixed with 25% formaldehyde for 30 min. Finally, the biomass of adherent bacteria were investigated by using the crystal violet staining assay as described above method of 2.3. The relative percentage of adhesion was calculated according to the following formula.| Relative% adhesion = Treated OD570 nm/control OD570 nm × 100 |
2.6. Assessment of metabolic activity of S. aureus cells in biofilm
The metabolic activity changes of S. aureus biofilm cells induced by QA were detected using 2,3-bis (2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide sodium salt (XTT) reduction assay as described previously method with minor modifications.22 After incubation with QA and washing progress, biofilm cells in the microplate were further incubated with 120 μL of the mixed solution of XTT (Sigma Aldrich Co., St. Louis, USA) and phenazine methosulphate (Sigma Aldrich Co., St. Louis, USA) dissolved in PBS (0.01 M, pH 7.2). The final concentration of XTT in each well was set at 100 μg mL−1 and phenazine methosulphate was 10 μg mL−1. Following incubation in the darkness at 37 °C for 3 h, the biofilm metabolic activity was determined through the measurement of the absorbance value at 492 nm in each well using the above microplate reader.2.7. Confocal laser scanning microscopic (CLSM)
The biofilm cell viability changes induced by QA were determined using LIVE/DEAD BacLight bacterial viability kit (Life Technologies, MA, USA) according to the manufacturer's instructions. Briefly, the biofilms in the presence and absence of QA were grown on the glass slides as described above method. After incubation and washing procedures, the biofilms on slides were stained with 500 μL of a combined dye solution of SYTO 9 and propidium iodide (PI) diluted with PBS (0.01 M, pH 7.2) keeping a final concentration of 5 μM of SYTO 9 and 30 μM of PI. Following treatment at 25 °C for 15 min in the darkness, the stained biofilms were observed by a confocal laser scanning microscope (SP8, Leica, Germany).2.8. Isolation of RNA and qRT-PCR analysis
The changes of the transcription levels of the related genes during S. aureus biofilm formation were evaluated by qRT-PCR analysis. S. aureus cells were performed according to the previously reported method with minor modifications.23 Briefly, 1 mL of logarithmic phase S. aureus cells (1 × 106 CFU mL−1) were cultured at 37 °C with shaking at 120 rpm. After 3 h, QA was added to a concentration of 1.25 mg mL−1 and the bacterial suspensions were further incubated for 5 h. Bacterial suspensions were then centrifuged at 3000 rpm for 5 min. RNA from S. aureus cells was isolated using UNIQ-10 Trizol (Sangon Biotech, Shanghai, China) according to the manufacturer's protocol. The isolated RNAs were reverse-transcribed into cDNA by using the RevertAid Premium Reverse Transcriptase (Thermo Fisher Scientific, MA, USA). Continuously, the expressions of the 5 selected genes by qRT-PCR were measured. The 5 selected genes were polysaccharide intercellular adhesion locus genes (icaA, icaR), RNA polymerase sigma factor (sigB), quorum sensing gene (agrA), and staphylococcal accessory regulator A (sarA), respectively. Gene-specific primers and 16s rRNA housekeeping gene primer were presented in Table 1.23 qRT-PCR was determined by using the 2xSG Fast qPCR Master Mix Kit (Sangon Biotech, Shanghai, China) and StepOne Real-Time PCR system (Applied Biosystems, CA, USA).| Gene | Primer |
|---|---|
| icaA | Forward 5′-CTG GCG CAG TCA ATA CTA TTT CGG GTG TCT-3′ |
| Reverse 5′-GAC CTC CCA ATG TTT CTG GAA CCA ACA TCC-3′ | |
| icaR | Forward 5′ TGC TTT CAA ATA CCA ACT TTC AAG A 3′ |
| Reverse 5′ ACG TTC AAT TAT CTA ATA CGC CTG A 3′ | |
| sigB | Forward 5′-AAG TGA TTC GTA AGG ACG TCT-3′ |
| Reverse 5′-TCG ATA ACT ATA ACC AAA GCC T-3′ | |
| agrA | Forward 5′-TGA TAA TCC TTA TGA GGT GCT T-3′ |
| Reverse 5′-CAC TGT GAC TCG TAA CGA AAA-3′ | |
| sarA | Forward 5′-CAA ACA ACC ACA AGT TGT TAA AGC-3′ |
| Reverse 5′-TGT TTG CTT CAG TGA TTC GTT T-3′ | |
| 16s rRNA | Forward 5′-ACTCCTACGGGAGGCAGCAG-3′ |
| Reverse 5′-ATTACCGCGGCTGCTGG-3′ |
2.9. Stainless steel model assay
Biofilm assay on stainless steel was performed as previously described method with minor modifications.6 AISI 304 grade annealed stainless steel was used for making coupons. Stainless steel coupons (2 cm × 2 cm × 1 mm thickness) were cleaned with a neutral detergent followed by distilled water, autoclaved at 121 °C for 20 min, and dried at 80 °C prior to use. An amount of 10 mL of bacterial suspensions of logarithmic phase S. aureus (1 × 108 CFU mL−1) was dispensed onto the stainless steel coupons submerged in separate tubes at 37 °C for 2 h. Continuously, the steel coupons were gently washed with 0.1% peptone water to remove planktonic cells, and then immersed in 10 mL of QA solutions (dissolved in 0.1% peptone water) at the concentrations of 5, 10, 15 mg mL−1, respectively, for 10 min. The negative control was performed with 0.1% peptone water. After washed with 0.1% peptone water to remove excess QA, each steel coupon was immersed in 10 mL of 0.1% peptone water and sonicated at 55 kHz for 10 min in a bath sonicator (AS3120B, Aote Saiensi Instrument Co., Tianjin, China). The viable bacterial counts in 0.1% peptone water from each tube were detected after serial dilution and spread on nutrient agar plates. After incubation of the inoculated nutrient agar plates at 37 °C for 24 h, the density of biofilm cells was calculated according to the following formula.log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU cm−2 = log[CFU mL−1 × 10/steel coupon area (cm2)] CFU cm−2 = log[CFU mL−1 × 10/steel coupon area (cm2)] |
2.10. Statistical analysis
All experiments were performed in independent triplicate. Statistical analyses were conducted with the SPSS software (version 20.0; SPSS, I., Chicago, IL). Data differences were considered statistically significant at P < 0.05.3. Results
3.1. QA inhibited the biofilm formation of S. aureus without affecting the bacterial growth
The biofilm biomass assay using crystal violet staining is a rapid and convenient method for testing the biofilm formation. To evaluate the effect of QA on S. aureus biofilm formation, S. aureus and QA at sub-MICs were co-cultured using the microdilution method. As shown in Fig. 1, when treated with QA ranging from 0.3125 to 1.25 mg mL−1 for 24 h, the biofilm biomass of S. aureus was significantly reduced ranging from 55% to 70% compared with the control biofilm (P < 0.01). To determine whether the effect of QA on biofilm was dependent on the growth inhibition of planktonic bacteria, the growth of S. aureus was detected by measuring absorbance values at 600 nm after incubation for 24 h. When treated with QA at 1.25 mg mL−1, the growth of S. aureus was decrease slightly by approximately 20% in (Fig. 1). So, it was suggested that QA did not influence (P > 0.05) the growth of S. aureus at its sub-MICs but could significantly inhibit the formation of S. aureus biofilm (P < 0.01).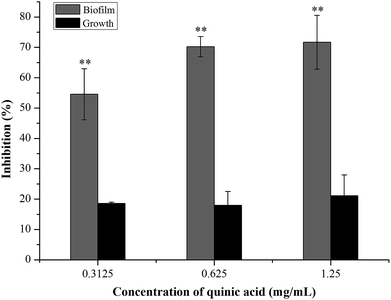 | ||
| Fig. 1 Effect of quinic acid on the planktonic growth and biofilm biomass of S. aureus ATCC 29213. Data were expressed as mean ± standard deviation (n = 3). **P < 0.01, compared with control. | ||
3.2. Biofilms treated with QA had loosely extracellular polymeric matrix
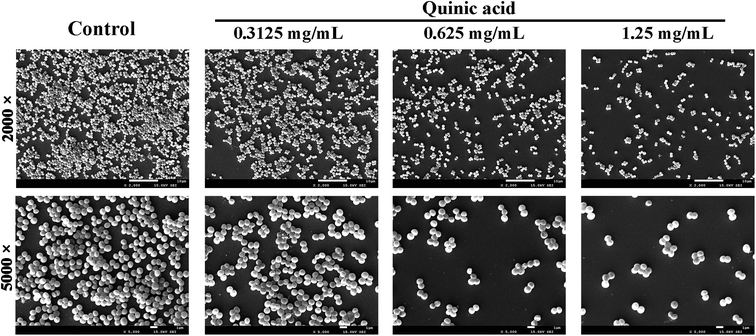
To visualize the morphology and ultrastructural structure of S. aureus biofilms treated with QA, the distinctions were observed in biofilm architecture at high resolution using SEM at 2000 and 5000 times magnification (Fig. 2). Biofilms exhibited monolayer and slackened cell cluster in the presence of 0.3125 mg mL−1 of QA. When biofilms were treated with higher concentration of QA, there were fewer clumps of attached microcolonies and even individual cells on the glass slides. In comparison, the untreated biofilm presented continuous clumps and large aggregates of cells.3.3. QA decreased the adhesion of S. aureus to fibrinogen
Fibrinogen, a kind of plasma protein, can be used as a substrate for staphylococcal adhesion.24 In order to examine the effect of QA on S. aureus adhesion, fibrinogen binding assay was conducted using microtitre plate coated with fibrinogen. The result was shown in Fig. 3. Compared to the control group, the relative percentage adhesion was significantly (P < 0.01) decreased after treatment with QA at 0.3125, 0.625 and 1.25 mg mL−1. The results suggested that QA reduced the S. aureus bacteria adhesion to fibrinogen.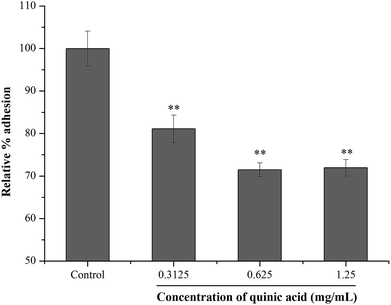 | ||
| Fig. 3 Effect of quinic acid on the relative percentage adhesion of S. aureus ATCC 29213 to fibrinogen. Data were expressed as mean ± standard deviation (n = 3). **P < 0.01, compared with control. | ||
3.4. QA decreased metabolic activity and viability of S. aureus biofilm cells
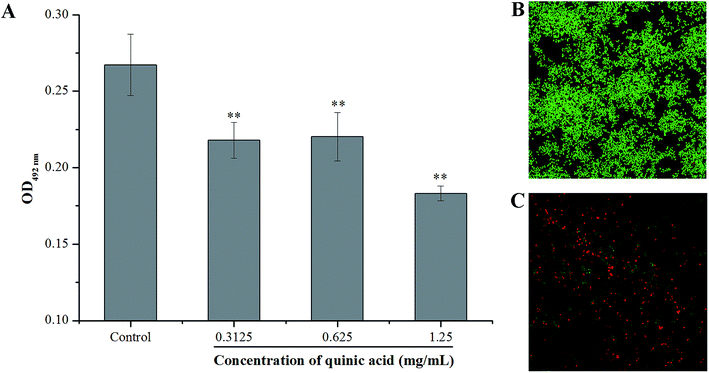
XTT reduction assay was carried out to evaluate the effect of QA on the metabolic activity of S. aureus biofilm cells. The absorbance values at 492 nm can indirectly reflect the total metabolic activity of cells, as the metabolically active cells are able to decrease the XTT into orange colored water soluble formazan.21 As shown in Fig. 4, QA significantly reduced the metabolic activity of S. aureus biofilm cells (P < 0.01). The absorbance values at 492 nm of S. aureus biofilm cells were detected to be 0.22 ± 0.01, 0.22 ± 0.02 and 0.18 ± 0.05 in the presence of QA at 0.3125, 0.625 and 1.25 mg mL−1, respectively, whereas the absorbance value of the untreated control cells was 0.27 ± 0.02.The LIVE/DEAD BacLight bacterial viability kit and CLSM were also used to detect whether QA can interfere with the viability of cells in the S. aureus biofilm. SYTO 9, a kind of nucleic acid dye, can penetrate both live and damaged bacterial cells, and emits strong green fluorescence. On the other hand, another kind of nucleic acid dye PI is only able to penetrate damaged or dead bacterial cells, and emits red fluorescence.8 The CLSM images of bacteria labeled with SYTO 9 and PI at 620 times magnification were presented in Fig. 4B and C. Obviously, the CLSM image of the negative control samples (Fig. 4B) with no QA treatment revealed that the formation of a dense biofilm viewed as almost green cells, and the green fluorescence intensity was strong, indicating that the untreated cells were viable and active. In comparison, a significant reduction of the green fluorescence was observed on the cells treated with QA at 1.25 mg mL−1 (Fig. 4C), and the green fluorescence intensity was very weak. Furthermore, the red fluorescence intensity was enhanced in Fig. 4C compared to the control, suggesting that the biofilm treated with 1.25 mg mL−1 of QA was composed of damaged and dead cells.
3.5. QA modulated the expression of related genes during S. aureus biofilm formation
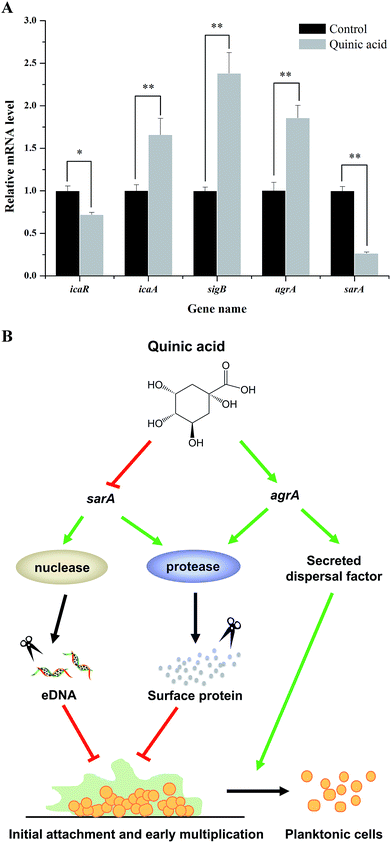
To further investigate the molecular mechanism of biofilm formation inhibition of QA, qRT-PCR was performed to detect the expression of several genes related to the biofilm formation. As shown in Fig. 5A, when 1.25 mg mL−1 of QA was used, the gene sarA which encodes staphylococcal accessory regulator A (SarA) was significantly repressed by 76% (P < 0.01). The polysaccharide intercellular adhesion locus gene icaA was activated, whereas the ica operon repressor icaR was inhibited by 47% compared to control. In addition, QA treatment significantly up-regulated agrA and sigB (P < 0.05), which encode quorum sensing gene regulator (AgrA) and RNA polymerase sigma factor (SigB), respectively.3.6. QA inhibited the S. aureus cell adhered to stainless steel
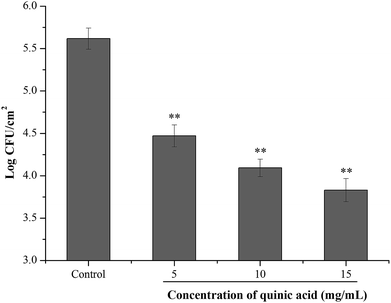
To investigate the effect of QA on the S. aureus cells adhered to stainless steel, initial planktonic populations of approximately 8.0 log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU mL−1 of S. aureus cells were allowed to adhere on the stainless steel for 2 h. After bacterial adhesion to stainless steel, the inhibitory activity of QA at different concentrations was evaluated using 10 min contact time. As shown in Fig. 6, the initial population of the adherent cells in S. aureus biofilm on the stainless steel was 5.82 log
CFU mL−1 of S. aureus cells were allowed to adhere on the stainless steel for 2 h. After bacterial adhesion to stainless steel, the inhibitory activity of QA at different concentrations was evaluated using 10 min contact time. As shown in Fig. 6, the initial population of the adherent cells in S. aureus biofilm on the stainless steel was 5.82 log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU cm−1.2 After 10 min contact time, treatment with QA at 5, 10 and 15 mg mL−1 significantly reduced the populations of the adherent cells by 1.05, 1.47 and 1.77 log
CFU cm−1.2 After 10 min contact time, treatment with QA at 5, 10 and 15 mg mL−1 significantly reduced the populations of the adherent cells by 1.05, 1.47 and 1.77 log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU cm−1,2 respectively (P < 0.01).
CFU cm−1,2 respectively (P < 0.01).
4. Discussion
Currently, the majority of microbial contamination in the food industry is attributed to biofilm formation and growth on food plants or during the processing processes.9 The biofilm cells are highly tolerant to harmful factors in the environment, such as desiccation, antimicrobial agents and the body's immune system, causing large economic losses.9,21 Along with economic loss problems, biofilm formation on food contact surfaces possesses a high health risk to consumers.6 Therefore, a high-efficiency way to prevent biofilm formation and eradicate the biofilm adhesion still remains challenging. The present study focused on an antibiofilm agent QA that could effectively inhibit biofilm formation of S. aureus without exerting any pressure over the planktonic cell growth.Our results of biofilm biomass measurement showed that QA at sub-MICs significantly inhibited the biofilm formation (P < 0.01) without interfering with the planktonic growth of S. aureus (P > 0.05). It revealed that the reduction in biofilm biomass was mainly attributed to the antibiofilm potential of QA rather than the decreased growth rate of S. aureus cells. Several studies have proved that organic acids had antibiofilm activity against S. aureus. For instance, citric acid, could significantly inhibit the biofilm formation of S. aureus.11 In addition, gallic acid, caffeic acid and chlorogenic acid had obviously inhibitory action on the biofilm formation of S. aureus.14 These findings suggested that these organic acids could be as S. aureus biofilm inhibitor candidates in food industry. However, the mechanism of their antibiofilm activity remains challenging.
In order to visually confirm the inhibitory action of QA against biofilm formation of S. aureus, the micrographs of SEM illustrated the details of the structurally complex matrix architecture and the bacteria in that matrix. QA treatment led to a huge collapse on the extracellular matrix architecture, resulting in the individual cells and loose microcolonies attached on the glass steel. The images correlated well with the quantitative results of crystal violet staining assay, which indicated that QA possessed a highly antibiofilm activity against S. aureus.
During the biofilm formation process, initial adhesion was the first crucial stage, which is mediated by the binding of S. aureus of surface anchored protein and host matrix proteins.25 Our results showed that QA significantly decreased the initial adhesion of S. aureus, which finally resulted in the reduction of biofilm biomass. One possible reason of this action was that QA might destroy the binding proteins and inhibit their activity.20 Notably, when treating with 1.25 mg mL−1, the relative percentage adhesion was decreased by approximately 30%, whereas the biofilm biomass was reduced by 72%. This finding suggested that the inhibition of biofilm formation was not absolutely dependent on the inhibition of initial adhesion.
In addition the adhesion ability, the free-floating cells aggregate to form biofilm is influenced by many other physiological factors, such as cell metabolic activity, bacterial viability, cell proliferation and accumulation of multilayer cell clusters.21 So, XTT reduction assay was carried out evaluating the effect of QA on the metabolic activity of cells in the S. aureus biofilm. As shown in Fig. 4A, QA significantly decreased the metabolic activity of S. aureus biofilm cells. Some studies have proved that the inhibition of biofilm formation was coupled with the decrease of the cell metabolic activity of biofilm.14,26 A possible reason for such action was that the decrease in metabolic activity was responsible for the resistance of biofilm to antibacterial compounds.27 Although XTT assay is a good predictor of the efficacy of antibiofilm compounds, it may have some limitations that it does not always ensure equivalence with cell death.28 Hence, we further investigated the cell viability of biofilm by using the LIVE/DEAD Baclight bacterial viability kit. It was found that QA not only obviously decreased biofilm biomass but also influenced cell viability of biofilm. When treated with 1.25 mg mL−1 of QA, S. aureus biofilm cells showed high number of dead cells. These results revealed that QA was likely to interfere with cell metabolic activity and cell viability of biofilm, and increase the sensitivity of biofilm cells, resulting in the prevention of biofilm formation.
Taken together, the above results suggested that the mechanism of QA against biofilm formation of S. aureus may be regulated by multiple pathways. To better understand the mechanism of biofilm formation of S. aureus and explore the key regulatory genes in biofilm cells exposed to QA, the transcription levels of genes related to biofilm formation were detected by using qRT-PCR. In S. aureus, the SarA, encoded by the gene sarA, is a staphylococcal accessory regulator that is closely related to biofilm adhesion and formation.29 As shown in Fig. 5A, the transcription level of sarA was repressed by QA. The gene sarA is necessary during the initial biofilm formation in S. aureus and the expression of sarA allows the development of an immature biofilm.30,31sarA mutant was reported to have no ability to form biofilm with increased protease and nuclease activities.30 The enhanced activities of protease and nuclease resulted in decreasing production of fibrinogen binding protein and release of eDNA, so as to lead to the inhibition of initial interactions between cell and cell/surface.32 This point was also well clarify the reason that QA significantly reduced the adhesion of S. aureus to fibrinogen (Fig. 3). In general, the inhibition of QA against biofilm formation was not completely due to the inactivation of sarA, and QA also up-regulated gene agrA, which is a dominant accessory regulator gene.32 Previous studies reported that gene agr activation was able to induce dispersion of S. aureus cells to planktonic state due to the enhancement of secreted dispersal factors.30,33 Moreover, gene agr can prevent the biofilm initiation, adhesion and maturation through activation of protease and decrease of extracellular matrix proteins.34 These phenotypes supported our observation of the relationships between repression of sarA, activation of agrA, and biofilm formation inhibition by QA.
However, in the present study, the biofilm biomass reduction was along with the repression of gene icaR and activation of gene icaA and sigB. It was contradictory with some previous literatures, which documented that down-regulated of gene icaR could activate gene icaA, leading to the activation of polysaccharide intercellular adhesion and the increase of biofilm biomass,1,32 and S. aureus mutants in sigB are unable to form a biofilm.33 These contradictory results might be due to the response of different types of S. aureus cells to environmental stress caused by various antibacterial agents. A previous study reported that S. aureus ica mutants had no reduction in biofilm formation, suggesting that ica was not the crucial factor in certain conditions of biofilm formation.29,32 Though gene sigB was important in the process of biofilm formation, SigB was reported to be a dispensable factor of RNA polymerase that might be activated in stress response.35 So, it was suggested that QA could inhibit biofilm formation of S. aureus via an ica/sigB-independent pathway.
In light of these results, we concluded that QA was likely to inhibit the initial biofilm formation by acting on the expression of gene sarA and agrA. An inferred model for this regulatory pathway was shown in Fig. 5B, which illustrated that key regulatory genes of QA against S. aureus biofilm formation were genes sarA and agrA.
Considering the potential application of QA in the inhibition of S. aureus biofilm formation in food processing, the inhibitory effect of QA against S. aureus biofilm formation on stainless steel was assessed. Stainless steel is a kind of widely used equipment material in the food industry because of its excellent physicochemical properties, relatively low cost and high resistance to corrosion.36 However, the hydrophilic properties of stainless steel provide an advantage for the initial attachment and biofilm formation of bacterial cells.6 As shown in Fig. 6, S. aureus could adhere in large numbers to stainless steel after a 2 h of contact. QA at MIC value and higher concentrations were able to significantly decrease the adhered populations of S. aureus (P < 0.01) after 10 min treatment. It was noticed that higher concentrations of QA were required to achieve the inhibitory effective against adhered S. aureus on stainless steel compared with that used in the biofilm biomass assays. One possible reason is that the population reduction experiment of the adhered bacteria on stainless steel was evaluated using much shorter contact time. Moreover, the sensitivity of planktonic bacteria to antibiofilm compounds is higher than that of adhered bacteria, and the reduction or removal of established biofilm is more difficult than the prevention of biofilm formation.37 Hence, strategies to prevent bacterial adhesion and biofilm formation on food or food contact surfaces are important for food industries.
5. Conclusion
In conclusion, this was the first time to evaluate the inhibitory activity of QA against biofilm formation of S. aureus. QA could effectively inhibit biofilm formation of S. aureus at sub-MIC values. QA could prevent the initial biofilm formation by inducing the expression of genes sarA and agrA. The present study suggested that QA was possible to be used on food devices as an antibiofilm agent to prevent biofilm contamination.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (no. 31701574).References
- M. H. Liu, X. X. Wu, J. K. Li, L. Liu, R. G. Zhang, D. Y. Shao and X. D. Du, Food Control, 2017, 73, 613–618 CrossRef CAS.
- E. Mataraci and S. Dosler, Antimicrob. Agents Chemother., 2012, 56, 6366–6371 CrossRef CAS.
- K. B. Ahn, J. E. Baik, C. H. Yun and S. H. Han, Front. Microbiol., 2018, 9, 327 CrossRef.
- W. Rozemeijer, P. Fink, E. Rojas, C. H. Jones, D. Pavliakova, P. Giardina, E. Murphy, P. Liberator, Q. Jiang, D. Girgenti, R. P. H. Peters, P. H. M. Savelkoul, K. U. Jansen, A. S. Anderson and J. Kluytmans, PLoS One, 2015, 10, e0116945 CrossRef PubMed.
- J. Kadariya, T. C. Smith and D. Thapaliya, Biomed Res. Int., 2014, 2014, 827965 CrossRef.
- T. Stern, E. Zelinger and Z. Hayouka, Chem. Commun., 2016, 52, 7102–7105 RSC.
- C. Fuente-Núñez, F. Reffuveille, E. F. Haney, S. K. Straus and R. E. W. Hancock, PLoS Pathog., 2014, 10, e1004152 CrossRef.
- S. H. Lee, L. P. Cappato, C. H. Corassin, A. G. Cruz and C. A. Oliveira, J. Dairy Sci., 2016, 99, 2384–2390 CrossRef CAS.
- N. A. Al-Shabib, F. M. Husain, I. Ahmad, M. S. Khan, R. A. Khan and J. M. Khan, Food Control, 2017, 79, 325–332 CrossRef CAS.
- M. M. Theron and F. R. J. Lues, Food Rev. Int., 2007, 23, 141–158 CrossRef CAS.
- M. Y. Akbas and T. Kokumer, Int. J. Food Sci. Technol., 2015, 50, 1666–1672 CrossRef CAS.
- C. L. Quave, M. Estévezcarmona, C. M. Compadre, G. Hobby, H. Hendrickson, K. E. Beenken and M. S. Smeltzer, PLoS One, 2012, 7, e28737 CrossRef CAS.
- J. H. Lee, Y. G. Kim, S. Y. Ryu, M. H. Cho and J. Lee, Int. J. Food Microbiol., 2014, 174, 47–55 CrossRef CAS.
- Ã. Luís, F. Silva, S. Sousa, A. P. Duarte and F. Domingues, Biofouling, 2014, 30, 69–79 CrossRef.
- N. Cinkilic, S. K. Cetintas, T. Zorlu, O. Vatan, D. Yilmaz, T. Cavas, S. Tunc, L. Ozkan and R. Bilaloglu, Food Chem. Toxicol., 2013, 53, 359–363 CrossRef CAS.
- S. A. Jang, D. W. Park, J. E. Kwon, H. S. Song, B. Park, H. Jeon, E. H. Sohn, H. J. Koo and S. C. Kang, Biomed. Pharmacother., 2017, 96, 563–571 CrossRef CAS.
- J. R. Bai, Y. P. Wu, X. Y. Wang, X. Y. Liu, K. Zhong, Y. N. Huang, Y. T. Chen and H. Gao, J. Food Saf., 2018, 38, e12416 CrossRef.
- J. R. Bai, Y. P. Wu, K. Zhong, K. Xiao, L. J. Liu, Y. N. Huang, Z. S. Wang and H. Gao, J. Food Prot., 2018, 81, 1187–1192 CrossRef CAS.
- S. Dosler and E. Mataraci, Peptides, 2013, 49, 53–58 CrossRef CAS PubMed.
- D. Ming, D. C. Wang, F. J. Cao, H. Xiang, D. Mu, J. J. Cao, B. B. Li, L. Zhong, X. Y. Dong, X. B. Zhong, L. Wang and T. D. Wang, Front. Microbiol., 2017, 8, 2263 CrossRef PubMed.
- M. Sivaranjani, S. Gowrishankar, A. Kamaladevi, S. K. Pandian, K. Balamurugan and A. V. Ravi, Int. J. Food Microbiol., 2016, 237, 73–82 CrossRef CAS.
- E. M. Costa, S. Silva, F. K. Tavaria and M. M. Pintado, J. Appl. Microbiol., 2017, 122, 1547–1557 CrossRef CAS.
- K. Lee, J. H. Lee, S. I. Kim, M. H. Cho and J. Lee, Appl. Microbiol. Biotechnol., 2014, 98, 9447–9457 CrossRef CAS.
- J. M. Patti, B. L. Allen, M. J. Mcgavin and M. Höök, Annu. Rev. Microbiol., 1994, 48, 585–617 CrossRef CAS.
- F. Gotz, Mol. Microbiol., 2002, 43, 1367–1378 CrossRef CAS PubMed.
- S. N. Khan, S. Khan, J. Iqbal, R. Khan and A. U. Khan, Front. Microbiol., 2017, 8, 1641 CrossRef.
- M. M. Bazargani and J. Rohloff, Food Control, 2016, 61, 156–164 CrossRef CAS.
- R. L. Kovács, Z. Tóth, F. Nagy, L. Daróczi, A. Bozó and L. Majoros, J. Appl. Microbiol., 2017, 122, 1529–1536 CrossRef.
- K. E. Beenken, P. M. Dunman, F. Mcaleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins and M. S. Smeltzer, J. Bacteriol., 2004, 186, 4665–4684 CrossRef CAS.
- B. R. Boles and A. R. Horswill, PLoS Pathog., 2008, 4, e1000052 CrossRef PubMed.
- A. I. Doulgeraki, P. Di Ciccio, A. Ianieri and G.-J. E. Nychas, Res. Microbiol., 2017, 168, 1–15 CrossRef CAS.
- A. E. Paharik and A. R. Horswill, Microbiol. Spectrum, 2016, 4, 1–27 CAS.
- K. J. Lauderdale, B. R. Boles, A. L. Cheung and A. R. Horswill, Infect. Immun., 2009, 77, 1623–1635 CrossRef CAS.
- J. M. Mootz, C. L. Malone, L. N. Shaw and A. R. Horswill, Infect. Immun., 2013, 81, 3227–3238 CrossRef CAS.
- I. I. Kullik and P. Giachino, Arch. Microbiol., 1997, 167, 151–159 CrossRef CAS.
- H. R. Van and C. W. Michiels, J. Appl. Microbiol., 2010, 109, 1117–1131 CrossRef.
- J. Parraruiz, C. Vidaillac, W. E. Rose and M. J. Rybak, Antimicrob. Agents Chemother., 2010, 54, 4329–4334 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2019 |